Abstract
Convalescent serum with a high abundance of neutralization IgG is a promising therapeutic agent for rescuing COVID-19 patients in the critical stage. Knowing the concentration of SARS-CoV-2 S1-specific IgG is crucial in selecting appropriate convalescent serum donors. Here, we present a portable microfluidic ELISA technology for rapid (15 min), quantitative, and sensitive detection of anti-SARS-CoV-2 S1 IgG in human serum with only 8 μL sample volume. We first identified a humanized monoclonal IgG that has a high binding affinity and a relatively high specificity towards SARS-CoV-2 S1 protein, which can subsequently serve as the calibration standard of anti-SARS-CoV-2 S1 IgG in serological analyses. We then measured the abundance of anti-SARS-CoV-2 S1 IgG in 16 convalescent COVID-19 patients. Due to the availability of the calibration standard and the large dynamic range of our assay, we were able to identify “qualified donors” for convalescent serum therapy with only one fixed dilution factor (200 ×). Finally, we demonstrated that our technology can sensitively detect SARS-CoV-2 antigens (S1 and N proteins) with pg/mL level sensitivities in 40 min. Overall, our technology can greatly facilitate rapid, sensitive, and quantitative analysis of COVID-19 related markers for therapeutic, diagnostic, epidemiologic, and prognostic purposes.
Keywords: COVID-19, Antibody detection, Immunoassay, Microfluidics
Highlights
-
•
Identified a calibrator antibody for anti-SARS-CoV-2 S1 IgG quantification.
-
•
Rapid (15 min) and sensitive detection of anti-SARS-CoV-2S1 IgG in serum.
-
•
Quantitative evaluation of COVID-19 convalescent serum with a single dilution factor (200X).
-
•
Sensitive detection of SARS-CoV-2 N- and S-protein in serum down to ~10 pg/mL with 3-4 orders of magnitude dynamic range.
-
•
8 μL sample/reagent volume.
1. Introduction
The disease (COVID-19) related to novel coronavirus (SARS-CoV-2) has caused more than half a million of deaths and remains a severe threat to global health (WHO, 2020c). The World Health Organization has indicated that the mortality rate for the critical care COVID-19 cases may be higher than 50% (WHO, 2020a, b). Unfortunately, to date there is no standardized therapy for treating COVID-19 patients, especially those in the critical stage (WHO, 2020a).
Recent clinical researches have demonstrated that the convalescent serum therapy is a promising approach to improve the survival rate in the severe cases (Bloch et al., 2020; Casadevall and Pirofski, 2020; Chen et al., 2020; Duan et al., 2020; Shen et al., 2020). SARS-CoV-2 neutralizing IgG (S1-specific IgG), the major active component in the convalescent serum is developed by the adaptive immune system approximately 7–10 days after SARS-CoV-2 infection (Padoan et al., 2020; Sun et al., 2020). Due to its high affinity and high abundance, neutralizing IgG can bind and block the binding epitopes, (e.g., receptor binding domains (RBDs) and other related domains, on the S1 protein on SARS-CoV-2), preventing it from invading human cells (Long et al., 2020; Shen et al., 2020). Due to the variations in the strength of the adaptive immune response, the abundance of SARS-CoV-2 S1 specific IgG varies significantly from patient to patient (Amanat et al., 2020; Ju et al., 2020). To ensure a high therapeutic efficacy, only the convalescent serum from recovered donors with a high level of SARS-CoV-2 S1-specific IgG should be selected as the therapeutic agent. Thus, knowing the concentration of SARS-CoV-2 S1-specific IgG is essential for selecting appropriate convalescent serum donors.
Yet, existing antibody detection methods are still far from adequate. The gold nanoparticle-based lateral flow assay (e.g., paper-based test strips) is popular for rapid detection of IgG/IgM antibodies (especially for point-of-care diagnostics). Although fast (5–20 min), it provides only binary (i.e., yes/no) information with very limited sensitivities. Therefore, it cannot be used for the quantitative evaluation of the convalescent serum. Conventional ELISA (enzyme-linked immunosorbent assay), on the other hand, can provide accurate and sensitive results, but it involves complicated and expensive instruments and long assay time (~3 h) (Amanat et al., 2020; Wang et al., 2020). Given the narrow dynamic range (<2 orders of magnitude), multiple dilution factors are required for performing the serological analysis of SARS-CoV-2 S1-specific IgG, which increases the cost and decreases the assay throughput. Moreover, due to the lack of an internal calibration standard, conventional ELISA cannot measure the effective concentration of the circulating anti-S1 IgG, making quality control of the convalescent serum harder (Ju et al., 2020).
