Abstract
Canine Cachavirus was novel parvovirus species has been firstly identified in dogs in USA and was classified within the proposed Chaphamaparvovirus genus. To investigate Cachavirus infection in dogs in China, 408 rectal swabs from healthy and diarrheic dogs obtained during 2018–2019 were screened. The rate of Cachavirus positivity was 0% and 1.55% in healthy or diarrheic dogs, respectively. However, statistical analysis suggested no association between the presence of the virus and clinical signs (p > 0.05). Nucleotide identity was 98.2%–98.9% for NS1 and 98.6%–99.1% for VP1, and amino acid identity was 97.9%–98.7% for NS1 and 98.8%–99.6% for VP1 between the five Chinese strains and Cachavirus-1A and Cachavirus-1B detected in the United States. Phylogenetic analysis also indicated that these Cachavirus strains are genetically related to Cachavirus-1A and Cachavirus-1B. This study confirms the presence of Cachavirus in pet dogs in China and provides novel findings on its molecular characteristics.
Keywords: Cachavirus, Chapparvovirus, Nonstructural protein 1, Capsid protein, Phylogenetic analysis, Mutation analysis
1. Introduction
Viruses belonging to the Parvoviridae family include 4–6-kb-long linear, single-stranded DNA genomes (Pénzes et al., 2019). Recently, Parvoviridae family has been re-organized by International Committee on Taxonomy of Viruses (ICTV). The novel subfamily Hamaparvovirinae comprises the current genera Hepandensoviruses, Penstyldensovirus, and Brevidensovirus, together with the yet unclassified chapparvoviruses (ChPV) (Cotmore et al., 2019). Phylogeny evidence suggested the ChPV detected in many kinds of vertebrates, harbored close relation to members of other genera of subfamily Hamaparvovirinae, which evidently derive from ancient members of this lineage, have been identified in several arthropod genomes. (Pénzes et al., 2019; Souza et al., 2017). These reports suggested that ChPV lineage infects both vertebrates and invertebrates, and transmission of ChPVs between distantly related host species may have occurred in the past.
Recently, cases of ChPV infection in various organisms have been reported. In addition to rat parvovirus 2 (RPV2), ChPV include fruit bat (Eidolon helvum) parvovirus 1 (Yinda et al., 2018), E. helvum parvovirus 2 (Baker et al., 2013), common vampire bat (Desmodus rotundus) parvovirus (Souza et al., 2017), murine kidney parvovirus (Roediger et al., 2018), murine ChPV (Williams et al., 2018), simian parvo-like virus 3 (Kapusinszky et al., 2017), turkey parvovirus 1 (Reuter et al., 2014), porcine parvovirus 7 (Palinski et al., 2016), Tasmanian devil-associated ChPV strains (Chong et al., 2019), red-crowned crane-associated parvovirus (Wang et al., 2019), chicken ChPV 1 and 2 (Lima et al., 2019), and peafowl parvovirus (PePV) 1 and PePV2 (Liu et al., 2020). Meanwhile, Cachavirus, which is very similar to ChPV, was first detected in dog feces in the United States in 2019 (Fahsbender et al., 2019), raising concerns regarding the emergence and coinfection of Cachavirus in dogs. The Cachavirus was shown to harbor two major open reading frames (ORFs) encoding NS1 and VP1. Therefore, Cachavirus may show an extensive geographic distribution and may infect diverse hosts (Souza et al., 2017). To obtain detailed understanding of the prevalence and evolution of Cachavirus in dogs, we performed a large-scale investigation using rectal swabs from healthy and diarrheic dogs.
2. Materials and methods
2.1. Sample collection and DNA extraction
Rectal swabs from 85 healthy and 323 diarrheic dogs were collected from veterinary hospitals in Henan, Hubei, and Zhejiang Provinces, China, during 2018–2019. Viral DNA and RNA were extracted using the EasyPure Viral DNA/RNA Kit (TransGen Biotechnology, Inc., Beijing, China) in accordance with the manufacturer's instructions. The extracted DNA and RNA sample were stored at −80 °C until use.
