Abstract
Autophagy is an intracellular degradation system that breaks down damaged organelles or damaged proteins using intracellular lysosomes. Recent studies have also revealed that various forms of selective autophagy play specific physiological roles under different cellular conditions. Lipid droplets, which are mainly found in adipocytes and hepatocytes, are dynamic organelles that store triglycerides and are critical to health. Lipophagy is a type of selective autophagy that targets lipid droplets and is an essential mechanism for maintaining homeostasis of lipid droplets. However, while processes that regulate lipid droplets such as lipolysis and lipogenesis are relatively well known, the major factors that control lipophagy remain largely unknown. This review introduces the underlying mechanism by which lipophagy is induced and regulated, and the current findings on the major roles of lipophagy in physiological and pathological status. These studies will provide basic insights into the function of lipophagy and may be useful for the development of new therapies for lipophagy dysfunction-related diseases.
Keywords: adipose, lipid droplets, lipophagy, liver, metabolic disorders, selective autophagy
INTRODUCTION
Lipid droplet (LD) is a unique structure surrounded by a monolayer phospholipid membrane that separates neutral lipids from the cytoplasmic environment. In white adipocytes, triacylglycerol (TG) is mainly stored as lipid esters in LD. In mammalian cells, the major component of the LD monolayer membrane is phosphatidylcholine (about 60%) (Wilfling et al., 2014). Changes in the phospholipid ratio of the LD membrane compositions mainly affect the synthesis, maturation, and degradation of LD (Liu and Czaja, 2013).
LD contributes to the synthesis of membrane components, signal ligands, formation of specific macromolecules, and the storage of lipids that can be used for energy production. Such lipid storage protects the cells from exposure to excess free fatty acids (FFA) and sterols, which may disrupt the composition of cell membranes, signaling pathways, and metabolic homeostasis (Kimmel and Sztalryd, 2016).
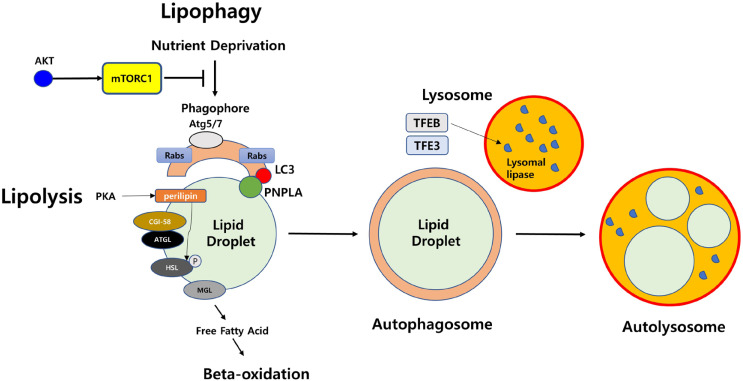
Surface proteins present in LD play an important role in regulating homeostasis and intracellular interactions of LD. In particular, the perilipin, which is part of the PAT (perilipin/ADRP/TIP47) family, regulates lipid homeostasis by regulating the access of lipases to neutral lipids in LD. Perilipin is phosphorylated by protein kinase A under starvation, and its phosphorylation initiates lipolysis (Hansen et al., 2017). A previous study revealed that the level of basal lipolysis is significantly elevated in adipocytes derived from a perilipin knockout mouse model (Tansey et al., 2001). Many catabolic enzymes also exist on the surface of LD. At the basal level, perilipin generally binds to CGI-58. Upon activation of lipolysis, CGI-58 is dissociated from phosphorylated perilipin and binds to triglyceride lipase (ATGL), which initiates TG hydrolysis. Hormone-sensitive lipase (HSL) is phosphorylated by phosphorylated perilipin and then moves to LD. HSL hydrolyzes diacylglycerol (DG) into monoacylglycerol (MG) (Ducharme and Bickel, 2008). Monoacylglycerol lipase (MGL) finally hydrolyzes MG into FFA and glycerol (Fredrikson et al., 1986) (Fig. 1).
Fig. 1. Overview of major proteins in lipolysis and lipophagy.
Lipolysis is processed into three separate steps. First, TG is hydrolyzed to make DG by adipose triglyceride lipase (ATGL). Second, DG is hydrolyzed to make MG by HSL, which is phosphorylated by perilipin. Finally, MG is hydrolyzed to make glycerol and FFAs by MGL. One the other hand, lipophagy is defined as selective autophagic degradation of LDs. Under nutrient starvation, lipophagy forms phagophore, which is composed of Atg5, Atg7, LC3, and Rab family. PNPLA family has specific molecular motifs to associate with LDs and play key roles in LD breakdown. Autophagosomes engulf LDs and fuse with a lysosome to form an autolysosome. Then, neutral lipids in LDs are hydrolyzed by lysosomal lipases, which are expressed by TFEB.
In addition to this lipolysis process, breakdown of lipids can also occur through lipophagy, a type of selective autophagy that targets LD. A previous study demonstrated that the breakdown of LD and TG occurs through lipophagy in hepatocytes (Singh et al., 2009a). While the importance and mechanism of lipolysis has been relatively well studied, mechanisms of lipophagy remain largely unknown. Recent findings on the molecular mechanisms and pathological implications of lipophagy that mainly occur in the liver and adipose tissues are reviewed in this paper.
MOLECULAR MECHANISMS OF LIPOPHAGY
Selective degradation of LD by lipophagy involves the use of these lipids as energy sources. Recent studies on the mechanism by which autophagic-related proteins mediate membrane fusion and subsequent degradation processes have been identified. Lipophagy begins with the recognition of cargo by the autophagosomal membrane through interaction with the microtubule-associated protein 1 light chain 3 (LC3) (Singh and Cuervo, 2012; Wang, 2016). ATGL, which is required for lipolysis, also plays an important role in lipophagy. LC3 promotes the movement of cytoplasmic ATGL to LD through interaction with the LIR domain of ATGL and induces lipophagy. ATGL facilitates lipophagy to regulate the catabolism of hepatic LD through SIRT1 activity (Sathyanarayan et al., 2017).