In this work, we present a portable microfluidic chemiluminescent ELISA technology for rapid (15 min), quantitative, and sensitive detection of SARS-CoV-2 S1 specific IgG. We first characterized four humanized (chimeric) monoclonal IgG and identified a suitable candidate (D006) with a high binding affinity and specificity towards SARS-CoV-2 S1 protein that can subsequently serve as the calibration standard of anti-SARS-CoV-2 S1 IgG in serological analyses. To evaluate clinical applicability of our technology, we conducted measurements with the serum samples collected from 16 recovered COVID-19 patients and 3 healthy donors. Because of the availability of a calibration standard and large dynamic range of our assay, we successfully converted the measured results into effective D006 concentrations and identified the best donor candidates for the convalescent serum therapy with only one fixed serum dilution factor (200 ×). Therefore, our technology can greatly accelerate and improve the on-site screening process of potential convalescent serum donors. To further extend the ability of the portable microfluidic technology, we also demonstrated sensitive detection of SARS-CoV-2 antigens (S1 and N proteins) with a lower limit of detection (LLOD) of ~10 pg/mL in 40 min using spiked serum as the model system.
2. Materials and methods
2.1. Microfluidic chemiluminescent ELISA system
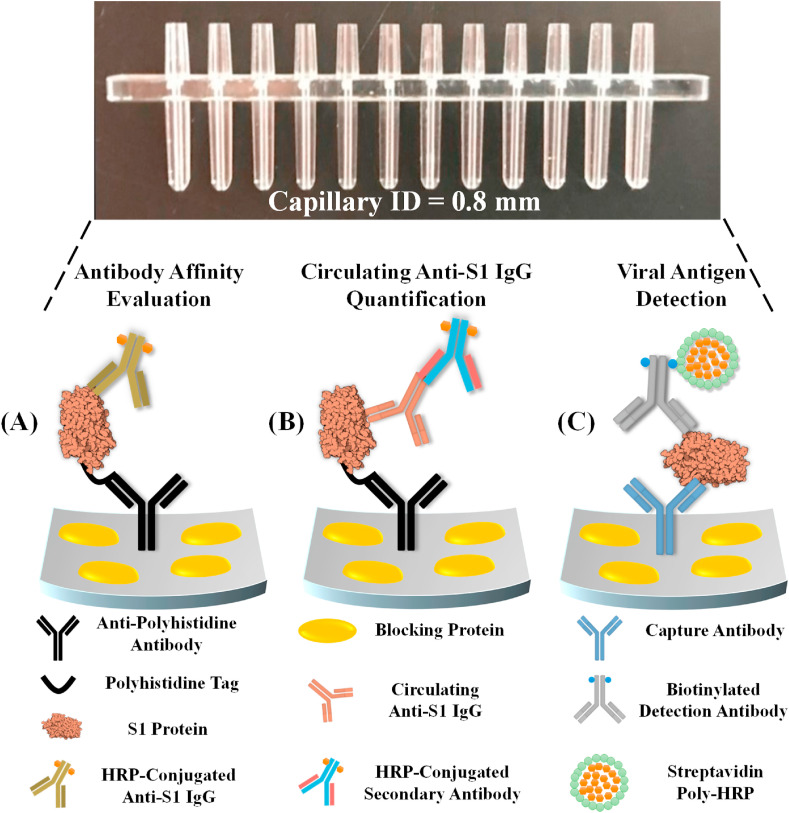
A detailed description of the microfluidic chemiluminescent ELISA system and the corresponding capillary sensor arrays can be found in our previous publications (Tan et al., 2018, 2020). A photo of the capillary sensor array can be found in Fig. 1 . It is made of polystyrene using the injection molding method. The sensor array has 12 channels, each of which has an inner diameter of 0.8 mm and approximately 8 μL volume, and acts as an ELISA reactor.
Fig. 1.
Graphical illustrations of the COVID-19 related immunoassays that were performed with our microfluidic chemiluminescent ELISA platform, including (A) affinity evaluation of calibration antibodies, (B) detection of circulating anti-SARS-CoV-2 S1 IgG in serum samples, and (C) detection of SARS-CoV-2 antigens such as S1 and N protein.