2.2. Virus screening and isolation
The extracted DNA and RNA samples were tested for Cachavirus (Fahsbender et al., 2019), canine coronavirus (CCV) (Nguyen et al., 2017), canine distemper virus (CDV) (Wang et al., 2017), and canine parvovirus (CPV) (Han et al., 2015) using RT-PCR as previously described.
Supernatants from the diluted Cachavirus -positive samples were filtered through a 0.22-μm membrane and inoculated onto a monolayer of Crandell–Rees feline kidney (CRFK, originating from cats) cells and Madin–Darby canine kidney (originating from dogs) cells. After 1-h incubation, the inocula were removed, placed in Dulbecco's modified Eagle's medium (Gibco, USA) supplemented with 2% fetal bovine serum, and incubated at 37 °C with 5% CO2. Cultures were directly inspected daily by microscopy for cytopathic effect (CPE) until 4 days after inoculation. The harvested supernatant was subjected to 3 freezing and thawing cycles, followed by inoculation on a new monolayer of each cell line until the fifth generation. Culture lysates and supernatant were also collected for pathogen screening as described above.
2.3. Amplification, cloning, and sequencing of partial Cachavirus genomes
Four specific primer pairs for genomic amplification covering fragments of ORFs encoding NS1 and VP1 were designed based on Cachavirus sequences [Cachavirus-1A (accession no.: MH893826) and Cachavirus-1B (accession no.: MK448316)] deposited in GenBank. Primer sequences are listed in Table S1. Gene amplification was performed via PCR using a 20-μL reaction mixture containing DNA template (>100 ng/μL), 6-pmol upstream/downstream primers, PrimeSTAR HS DNA polymerase, and supporting reaction buffer (TaKaRa Biotechnology Co., Ltd., Dalian, China). The following cycling conditions were used: initial denaturation at 95 °C for 3 min; 30 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 1 min; and final extension at 72 °C for 10 min. The obtained amplicons were cloned into pMD18-T Easy Vector (TaKaRa Biotechnology Co., Ltd.) for subsequent sequencing (Syn-Biotechnology, Suzhou, China). PCR and partial-genome sequencing were performed at least three times.
2.4. Identity and phylogenetic analyses
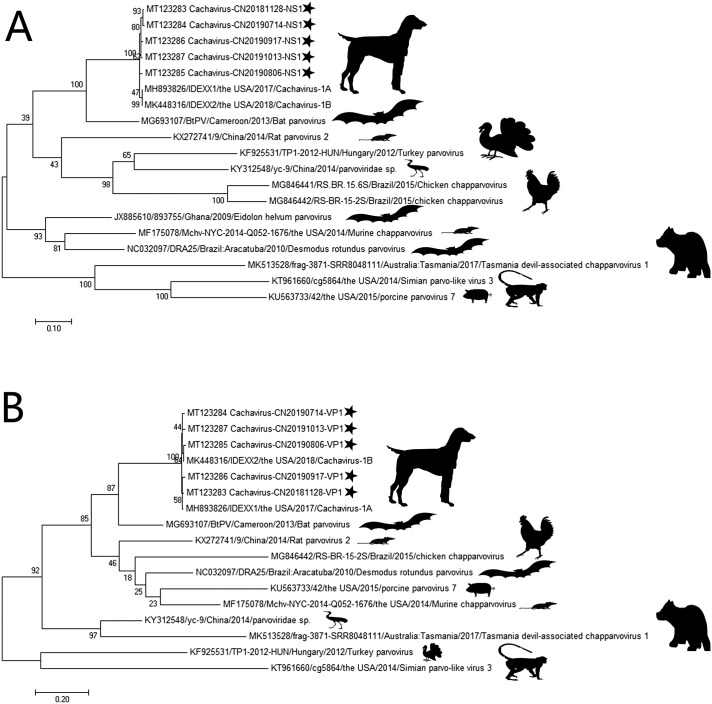
The ClustalW method was employed using MEGA 7.0 to analyze the identity between the obtained and reference strains. Moreover, phylogenetic trees were constructed based on NS1 and VP1 sequences to analyze the relationships between the tested Chinese strains and reference strains. Specific information of each viral strain is presented in Fig. 1A and B. Specifically, the maximum likelihood method was used. MEGA 7.0 was used with the settings of pairwise deletion option and 1000 bootstrap replicates.