Small regulatory Rab GTPase (Rab) molecular switch families are also found in LDs (Kiss and Nilsson, 2014). Rab7 (Schroeder et al., 2015) and Rab10 (Li et al., 2016) are found to be essential for lipophagy in hepatocytes under certain conditions. A previous study demonstrated that small GTPase Rab7 plays an important role in regulating autolysosome-mediated lipid degradation in adipocytes (Lizaso et al., 2013). Besides, Rab7 is activated under nutrient deprivation and facilitates the recruitment of multi-vesicular bodies and lysosomes to the surface of LD during lipophagy. The deletion of Rab7 decreases hepatocellular lipophagy by causing morphological alterations of multi-vesicular bodies, lysosomes, and autophagosomes (Schroeder et al., 2015). Rab10 forms a complex with the adapter protein EHBP1 (EH domain-binding protein 1) and the membrane-modified adenosine triphosphatase EHD2 (EH domain containing 2) and promotes the migration of LC3-positive autophagic membranes to the LD surface. The deletion of Rab 10 function causes the accumulation of LD (Li et al., 2016).
Lipases such as PNPLA5 (patatin-like phospholipase domain-containing enzyme 5) present in LD have been shown to contribute to lipophagy and autophagic proteolysis (Dupont et al., 2014). In addition to their role in the recognition of LD, these lipases play an important role in initiating lipophagy by inducing the recruitment of triglycerides and sterol esters, directly contributing to the formation of autophagosome (Shpilka et al., 2015; Ward et al., 2016). PNPLA8 mediates SREBP-2 driven lipophagy by interacting with LC3 in the hepatocytes of high-fat diet (HFD)-fed mice (Kim et al., 2016). PNPLA3 plays an essential role in the formation of autophagosomes during the lipophagy process in starved human hepatocytes (Negoita et al., 2019).
Perilipin, which exists on the surface of LD, is removed before the degradation of LD by lipophagy. The chaperone-mediated autophagy of perilipin is processed through AMP-activated protein kinase (AMPK) (Kaushik and Cuervo, 2015). Lipophagy is regulated by the nutritional status of the cells through farsonoid X receptor (FXR), peroxisome proliferator-activated receptor alpha (PPARα), cAMP response element-binding protein (CREB), mTOR, or AMPK (Li et al., 2019; Seok et al., 2014; Zhang et al., 2018a; 2018b). If the cells are in a nutrient-rich status that does not require FFA as an energy source, lipophagy is inhibited. However, in the case of dietary restrictions, lipophagy leads to a breakdown of triglycerides in LDs. Depending on the size, the LD may be targeted by macroautophagy, an autophagosome in which the entire small LD is captured (Singh et al., 2009a). In the lysosome, the LDs are broken down by lysosomal acid lipase (function at acidity; pH = 4.5-5), which can degrade TG, DG, cholesteryl esters, and retinyl esters (Grumet et al., 2016; Schulze et al., 2017c; Zechner et al., 2017). The expression of lysosomal lipases is regulated by lysosomal biogenesis transcription factor EB (TFEB) under nutrient-deficient conditions in Caenorhabditis elegans and mice hepatocytes (Settembre et al., 2013). TFE3 can induce lipophagy in hepatocytes (Xiong et al., 2016). The forkhead homeobox transcription factor (FOXO1) is related to lysosomal lipase and triggers lipophagy in adipocytes under fasting conditions (Barbato et al., 2013). The functions of core proteins in lipophagy are briefly shown (Fig. 1).
Interestingly, Tatsumi et al. (2018) demonstrated that a forced lipophagy system using a fusion of LD-binding domain and p62 significantly reduces the number of LD, decreases the level of TG throughout embryonic development, and eventually causes developmental retardation in mouse embryos. In addition, lipophagy-induced embryos show the removal of excessive LD and are resistant to lipotoxicity (Tatsumi et al., 2018). This data suggests that lipophagy can play an important role in the development.
LIPOPHAGY-RELATED METABOLISM DISORDERS
Lipophagy in obesity
The activity of autophagy is generally lowered in HFD models (Koga et al., 2010; Rodriguez-Navarro et al., 2012). Several studies suggest that autophagy is important in the process of adipocyte differentiation. Mouse embryonic fibroblasts derived from Atg5 knockout mice exhibit a severe defect in the process of adipocyte differentiation (Baerga et al., 2009). The knockdown of Atg7 revealed that preadipocytes cannot differentiate into mature adipocytes and show a lean phenotype in mice (Singh et al., 2009b). Similar to this previous study, Fu et al. (2019) recently demonstrated that miR-129-5p, which targets ATG7, can significantly suppress adipocyte differentiation by decreasing the level of specific adipogenic markers, such as FABP4 and PPARγ, in mature white adipocytes. Deficiency of Bif-1, which associates with Beclin 1 through UVRAG and control autophagy, leads to the expansion of fat mass, down-regulates the basal lipolysis level of adipose, and causes obesity. Bif-1 deficiency also significantly decreases the expression of the autophagy lysosomal proteins, Atg9 and Lamp1 (Liu et al., 2016). SIRT3 activates lipophagy by stimulating the AMPK-unc-51-like kinase 1 (ULK1) pathway and induces smaller LD size and decreased lipid accumulation in mature adipocytes (Zhang et al., 2020).
Xu et al. (2013) demonstrated that increased lipid retention and a markedly decreased lipolysis rate have occurred in the white adipose tissue when a lysosomal function is inhibited in macrophages. In the brown adipose tissue, lipophagy may play a key role in lipid homeostasis. Interestingly, when the mouse is exposed to cold, the LD and LC3 are co-localized, which implies that cytoplasmic lipases move to the LD for conventional lipolysis (Martinez-Lopez et al., 2016). Recently, Lim et al. (2018) reported that a newly screened autophagy enhancer induces nuclear localization of TFEB and accelerates the removal of intracellular lipid in vitro. It also improves the metabolic profile and reduces inflammasome activity in ob/ob mice (Lim et al., 2018). Therefore, in addition to the stimulation of lipolysis, strategies to promote the activation of lipophagy can be one of the therapeutic strategies to improve obesity (Table 1).