Fig. 1.
Phylogenetic tree based on NS1 and VP1 in Cachavirus. A: Phylogenetic tree of NS1. B: Phylogenetic tree of VP1. “★” indicates the virus identified in dogs in China.
2.5. Mutation and protein structure prediction for NS1 and VP1
Illustrator for Biological Sequences was used to obtain a visual overview of NS1 and VP1 from the five Chinese strains and Cachavirus-1A. Based on the sequence alignment results, mutated amino acid sequences were marked on SWISS-MODEL (https://swissmodel.expasy.org/interactive), and post-modeling Pdb files were constructed using PyMOL for collation and preservation.
2.6. Statistical analysis
Chi square or fisher's exact test was used to compare the frequency of Cachavirus between healthy and diarrheic dogs. Statistical analyses were performed using GraphPad Prism 8.0 (San Diego, CA, USA). A p < 0.05 was considered statistically significant.
3. Results
3.1. Positivity and coinfection rates
Upon viral screening, five Cachavirus-positive samples were collected from diarrheic dogs in Henan and Hubei Provinces and no infected samples were identified from healthy dogs. Moreover, CDV and CPV coinfection with the five Chinese strains was identified (Table 1 ). The positivity rate in healthy or diarrheic dogs was 0% and 1.55%, respectively. Statistical analysis showed no association between the presence of the virus and clinical signs (p > 0.05).
Table 1.
Details the five Chinese Cachavirus strains identified in this study.
| Sample name | Health status | Age (months) | Province | Date | Coinfection |
|---|---|---|---|---|---|
| Cachavirus-CN20181128 | Diarrhea | 11 | Henan | November 2018 | – |
| Cachavirus-CN20190714 | Diarrhea | 6 | Henan | July 2019 | – |
| Cachavirus-CN20190806 | Diarrhea | 5 | Henan | August 2019 | – |
| Cachavirus-CN20190917 | Diarrhea | 6 | Hubei | September 2019 | CPV, CDV |
| Cachavirus-CN20191013 | Diarrhea | 4 | Hubei | October 2019 | CPV, CCV |
The positive rates of CPV, CDV, and CCV were 60.1% (194/323), 32.2% (104/323), and 44.0% (142/323), respectively, and these were only detected in diarrheic dogs. In positive samples, coinfections with two (CPV + CDV, n = 35; CPV + CCV, n = 41; or CDV + CCV, n = 29) or three (Cachavirus + CPV + CDV, n = 1; Cachavirus + CPV + CCV, n = 1; CPV + CDV + CCV, n = 14) pathogens were identified, indicating universal coinfection.
3.2. Virus isolation
Until the fifth generation, CPE was only observed in the cultured cells inoculated with the 3 Cachavirus-positive samples coinfected with CPV, and Cachavirus DNA was not detected in culture lysates and supernatant by PCR.
3.3. NS1 and VP1 identity
Partial genomes [covering ORFs encoding NS1 (length = 1992 nt; GC contents = ~37.7%) and VP1 (length = 1515 nt; GC contents = ~36.3%)] of the five Chinese strains were deposited in GenBank under the accession nos. MT123283–MT123287. Nucleotide identity was 98.2%–98.9% for NS1 and 98.6%–99.1% for VP1, and amino acid identity was 97.9%–98.7% for NS1 and 98.8%–99.6% for VP1. Compared with ChPV strains from other hosts, the five Chinese strains showed the highest NS1 nucleotide (73.4%) and amino acid (66.9%) identity with bat parvovirus and the highest VP1 nucleotide identity (65.3%) with bat parvovirus and VP1 amino acid identity (70.0%) with simian parvo-like virus 3.
3.4. Phylogenetic analysis of NS1 and VP1
In the phylogenetic tree based on NS1 and VP1 sequences (Fig. 1), the reference strains and the five Chinese strains were generally divided into two major branches. Intuitively, the five Chinese strains were more closely related to Cachavirus-1A and Cachavirus-1B.