Table 1.
Lipophagy and lipophagy-related metabolic disorders: Identified proteins and the possible therapeutic
| Metabolic disorder | Lipophagy status | Key protein | Possible therapeutic | Reference |
|---|---|---|---|---|
| Obesity | Decrease | Bif-1, CaMKIV, SIRT3, etc. | Upregulation | (Liu et al., 2016; 2020; Xu et al., 2013; Zhang et al., 2020) |
| Diabetes mellitus | Decrease | FGF21, etc. | Upregulation | (Byun et al., 2020; Kim et al., 2013) |
| Alcholic fatty liver disease | Decrease | AKT, FXR, Nrf2, mTOR, Rab7, etc. | Upregulation | (Schulze et al., 2017b; Wu et al., 2014; Zhang et al., 2018b; Zhao et al., 2018) |
| Non-alcoholic fatty liver disease | Decrease | AMPK, GNMT, FGF21, IRGM, Rubicon, SOD1, SREBP-2, etc. | Upregulation | (Deng et al., 2017; Levine and Kroemer, 2019; Kurahashi et al., 2015; Lin et al., 2016; Smith et al., 2016; Tanaka et al., 2016; Zhu et al., 2016; Zubiete-Franco et al., 2016) |
| Liver fibrosis | Increase | Perilipin 1, PNPLA3, Rab18, Rab25, etc. | Downregulation | (Bruschi et al., 2017; Miyamae et al., 2016; O'Mahony et al., 2015; Pirazzi et al., 2014; Zhang et al., 2017) |
Lipophagy in diabetes mellitus
Diabetes mellitus (DM) is closely linked to obesity and characterized by high blood sugar levels over long periods. If DM is left untreated, many complications such as cardiovascular disease and chronic kidney disease can arise (Kleinert et al., 2018). DM generally occurs when the pancreas does not produce enough insulin, or when the cells do not respond properly to insulin. The increased prevalence of type 2 diabetes worldwide can be due to a combination of excess weight gain and insufficient exercise (Kim et al., 2018). It begins with insulin resistance, a pathological condition in which cells fail to respond to insulin normally, and insulin deficiency may also develop as the disease progresses (Goldman et al., 2010; Kosacka et al., 2015).
A previous study demonstrated that Atg7 knockout mice exhibit structural and functional defects in the pancreatic β-cells and increases the incidence of glucose intolerance and diabetes under metabolic stress (Jung et al., 2008). Damage of lipophagy in mice can cause increased ER stress in the liver and aggravate insulin tolerance. Hepatic specific overexpression of Atg7 in obese mice restores insulin receptor signaling in liver tissue, reduces obesity-induced ER stress, and improves glucose tolerance and insulin sensitivity (Yang et al., 2010). A recent study demonstrated that post-developmental defects of autophagy genes such as Atg3 and Atg16L1 in fully differentiated adipocytes cause the dysfunction of mitochondria, inflammation, and insulin resistance (Cai et al., 2018). In mice with skeletal muscle-specific deletion of Atg7, the expression of fibroblast growth factor 21 (FGF21), which improves defective autophagy and hepatic steatosis in obese mice, is induced under high fat diet-induced obesity and insulin resistance conditions (Kim et al., 2013). A recent study demonstrated that FGF21 contributes to hepatic autophagy and lipid degradation by stimulating Jumonji-D3 (JMJD3/KDM6B) histone demethylase, which regulates global autophagy-network genes such as Atg1 and Atg7 (Byun et al., 2020). In addition, CaMKIV improves hepatic autophagic imbalance and alleviates impaired insulin sensitivity through phosphorylated CREB (Liu et al., 2020). These studies suggest that the regulation of lipophagy could be one of the important factors for insulin sensitivity and glucose homeostasis in DM (Table 1).
Alcoholic fatty liver disease
Alcoholic fatty liver disease indicates liver damage caused by excessive or chronic alcohol intake. Oxidative stress, mitochondrial damage, and apoptosis occur in the cytoplasm of hepatocytes. Alcohol oxidation induces excessive lipid accumulation called steatosis, in the liver (Ding et al., 2010; Lívero and Acco, 2016).
The role of lipophagy seems to be dependent on acute or chronic alcohol intake (Ding et al., 2011). Interestingly, acute drinking activates hepatic autophagy and limits fat accumulation by selectively removing excess LDs in vivo and primary hepatocytes (Ding et al., 2011; Thomes et al., 2012; Wang et al., 2015). On the other hand, chronic alcohol intake significantly reduces the expression of Atg3 and Atg7, while increasing the phosphorylation of mTOR (Thomes et al., 2015). Chronic alcohol exposure also decreases the expression levels of total and nuclear TFEB, which contribute to lysosomal biogenesis, and eventually leads to a decrease of both biogenesis and function of the lysosome (Chao et al., 2018; Schulze et al., 2017a). Another study demonstrated that chronic alcohol intake significantly increases the protein expressions of mTOR, ULK1, and p62, while reducing the protein expressions of light chain 3II (LC3II)/LC3I and Beclin1 (Lu et al., 2020a). Treatment with Torin-1, an mTOR inhibitor, restores lysosomal biogenesis and markedly improves steatosis and liver damage, indicating that such damage is likely to be caused by the activation of mTOR signal transduction and reduction of lysosomal biogenesis in mouse hepatocytes (Chao et al., 2018; Ni et al., 2012). ROS production by ethanol mainly occurs through ethanol metabolism, cytochrome P450 2E1 induction, and impaired mitochondria (Lu and Cederbaum, 2018). Chronic alcohol exposure inhibits the activation of Rab7 and leads to depletion of lysosomes for LD degradation, ultimately inhibiting lipophagy in hepatocytes (Schulze et al., 2017b). Chronic alcohol intake also interferes with the interaction of FXR with RXRα and inhibits the activity of FXR (Wu et al., 2014). However, activation of the nuclear factor erythroid-derived 2-like 2 (Nrf2), which is involved in stress-induced autophagy, protects against ethanol-induced liver disease (Zhao et al., 2018). Therefore, in patients with alcoholic fatty liver disease, restoring lipophagy function could be an important therapeutic approach (Table 1).
Non-alcoholic fatty liver disease
Non-alcoholic fatty liver disease (NAFLD) is caused by an increase in FFA levels and lipogenesis in hepatocytes. NAFLD exhibits insulin resistance and lipogenesis (Samuel and Shulman, 2018; Tavernarakis et al., 2019). Several previous reports have suggested that this NAFLD can be involved in autophagy. For example, Rubicon, which interacts with Beclin-1 as an autophagy repressor, is upregulated in the livers of NAFLD patients. Tanaka et al. (2016) demonstrated that Rubicon knockout mice exhibit improved autophagy flux and enhanced NAFLD. The knock-down of the immunity-related GTPase family M (IRGM) gene (an autophagy-related gene) suppresses autophagic flux and increases the LD content in HepG2 cells, which can be reversed with rapamycin treatment, an autophagy activator (Levine and Kroemer, 2019; Lin et al., 2016). Smith et al. (2016) suggested that activation of AMPK may significantly ameliorate NAFLD by suppressing de novo lipogenesis in the liver, increasing fatty acid oxidation in the liver, and enhancing mitochondrial function in the adipose tissue. Apoprotein E (ApoE) knockout mice show increased fat mass in the liver tissue compared to wild type mice with a HFD. These mice also exhibit the downregulation of p-AMPK, AMPK, Beclin1, and LC3 levels, while showing upregulation of ROS, p-mTOR, and mTOR. Treatment with AICAR and rapamycin, which are AMPK and autophagy activators, alleviate the symptoms of NAFLD (Lu et al., 2020b). The blockade of SREBP-2 activity increases the expression of autophagy-related genes and improves abnormal autophagic flux in the NAFLD model (Deng et al., 2017). Mice genetically deficient in superoxide dismutase 1 (SOD1) show a low visceral fat mass, but a significant increase in the size of LD during fasting, eventually leading to liver damage. Although LC3 II was activated in this mouse model, p62 was observed to accumulate abnormally, suggesting that abnormal lipophagy process may be responsible for liver damage and impaired lipid metabolism (Kurahashi et al., 2015). Treatment of FGF21 significantly improves the pathological phenotypes of NAFLD by increasing autophagic flux and the expression of autophagy-related genes such as LC3 II in monosodium L-glutamate (MSG)-induced obese mice (Zhu et al., 2016). The expression of glycine N-methyltransferase (GNMT) is reduced in human NAFLD patients. GNMT knockout mice show excess levels of serum methionine and abnormal lipophagy process, caused by defects at the lysosomal level. The methionine deficient diet can improve lipophagy, and lead to ameliorate liver steatosis in GNMT knockout mice (Zubiete-Franco et al., 2016). Thus, further studies on lipophagy activation may pave the way for new therapeutic strategies against NAFLD (Table 1).
Liver fibrosis
Liver fibrosis, characterized by the formation of abnormal scar tissues by excessive deposition of extracellular matrix, is a chronic or recurrent liver disease caused by viral infections, autoimmune conditions, or aging (Iredale et al., 2013; Schuppan et al., 2018). Liver fibrosis has been reported to be associated with hepatic stellate cells (HSCs) (Tsuchida and Friedman, 2017). In normal livers, HSCs are present in non-proliferative, quiescent stationary states. At the onset of liver injury, HSCs are activated and transdifferentiated into myofibroblasts. These cells possess proliferative, contractile, and inflammatory characteristics and express alpha-smooth muscle actin protein and various fibrogenic factors (Schon et al., 2016; Zhang et al., 2018b).
Perilipin is co-localized with LC3 in HSCs, indicating that LD degradation is processed during HSC activation (Miyamae et al., 2016). The knockdown of PNPLA3 significantly reduces the level of α-SMA in the process of HSC activation (Bruschi et al., 2017; Pirazzi et al., 2014). Loss of LD in HSCs is regulated by Rab18 GTPase activity (O’Mahony et al., 2015). The small GTPase, Rab25, also plays a role in the turnover of LDs. The production of ROS during the HSC activation process increases the expression level of Rab25. Lipophagy in HSC is partially mediated through Rab25 in a ROS-dependent manner (Zhang et al., 2017). Autophagy is activated in HSCs after liver injury with CCl4 in vivo. In mice, HSCs that specifically lack Atg7, fibrogenesis, and the accumulation of ECM are significantly decreased in response to CCl4 (Hernandez-Gea et al., 2012). Accumulation of LDs or inhibition of mitochondrial beta-oxidation of fatty acids significantly prevents fibrosis (Hernandez-Gea et al., 2012). Blocking autophagy with bafilomycin A1 results in the decrease of fibrosis formation and the accumulation of extracellular matrix in HSC (Thoen et al., 2011). Therefore, selectively blocking lipophagy in HSCs may be an attractive therapeutic for liver fibrosis (Table 1). However, further investigations are required to examine the relationship between lipophagy and the activation of HSCs.
CONCLUSION
Over the last few decades, autophagy research has revealed several key proteins and signal transduction networks that control autophagy pathways. However, lipophagy, a selective autophagy, is yet to be elucidated compared to general autophagy, including macroautophagy. Several important findings on lipid metabolism and energy homeostasis have been conducted in the field of lipophagy. The identification of key proteins for lipophagy has facilitated a better understanding of the mechanism by which autophagic machinery recognizes and degrades LDs. The sequential downstream in cell signaling networks of lipophagy also remain important. Since the accumulation of LDs is related to the etiology of several metabolic disorders, further research on the precise mechanisms of lipophagy would reveal valuable new targets and ultimately provide therapeutic approaches for the treatment of obesity, diabetes, and various liver diseases such as liver fibrosis, and NAFLD.
ACKNOWLEDGMENTS
This work was supported by the research fund from Konkuk University (2019-A019-0401).