3.5. Mutation and protein structural analyses
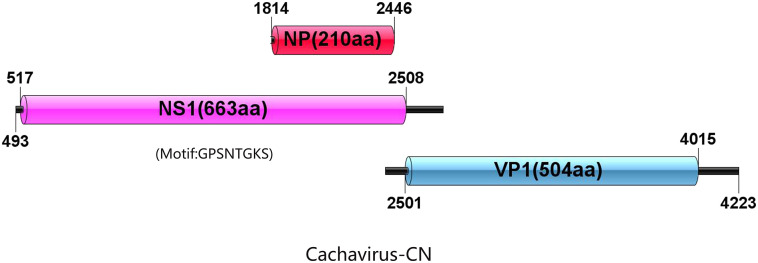
Cachavirus-1A was used as a reference to display the putative structure of proteins in the five Chinese strains (Fig. 2 ). The genomic structure of the five strains was consistent with that of Cachavirus-1A, with two major ORFs encoding NS1 and VP1 (Fig. 2); the putative nucleoprotein (NP) was 210-amino acid long. The five strains shared 99.4% NP nucleotide identity and 99.5% amino acid identity with Cachavirus-1A. The ATP-binding Walker loop motif GPSNTGKS was also present in NS1 ORF of the five strains similar to that of Cachavirus-1A.
Fig. 2.
Structures of open reading frames encoding NS1 and VP1 of Cachavirus-1A and Cachavirus-CN.
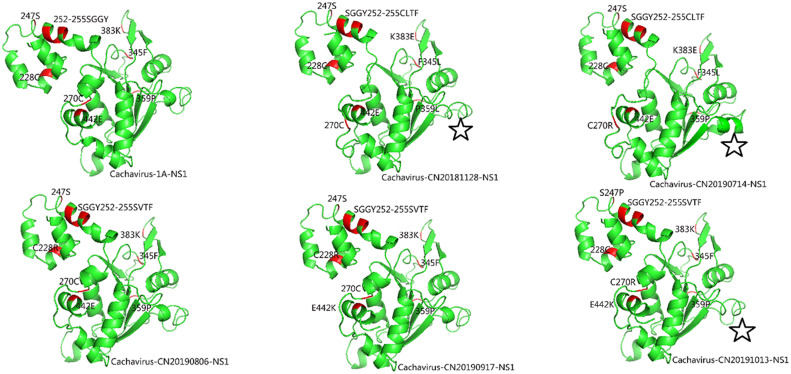
The site analysis of VP1 of Cachavirus-1A revealed the following mutated sites in the five Chinese strains: Ala(A)2Pro(P) (40%) and Val(V)265Ile(I) (40%). Most mutations were found in NS1 of Chinese Cachavirus strains compared with the two strains from the United States, and the mutated sites of the five Chinese strains were displayed in Table 2 . Due to these mutations, predict changes based on tertiary structure modeling with NS1 structure in Cachavirus-1A as a reference were shown in Fig. 3 .
Table 2.
Main amino acid mutation sites in NS1 of the five Chinese Cachavirus strains and reference strains.
| Strains | Substitution of amino acid residues in NS1 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 228 | 247 | 252 | 253 | 254 | 255 | 270 | 345 | 359 | 383 | 442 | 663 | |
| Cachavirus-1A | C | S | S | G | G | Y | C | F | p | K | E | G |
| Cachavirus-CN20181128 | C | S | C | L | T | F | C | L | L | E | E | G |
| Cachavirus-CN20190714 | C | S | C | L | T | F | R | L | P | E | E | G |
| Cachavirus-CN20190806 | R | S | S | V | T | F | C | F | P | K | E | A |
| Cachavirus-CN20190917 | R | S | S | V | T | F | C | F | P | K | K | A |
| Cachavirus-CN20191013 | C | P | S | V | T | F | R | F | P | K | K | G |
Fig. 3.