Footnotes
AUTHOR CONTRIBUTIONS
D.W.S. conceived and wrote the manuscript.
CONFLICT OF INTEREST
The author has no potential conflicts of interest to disclose.
REFERENCES
- Baerga R., Zhang Y., Chen P.H., Goldman S., Jin S.V. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy. 2009;5:1118–1130. doi: 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbato D.L., Tatulli G., Aquilano K., Ciriolo M. FoxO1 controls lysosomal acid lipase in adipocytes: implication of lipophagy during nutrient restriction and metformin treatment. Cell Death Dis. 2013;4:e861. doi: 10.1038/cddis.2013.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruschi F.V., Claudel T., Tardelli M., Caligiuri A., Stulnig T.M., Marra F., Trauner M. The PNPLA3 I148M variant modulates the fibrogenic phenotype of human hepatic stellate cells. Hepatology. 2017;65:1875–1890. doi: 10.1002/hep.29041. [DOI] [PubMed] [Google Scholar]
- Byun S., Seok S., Kim Y.C., Zhang Y., Yau P., Iwamori N., Xu H.E., Ma J., Kemper B., Kemper J.K. Fasting-induced FGF21 signaling activates hepatic autophagy and lipid degradation via JMJD3 histone demethylase. Nat. Commun. 2020;11:807. doi: 10.1038/s41467-020-14384-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Pires K.M., Ferhat M., Chaurasia B., Buffolo M.A., Smalling R., Sargsyan A., Atkinson D.L., Summers S.A., Graham T.E., et al. Autophagy ablation in adipocytes induces insulin resistance and reveals roles for lipid peroxide and Nrf2 signaling in adipose-liver crosstalk. Cell Rep. 2018;25:1708–1717.:e5. doi: 10.1016/j.celrep.2018.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao X., Wang S., Zhao K., Li Y., Williams J.A., Li T., Chavan H., Krishnamurthy P., He X.C., Li L. Impaired TFEB-mediated lysosome biogenesis and autophagy promote chronic ethanol-induced liver injury and steatosis in mice. Gastroenterology. 2018;155:865–879.:e12. doi: 10.1053/j.gastro.2018.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Pan X., Cheng C., Liu B., Zhang H., Zhang Y., Xu K. Regulation of SREBP-2 intracellular trafficking improves impaired autophagic flux and alleviates endoplasmic reticulum stress in NAFLD. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862:337–350. doi: 10.1016/j.bbalip.2016.12.007. [DOI] [PubMed] [Google Scholar]
- Ding W.X., Li M., Chen X., Ni H.M., Lin C.W., Gao W., Lu B., Stolz D.B., Clemens D.L., Yin X.M. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W.X., Manley S., Ni H.M. The emerging role of autophagy in alcoholic liver disease. Exp. Biol. Med. (Maywood) 2011;236:546–556. doi: 10.1258/ebm.2011.010360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducharme N.A., Bickel P.E. Lipid droplets in lipogenesis and lipolysis. Endocrinology. 2008;149:942–949. doi: 10.1210/en.2007-1713. [DOI] [PubMed] [Google Scholar]
- Dupont N., Chauhan S., Arko-Mensah J., Castillo E.F., Masedunskas A., Weigert R., Robenek H., Proikas-Cezanne T., Deretic V. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr. Biol. 2014;24:609–620. doi: 10.1016/j.cub.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrikson G., Tornqvist H., Belfrage P. Hormone-sensitive lipase and monoacylglycerol lipase are both required for complete degradation of adipocyte triacylglycerol. Biochim. Biophys. Acta. 1986;876:288–293. doi: 10.1016/0005-2760(86)90286-9. [DOI] [PubMed] [Google Scholar]
- Fu X., Jin L., Han L., Yuan Y., Mu Q., Wang H., Yang J., Ning G., Zhou D., Zhang Z. miR-129-5p inhibits adipogenesis through autophagy and may be a potential biomarker for obesity. Int. J. Endocrinol. 2019;2019:5069578. doi: 10.1155/2019/5069578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman S., Zhang Y., Jin S. Autophagy and adipogenesis: implications in obesity and type II diabetes. Autophagy. 2010;6:179–181. doi: 10.4161/auto.6.1.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumet L., Eichmann T.O., Taschler U., Zierler K.A., Leopold C., Moustafa T., Radovic B., Romauch M., Yan C., Du H. Lysosomal acid lipase hydrolyzes retinyl ester and affects retinoid turnover. J. Biol. Chem. 2016;291:17977–17987. doi: 10.1074/jbc.M116.724054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J.S., de Mare S., Jones H.A., Goransson O., Lindkvist-Petersson K. Visualization of lipid directed dynamics of perilipin 1 in human primary adipocytes. Sci. Rep. 2017;7:15011. doi: 10.1038/s41598-017-15059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Gea V., Ghiassi-Nejad Z., Rozenfeld R., Gordon R., Fiel M.I., Yue Z., Czaja M.J., Friedman S.L. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iredale J.P., Thompson A., Henderson N.C. Extracellular matrix degradation in liver fibrosis: biochemistry and regulation. Biochim. Biophys. Acta. 2013;1832:876–883. doi: 10.1016/j.bbadis.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Jung H.S., Chung K.W., Kim J.W., Kim J., Komatsu M., Tanaka K., Nguyen Y.H., Kang T.M., Yoon K.H., Kim J.W. Loss of autophagy diminishes pancreatic β cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Kaushik S., Cuervo A.M. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 2015;17:759–770. doi: 10.1038/ncb3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lim Y.M., Lee M.S. The role of autophagy in systemic metabolism and human-type diabetes. Mol. Cells. 2018;41:11–17. doi: 10.14348/molcells.2018.2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., Jeong Y.T., Oh H., Kim S.H., Cho J.M., Kim Y.N., Kim S.S., Kim D.H., Hur K.Y., Kim H.K. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat. Med. 2013;19:83. doi: 10.1038/nm.3014. [DOI] [PubMed] [Google Scholar]
- Kim K.Y., Jang H.J., Yang Y.R., Park K.I., Seo J., Shin I.W., Jeon T.I., Ahn S.C., Suh P.G., Osborne T.F. SREBP-2/PNPLA8 axis improves non-alcoholic fatty liver disease through activation of autophagy. Sci. Rep. 2016;6:1–14. doi: 10.1038/srep37794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel A.R., Sztalryd C. The perilipins: major cytosolic lipid droplet-associated proteins and their roles in cellular lipid storage, mobilization, and systemic homeostasis. Ann. Rev. Nutr. 2016;36:471–509. doi: 10.1146/annurev-nutr-071813-105410. [DOI] [PubMed] [Google Scholar]
- Kiss R.S., Nilsson T. Rab proteins implicated in lipid storage and mobilization. J. Biomed. Res. 2014;28:169. doi: 10.7555/JBR.28.20140029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinert M., Clemmensen C., Hofmann S.M., Moore M.C., Renner S., Woods S.C., Huypens P., Beckers J., de Angelis M.H., Schurmann A., et al. Animal models of obesity and diabetes mellitus. Nat. Rev. Endocrinol. 2018;14:140–162. doi: 10.1038/nrendo.2017.161. [DOI] [PubMed] [Google Scholar]
- Koga H., Kaushik S., Cuervo A.M. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosacka J., Kern M., Kloting N., Paeschke S., Rudich A., Haim Y., Gericke M., Serke H., Stumvoll M., Bechmann I., et al. Autophagy in adipose tissue of patients with obesity and type 2 diabetes. Mol. Cell. Endocrinol. 2015;409:21–32. doi: 10.1016/j.mce.2015.03.015. [DOI] [PubMed] [Google Scholar]
- Kurahashi T., Hamashima S., Shirato T., Lee J., Homma T., Kang E.S., Fujii J. An SOD1 deficiency enhances lipid droplet accumulation in the fasted mouse liver by aborting lipophagy. Biochem. Biophys. Res. Commun. 2015;467:866–871. doi: 10.1016/j.bbrc.2015.10.052. [DOI] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Yang P., Zhao L., Chen Y., Zhang X., Zeng S., Wei L., Varghese Z., Moorhead J.F., Chen Y. CD36 plays a negative role in the regulation of lipophagy in hepatocytes through an AMPK-dependent pathway. J. Lipid Res. 2019;60:844–855. doi: 10.1194/jlr.M090969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Schulze R.J., Weller S.G., Krueger E.W., Schott M.B., Zhang X., Casey C.A., Liu J., Stöckli J., James D.E. A novel Rab10-EHBP1-EHD2 complex essential for the autophagic engulfment of lipid droplets. Sci. Adv. 2016;2:e1601470. doi: 10.1126/sciadv.1601470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H., Lim Y.M., Kim K.H., Jeon Y.E., Park K., Kim J., Hwang H.Y., Lee D.J., Pagire H., Kwon H.J. A novel autophagy enhancer as a therapeutic agent against metabolic syndrome and diabetes. Nat. Commun. 2018;9:1–14. doi: 10.1038/s41467-018-03939-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.C., Chang P.F., Lin H.F., Liu K., Chang M.H., Ni Y.H. Variants in the autophagy-related gene IRGM confer susceptibility to non-alcoholic fatty liver disease by modulating lipophagy. J. Hepatol. 2016;65:1209–1216. doi: 10.1016/j.jhep.2016.06.029. [DOI] [PubMed] [Google Scholar]
- Liu J., Li Y., Zhou X., Zhang X., Meng H., Liu S., Zhang L., He J., He Q., Geng Y. CaMKIV limits metabolic damage through induction of hepatic autophagy by CREB in obese mice. J. Endocrinol. 2020;244:353–367. doi: 10.1530/JOE-19-0251. [DOI] [PubMed] [Google Scholar]
- Liu K., Czaja M.J. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013;20:3–11. doi: 10.1038/cdd.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Takahashi Y., Desai N., Zhang J., Serfass J.M., Shi Y.G., Lynch C.J., Wang H.G. Bif-1 deficiency impairs lipid homeostasis and causes obesity accompanied by insulin resistance. Sci. Rep. 2016;6:20453. doi: 10.1038/srep20453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lívero F.A., Acco A. Molecular basis of alcoholic fatty liver disease: from incidence to treatment. Hepatol. Res. 2016;46:111–123. doi: 10.1111/hepr.12594. [DOI] [PubMed] [Google Scholar]
- Lizaso A., Tan K.T., Lee Y.H. β-adrenergic receptor-stimulated lipolysis requires the RAB7-mediated autolysosomal lipid degradation. Autophagy. 2013;9:1228–1243. doi: 10.4161/auto.24893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N.S., Chiu W.C., Chen Y.L., Peng H.C., Shirakawa H., Yang S.C. Fish oil up-regulates hepatic autophagy in rats with chronic ethanol consumption. J. Nutr. Biochem. 2020;77:108314. doi: 10.1016/j.jnutbio.2019.108314. [DOI] [PubMed] [Google Scholar]
- Lu W., Mei J., Yang J., Wu Z., Liu J., Miao P., Chen Y., Wen Z., Zhao Z., Kong H., et al. ApoE deficiency promotes non-alcoholic fatty liver disease in mice via impeding AMPK/mTOR mediated autophagy. Life Sci. 2020;252:117601. doi: 10.1016/j.lfs.2020.117601. [DOI] [PubMed] [Google Scholar]
- Lu Y., Cederbaum A.I. Cytochrome P450s and alcoholic liver disease. Curr. Pharm. Des. 2018;24:1502–1517. doi: 10.2174/1381612824666180410091511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez N., Garcia-Macia M., Sahu S., Athonvarangkul D., Liebling E., Merlo P., Cecconi F., Schwartz G.J., Singh R. Autophagy in the CNS and periphery coordinate lipophagy and lipolysis in the brown adipose tissue and liver. Cell Metab. 2016;23:113–127. doi: 10.1016/j.cmet.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamae Y., Nishito Y., Nakai N., Nagumo Y., Usui T., Masuda S., Kambe T., Nagao M. Tetrandrine induces lipid accumulation through blockade of autophagy in a hepatic stellate cell line. Biochem. Biophys. Res. Commun. 2016;477:40–46. doi: 10.1016/j.bbrc.2016.06.018. [DOI] [PubMed] [Google Scholar]