Predicted tertiary structure model of NS1 of Cachavirus. The red regions indicate mutation sites and the five-pointed star indicates altered protein structure without mutations. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Viral enteritis is a major threat to pet dogs that can lead to life-threating illness. Canine parvovirus (CPV-2), canine distemper virus (CDV), canine coronavirus (CCoV) and canine adenovirus (CAV) are the main pathogens that cause canine viral enteritis (Cardillo et al., 2020; Deng et al., 2018). In this study, Cachavirus, belonging to ChPV, was reported for the first time in diarrheic dogs in China; however, statistical analysis showed no association between the presence of the virus and clinical signs (p > 0.05). As previously reported, only 3 of the 203 fecal samples from healthy dogs tested positive, and statistically significant differences (p < 0.05) were not detected among 803 diarrhea samples from 2017 (4.0%; p = 0.08), but detected among 965 diarrhea samples from 2018 (4.66%, p = 0.037) with the increment positive rate (Fahsbender et al., 2019). Meanwhile, virus screening revealed Cachavirus coinfection with CPV, CDV and CCV in dogs, which should be studied further in the context of pathogenesis.
Some ChPVs and other viruses belonging to Parvoviridae family are mainly detected in diarrhea samples.(Pénzes et al., 2019; Souza et al., 2017). Pathogenicity of chicken Parvovirus have recently determined in specific pathogen-free chicks infected experimentally (Nuñez et al., 2020). However, report about pathogenicity of the ChPVs in vertebrate hosts is still very limited. However, a ChPV called mouse kidney parvovirus (MKPV) circulates among laboratory mice populations has been demonstrated to cause a kidney disease known as inclusion body nephropathy (Roediger et al., 2018). Meanwhile, murine ChPV, has been detected at a very high prevalence in murine liver tissue, suggesting it is a gastrointestinal agent (Williams et al., 2018). Herein, whether ChPV infection leads to gastroenteritis in dogs needs warrants further investigation.
Primers targeting the whole genome have been designed based on the genome sequences of Cachavirus-1A and Cachavirus-1B; however, these primers failed to amplify the 5′ untranslated region (1–493); this may be because of the special DNA structure or the Chinese strains may markedly differs from that of strains reported from the United States. Therefore, future research should explore the mechanism of virus propagation using second-generation sequencing to obtain accurate whole genome sequences. Unfortunately, Cachavirus replication was not detected in CRFK and MDCK cell line. The reason for the non-cultivatable nature of the viruses remains unclear.
According to identity and phylogenetic analyses based on NS1 and VP1 of the five Chinese strains, all strains are closely related to the two strains detected in the United States. Besides these two Cachavirus strains, the detected Chinese strains shared the highest NS1 and VP1 identity with bat parvovirus from Cameroon and simian parvo-like virus 3 from diarrheic rhesus macaques in the United States (Kapusinszky et al., 2017; Yinda et al., 2018). Frequent recombination between mouse parvoviruses (MPVs), Minute viruses of mice (MVMs) and hamster parvovirus (HaPV) has been predicted (Shackelton et al., 2007). Phylogenetic analysis also revealed the closer relationship between the five Chinese Cachavirus strains with bat parvovirus from Cameroon than with other ChPVs; however, no recombination events occurred based on recombination analysis (data not shown). Hence, the role of bats and primates in the transmission and evolution of ChPVs remains unknown. The five Chinese strains harbored increased number of mutations compared with the two strains from the United States. Previous report have highlighted the importance of obtaining the NS1 coding sequence in molecular epidemiology investigations of CPV-2 (Cotmore et al., 2014; Mira et al., 2019). NS1 of Cachavirus-1A, Cachavirus-CN20181128, and Cachavirus-CN20190714 shows the SGGY252-255CLTF mutation, whereas NS1 of the three Chinese strains showed the GGY253-255VTF mutation. However, these continuous mutations did not lead to remarkable changes in protein structure. In contrast, mutation at the 442 site led to significant structural changes in Cachavirus-CN20190917 and Cachavirus-CN20191013 (Glu442Lys). Meanwhile, Cachavirus-CN20181128, Cachavirus-CN20190714, and Cachavirus-CN20191013 showed different structural changes of the 406–416 loci without mutations, whether these changes are caused by the neighboring effect of adjacent mutations should be studied in future. Furthermore, these mutation sites may show different origins due to regional variations.