- Negoita F., Blomdahl J., Wasserstrom S., Winberg M.E., Osmark P., Larsson S., Stenkula K.G., Ekstedt M., Kechagias S., Holm C. PNPLA3 variant M148 causes resistance to starvation‐mediated lipid droplet autophagy in human hepatocytes. J. Cell. Biochem. 2019;120:343–356. doi: 10.1002/jcb.27378. [DOI] [PubMed] [Google Scholar]
- Ni H.M., Williams J.A., Yang H., Shi Y.H., Fan J., Ding W.X. Targeting autophagy for the treatment of liver diseases. Pharmacol. Res. 2012;66:463–474. doi: 10.1016/j.phrs.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Mahony F., Wroblewski K., O'Byrne S.M., Jiang H., Clerkin K., Benhammou J., Blaner W.S., Beaven S.W. Liver X receptors balance lipid stores in hepatic stellate cells through Rab18, a retinoid responsive lipid droplet protein. Hepatology. 2015;62:615–626. doi: 10.1002/hep.27645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirazzi C., Valenti L., Motta B.M., Pingitore P., Hedfalk K., Mancina R.M., Burza M.A., Indiveri C., Ferro Y., Montalcini T., et al. PNPLA3 has retinyl-palmitate lipase activity in human hepatic stellate cells. Hum. Mol. Genet. 2014;23:4077–4085. doi: 10.1093/hmg/ddu121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro J.A., Kaushik S., Koga H., Armi C., Shui G., Wenk M.R., Di Paolo G., Cuervo A.M. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E705–E714. doi: 10.1073/pnas.1113036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V., Dötsch V., Johansen T., Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Samuel V.T., Shulman G.I. Nonalcoholic fatty liver disease as a nexus of metabolic and hepatic diseases. Cell Metab. 2018;27:22–41. doi: 10.1016/j.cmet.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayan A., Mashek M.T., Mashek D.G. ATGL promotes autophagy/lipophagy via SIRT1 to control hepatic lipid droplet catabolism. Cell Rep. 2017;19:1–9. doi: 10.1016/j.celrep.2017.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon H.T., Bartneck M., Borkham-Kamphorst E., Nattermann J., Lammers T., Tacke F., Weiskirchen R. Pharmacological intervention in hepatic stellate cell activation and hepatic fibrosis. Front. Pharmacol. 2016;7:33. doi: 10.3389/fphar.2016.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder B., Schulze R.J., Weller S.G., Sletten A.C., Casey C.A., McNiven M.A. The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology. 2015;61:1896–1907. doi: 10.1002/hep.27667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze R.J., Drizyte K., Casey C.A., McNiven M.A. Hepatic lipophagy: new insights into autophagic catabolism of lipid droplets in the liver. Hepatol. Commun. 2017a;1:359–369. doi: 10.1002/hep4.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze R.J., Rasineni K., Weller S.G., Schott M.B., Schroeder B., Casey C.A., McNiven M.A. Ethanol exposure inhibits hepatocyte lipophagy by inactivating the small guanosine triphosphatase Rab7. Hepatol. Commun. 2017b;1:140–152. doi: 10.1002/hep4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze R.J., Sathyanarayan A., Mashek D.G. Breaking fat: the regulation and mechanisms of lipophagy. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017c;1862:1178–1187. doi: 10.1016/j.bbalip.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuppan D., Ashfaq-Khan M., Yang A.T., Kim Y.O. Liver fibrosis: direct antifibrotic agents and targeted therapies. Matrix Biol. 2018:68–69. doi: 10.1016/j.matbio.2018.04.006. 435-451. [DOI] [PubMed] [Google Scholar]
- Seok S., Fu T., Choi S.E., Li Y., Zhu R., Kumar S., Sun X., Yoon G., Kang Y., Zhong W. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516:108–111. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., De Cegli R., Mansueto G., Saha P.K., Vetrini F., Visvikis O., Huynh T., Carissimo A., Palmer D., Klisch T.J. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka T., Welter E., Borovsky N., Amar N., Mari M., Reggiori F., Elazar Z. Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. EMBO J. 2015;34:2117–2131. doi: 10.15252/embj.201490315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Cuervo A.M. Lipophagy: connecting autophagy and lipid metabolism. Int. J. Cell Biol. 2012;2012:282041. doi: 10.1155/2012/282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., Czaja M.J. Autophagy regulates lipid metabolism. Nature. 2009a;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Xiang Y., Wang Y., Baikati K., Cuervo A.M., Luu Y.K., Tang Y., Pessin J.E., Schwartz G.J., Czaja M.J. Autophagy regulates adipose mass and differentiation in mice. J. Clin. Invest. 2009b;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B.K., Marcinko K., Desjardins E.M., Lally J.S., Ford R.J., Steinberg G.R. Treatment of nonalcoholic fatty liver disease: role of AMPK. Am. J. Physiol. Endocrinol. Metab. 2016;311:E730–E740. doi: 10.1152/ajpendo.00225.2016. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Hikita H., Tatsumi T., Sakamori R., Nozaki Y., Sakane S., Shiode Y., Nakabori T., Saito Y., Hiramatsu N. Rubicon inhibits autophagy and accelerates hepatocyte apoptosis and lipid accumulation in nonalcoholic fatty liver disease in mice. Hepatology. 2016;64:1994–2014. doi: 10.1002/hep.28820. [DOI] [PubMed] [Google Scholar]
- Tansey J., Sztalryd C., Gruia-Gray J., Roush D., Zee J., Gavrilova O., Reitman M., Deng C.X., Li C., Kimmel A. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6494–6499. doi: 10.1073/pnas.101042998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi T., Takayama K., Ishii S., Yamamoto A., Hara T., Minami N., Miyasaka N., Kubota T., Matsuura A., Itakura E., et al. Forced lipophagy reveals that lipid droplets are required for early embryonic development in mouse. Development. 2018;145:dev161893. doi: 10.1242/dev.161893. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N., Kounakis K., Chaniotakis M., Markaki M. Emerging roles of lipophagy in health and disease. Front. Cell Dev. Biol. 2019;7:185. doi: 10.3389/fcell.2019.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoen L.F., Guimaraes E.L., Dolle L., Mannaerts I., Najimi M., Sokal E., van Grunsven L.A. A role for autophagy during hepatic stellate cell activation. J. Hepatol. 2011;55:1353–1360. doi: 10.1016/j.jhep.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Thomes P.G., Trambly C.S., Fox H.S., Tuma D.J. Acute and chronic ethanol administration differentially modulate hepatic autophagy and transcription factor EB. Alcohol. Clin. Exp. Res. 2015;39:2354–2363. doi: 10.1111/acer.12904. [DOI] [PubMed] [Google Scholar]
- Thomes P.G., Trambly C.S., Thiele G.M., Duryee M.J., Fox H.S., Haorah J., Donohue T.M. Proteasome activity and autophagosome content in liver are reciprocally regulated by ethanol treatment. Biochem. Biophys. Res. Commun. 2012;417:262–267. doi: 10.1016/j.bbrc.2011.11.097. [DOI] [PubMed] [Google Scholar]
- Tsuchida T., Friedman S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017;14:397–411. doi: 10.1038/nrgastro.2017.38. [DOI] [PubMed] [Google Scholar]
- Wang C.W. Lipid droplets, lipophagy, and beyond. Biochim. Biophys. Acta. 2016;1861:793–805. doi: 10.1016/j.bbalip.2015.12.010. [DOI] [PubMed] [Google Scholar]
- Wang L., Khambu B., Zhang H., Yin X.M. Autophagy in alcoholic liver disease, self-eating triggered by drinking. Clin. Res. Hepatol. Gastroenterol. 2015;39:S2–S6. doi: 10.1016/j.clinre.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C., Martinez-Lopez N., Otten E.G., Carroll B., Maetzel D., Singh R., Sarkar S., Korolchuk V.I. Autophagy, lipophagy and lysosomal lipid storage disorders. Biochim. Biophys. Acta. 2016;1861:269–284. doi: 10.1016/j.bbalip.2016.01.006. [DOI] [PubMed] [Google Scholar]
- Wilfling F., Haas J.T., Walther T.C., Farese R.V. Lipid droplet biogenesis. Curr. Opin. Cell Biol. 2014;29:39–45. doi: 10.1016/j.ceb.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Zhu B., Peng X., Zhou M., Jia D., Gu J. Activation of farnesoid X receptor attenuates hepatic injury in a murine model of alcoholic liver disease. Biochem. Biophys. Res. Commun. 2014;443:68–73. doi: 10.1016/j.bbrc.2013.11.057. [DOI] [PubMed] [Google Scholar]
- Xiong J., Wang K., He J., Zhang G., Zhang D., Chen F. TFE3 alleviates hepatic steatosis through autophagy-induced lipophagy and PGC1α-mediated fatty acid β-Oxidation. Int. J. Mol. Sci. 2016;17:387. doi: 10.3390/ijms17030387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Grijalva A., Skowronski A., van Eijk M., Serlie M.J., Ferrante A.W. Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18:816–830. doi: 10.1016/j.cmet.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Li P., Fu S., Calay E.S., Hotamisligil G.S. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner R., Madeo F., Kratky D. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat. Rev. Mol. Cell Biol. 2017;18:671–684. doi: 10.1038/nrm.2017.76. [DOI] [PubMed] [Google Scholar]
- Zhang H., Yan S., Khambu B., Ma F., Li Y., Chen X., Martina J.A., Puertollano R., Li Y., Chalasani N. Dynamic MTORC1-TFEB feedback signaling regulates hepatic autophagy, steatosis and liver injury in long-term nutrient oversupply. Autophagy. 2018a;14:1779–1795. doi: 10.1080/15548627.2018.1490850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Liu J., Tong Q., Lin L. SIRT3 acts as a positive autophagy regulator to promote lipid mobilization in adipocytes via activating AMPK. Int. J. Mol. Sci. 2020;21:E372. doi: 10.3390/ijms21020372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Yao Z., Chen Y., Qian L., Jiang S., Zhou J., Shao J., Chen A., Zhang F., Zheng S. Lipophagy and liver disease: new perspectives to better understanding and therapy. Biomed. Pharmacother. 2018b;97:339–348. doi: 10.1016/j.biopha.2017.07.168. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhao S., Yao Z., Wang L., Shao J., Chen A., Zhang F., Zheng S. Autophagy regulates turnover of lipid droplets via ROS-dependent Rab25 activation in hepatic stellate cell. Redox Biol. 2017;11:322–334. doi: 10.1016/j.redox.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N., Guo F.F., Xie K.Q., Zeng T. Targeting Nrf-2 is a promising intervention approach for the prevention of ethanol-induced liver disease. Cell. Mol. Life Sci. 2018;75:3143–3157. doi: 10.1007/s00018-018-2852-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Wu Y., Ye X., Ma L., Qi J., Yu D., Wei Y., Lin G., Ren G., Li D. FGF21 ameliorates nonalcoholic fatty liver disease by inducing autophagy. Mol. Cell. Biochem. 2016;420:107–119. doi: 10.1007/s11010-016-2774-2. [DOI] [PubMed] [Google Scholar]
- Zubiete-Franco I., Garcia-Rodriguez J.L., Martinez-Una M., Martinez-Lopez N., Woodhoo A., Juan V.G., Beraza N., Lage-Medina S., Andrade F., Fernandez M.L., et al. Methionine and S-adenosylmethionine levels are critical regulators of PP2A activity modulating lipophagy during steatosis. J. Hepatol. 2016;64:409–418. doi: 10.1016/j.jhep.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]