5. Conclusion
To sum up, this is the first report about the presence of Cachavirus in dogs in China. Phylogenetic analysis of NS1 and VP1 of the five Chinese strains revealed a close relationship with the two Cachavirus strains from the United States. These findings provide a reference for studying the transmission and evolution of ChPV. Nonetheless, large-scale studies are warranted to confirm the pathogenicity of this virus.
Ethics statement
Sampling was not harmful to the dogs. The research protocol was approved by the Animal Welfare and Ethics Committee of Nanyang Normal University (approval number: 18032, year: 2018). An ethical statement was not needed sample collection.
Author contributions
Conceived and designed the experiments: JJ. Performed the experiments: WH QL. Analyzed the data: QC. Contributed reagents/materials/analysis tools: WH QL. Wrote the paper: WH.
Declaration of Competing Interest
The authors have no competing interests to declare.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant no. 31870917), the Scientific and Technological Project of Henan Province (grant nos. 182107000040 and 182102110084), the Key Scientific and Technological Project of the Education Department of Henan Province (grant no. 18A230012), Key Scientific and Technological Project of Nanyang City (grant nos. KJGG2018144 and KJGG2018069), and the Technological Project of Nanyang Normal University (grant nos. 18046 and 2018CX014).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2020.104529.
Appendix A. Supplementary data
Primers for detecting CCV, CDV, CPV, and Cachavirus as well as primers for amplifying the Cachavirus genome
References
- Baker K.S., Leggett R.M., Bexfield N.H., Alston M., Daly G., Todd S. Metagenomic study of the viruses of African straw-coloured fruit bats: detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology. 2013;441:95–106. doi: 10.1016/j.virol.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardillo L., Piegari G., Iovane V., Viscardi M., Alfano F., Cerrone A., Pagnini U., Montagnaro S., Galiero G., Pisanelli G., Fusco G. Lifestyle as risk factor for infectious causes of death in young dogs: a retrospective study in Southern Italy (2015–2017) Vet. Med. Int. 2020 doi: 10.1155/2020/6207297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong R., Shi M., Grueber C.E., Holmes E.C., Hogg C.J. Fecal viral diversity of captive and wild Tasmanian devils characterized using virion-enriched metagenomics and metatranscriptomics. J. Virol. 2019;93(11) doi: 10.1128/JVI.00205-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore S.F., Agbandje-McKenna M., Chiorini J.A. The family parvoviridae. Arch. Virol. 2014;159(5):1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore S.F., Agbandje-McKenna M., Canuti M. ICTV virus taxonomy profile: parvoviridae. J. Gen. Virol. 2019;100(3):367–368. doi: 10.1099/jgv.0.001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Zhang J., Su J., Liu H. A multiplex PCR method for the simultaneous detection of three viruses associated with canine viral enteric infections. Arch. Virol. 2018;163:1233–1238. doi: 10.1007/s00705-018-3828-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahsbender E., Altan E., Seguin M.A., Young P., Estrada M., Leutenegger C., Delwart E. Chapparvovirus DNA found in 4% of dogs with diarrhea. Viruses. 2019;11 doi: 10.3390/v11050398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.C., Guo H.C., Sun S.Q., Shu L., Wei Y.Q., Sun D.H. Full-length genomic characterizations of two canine parvoviruses prevalent in Northwest China. Arch. Microbiol. 2015;197:621–626. doi: 10.1007/s00203-015-1093-4. [DOI] [PubMed] [Google Scholar]
- Kapusinszky B., Ardeshir A., Mulvaney U., Deng X., Delwart E. Case-control comparison of enteric viromes in captive rhesus macaques with acute or idiopathic chronic diarrhea. J. Virol. 2017;91(18) doi: 10.1128/JVI.00952-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima D.A., Cibulski S.P., Tochetto C., APM V., Finkler F., Teixeira T.F. The intestinal virome of malabsorption syndrome-affected and unaffected broilers through shotgun metagenomics. Virus Res. 2019;261:9–20. doi: 10.1016/j.virusres.2018.12.005. [DOI] [PubMed] [Google Scholar]
- Liu X., Wang H., Liu X., Li Y., Chen J., Zhang J. Genomic and transcriptional analyses of novel parvoviruses identified from dead peafowl. Virology. 2020;539:80–91. doi: 10.1016/j.virol.2019.10.013. [DOI] [PubMed] [Google Scholar]
- Mira F., Canuti M., Purpari G., Lastra A., Decaro N., Guercio A. Molecular characterization and evolutionary analyses of carnivore protoparvovirus 1 NS1 gene. Viruses. 2019;11(4):308. doi: 10.3390/v.11040308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D., Terada Y., Minami S., Yonemitsu K., Nagata N., Shimoda H., Maeda K. Characterization of canine coronavirus spread among domestic dogs in Vietnam. J. Vet. Med. Sci. 2017;79(2):343–349. doi: 10.1292/jvms.16-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez L.F., Santander-Parra S.H., De la Torre D.I., Sá L., Buim M.R., Astolfi-Ferreira C.S., Piantino F.A.J. Molecular characterization and pathogenicity of chicken parvovirus (ChPV) in specific pathogen free chicks infected experimentally. Pathogens. 2020;9 doi: 10.3390/pathogens9080606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palinski R.M., Mitra N., Hause B.M. Discovery of a novel parvovirinae virus, porcine parvovirus 7, by metagenomic sequencing of porcine rectal swabs. Virus Genes. 2016;52:564–567. doi: 10.1007/s11262-016-1322-1. [DOI] [PubMed] [Google Scholar]
- Pénzes J.J., de Souza W.M., Agbandje-McKenna M., Gifford R.J. Vol. 11. 2019. An Ancient Lineage of Highly Divergent Parvoviruses Infects both Vertebrate and Invertebrate Hosts; p. 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G., Boros A., Delwart E., Pankovics P. Novel circular single-stranded DNA virus from Turkey faeces. Arch. Virol. 2014;159:2161–2164. doi: 10.1007/s00705-014-2025-3. [DOI] [PubMed] [Google Scholar]
- Roediger B., Lee Q., Tikoo S., JCA C., Henderson J.M., Jormakka M. An atypical parvovirus drives chronic tubulointerstitial nephropathy and kidney fibrosis. Cell. 2018;175:530–543. doi: 10.1016/j.cell.2018.08.013. e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelton, L.A., Hoelzer, K., Parrish, C.R., Holmes, E.C., 2007. Comparative analysis reveals frequent recombination in the parvoviruses. J. Gen. Virol. 88(Pt 12): 3294–301.doi: 10.1099/vir.0.83255-0. [DOI] [PMC free article] [PubMed]
- Souza W.M., Romeiro M.F., Fumagalli M.J., Modha S., de Araujo J., Queiroz L.H. Chapparvoviruses occur in at least three vertebrate classes and have a broad biogeographic distribution. J. Gen. Viro.1. 2017;98:225–229. doi: 10.1099/jgv.0.000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wang J., Li R., Liu L., Yuan W. Rapid and sensitive detection of canine distemper virus by real-time reverse transcription recombinase polymerase amplification. BMC Vet. Res. 2017;13:241. doi: 10.1186/s12917-017-1180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yang S., Liu D., Zhou C., Li W., Lin Y. The fecal virome of red-crowned cranes. Arch. Virol. 2019;164:3–16. doi: 10.1007/s00705-018-4037-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.H., Che X., Garcia J.A., Klena J.D., Lee B., Muller D. Viral diversity of house mice in new York City. mBio. 2018;9(2) doi: 10.1128/mBio.01354-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yinda C.K., Ghogomu S.M., Conceicao-Neto N., Matthijnssens J. Cameroonian fruit bats harbor divergent viruses, including rotavirus H, bastroviruses, and picobirnaviruses using an alternative genetic code. Virus Evol. 2018;4(1) doi: 10.1093/ve/vey008. vey008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers for detecting CCV, CDV, CPV, and Cachavirus as well as primers for amplifying the Cachavirus genome