2.2. Chemical reagents
The chemiluminescent substrate (SuperSignal™ ELISA Femto Substrate, 37075), the UltraPure™ DNase/RNase-free distilled water (10977023), and the SuperBlock™ (PBS) blocking buffer (37515) were purchased from Thermo Fisher. The ELISA coating buffer (1 × PBS, DY006) and concentrated wash buffer (WA126) were purchased from R&D Systems. The concentrated reagent diluent (10% BSA in 1 × PBS) was purchased from Sigma-Aldrich (SRE0036-250ML). The normal human serum (H4522-20ML), which was used as the dilution buffer and as one of the negative controls in IgG detection experiments, and the heat-inactivated normal human serum (H5667-20ML), which was used as another negative control in IgG detection experiments, were both purchased from Millipore Sigma. Human-cell-expressed SARS-CoV-2 Spike S1-His recombinant protein (40591-V08H), human-cell-expressed SARS-CoV-2 Spike RBD-His recombinant protein (40592-V08H) and insect-cell-expressed SARS-CoV Spike S1-His recombinant protein (40150-V08B1) were provided by Sino Biological. The recombinant CR3022 therapeutic antibody was purchased from Creative Biolabs (MRO-1214LC). The humanized chimeric antibodies D001, D003, and D006 were developed and provided by Sino Biological (Catalog number: 40150-D001, 40150-D003, and 40150-D006). The anti-polyhistidine antibody that was used in polyhistidine-mediated S1 protein immobilization (see Fig. S1) was purchased from Thermo Fisher (MA1-21315). The horseradish peroxide (HRP) conjugated secondary antibody used in the IgG detection experiment was from the detection antibody in Thermo Fisher's human total IgG ELISA kit (88-50550-22). The HRP conjugation of CR3022, D001, D003, and D006 antibodies were carried out with Abcam's HRP conjugation kit (ab102890) with a molar ratio of antibody: HRP = 1 : 4.
The antibody pairs for viral antigen detection were developed and provided by Sino Biological. 40150-D006 (capture) and 40591-MM43 (detection) were used for S1 detection. 40143-R001 (capture) and 40143-MM05 (detection) were used for N detection. The biotinylation of the detection antibodies were performed with Thermo Fisher's EZ-Link™ Micro NHS-PEG4-Biotinylation Kit (21955), following the manufacturer's conjugation and purification protocols. The yields of the conjugation were determined with Thermo Fisher's NanoDrop™ One microvolume UV-Vis spectrophotometer. The streptavidin poly-HRP solution (21140) and the poly-HRP dilution buffer (1% casein in 1 × PBS, N500) were purchased from Thermo Fisher.
2.3. ELISA protocols
The schematic diagrams for the all three ELISA assays can be found in Fig. 1. The ELISA protocols (especially the incubation time for each steps) can be found in Fig. S1. For all three assays, the working solution of the wash buffer was diluted with UltraPure™ DNase/RNase-free distilled water to achieve 1 × working concentration. The working solution of the reagent diluent (2.5% BSA in 1 × PBS) was prepared by diluting the 10% BSA four times with the ELISA coating buffer (1 × PBS).
In the first step of the reactor preparation process (Fig. S1(A)), the working solution of the capture antibody (i.e., D006 in S1 detection experiments) or the anti-His antibody (in antibody affinity assessment and IgG detection experiments) were prepared by diluting the stock solution with the ELISA coating buffer (1 × PBS, pH = 7.4) to achieve a final concentration of 10 μg/mL. Note that for the antibody affinity experiments and the actual detection of anti-S1 IgG, the second incubation step was used for blocking, plus S1 protein immobilization (with 2 μg/mL of S1-His protein dissolved in 2.5% BSA). For the S1 protein detection, this step was used for blocking only (with 1% BSA in 1 × PBS).
For the antibody affinity experiments, various concentrations of HRP-conjugated monoclonal antibodies were prepared by diluting the stock solution in 2.5% BSA in 1 × PBS. For the anti-S1 IgG detection experiments (see Fig. S1(C) for the detailed protocol), various concentrations of monoclonal antibodies were prepared by diluting the stock solutions with 50 times diluted human serum (the serum was diluted with 1 × reagent diluent, which correlates to 1% BSA). The working solution of the detection antibody (in this case, the HRP-conjugated secondary antibody) was prepared by diluting the stock solution 250 times in 1 × reagent diluent.
For the viral antigen detection experiments, various concentrations of the viral antigen proteins (S1 or N) were prepared by diluting the stock solution with 10 times diluted human serum. The working solution of the biotinylated detection antibodies (40591-MM43 for S1 detection, 40143-MM05 for N detection) were prepared by diluting the stock solution in 2.5% BSA in PBS. The final concentrations were 20 ng/mL for S1 detection and 30 ng/mL for N protein detection. The working solution for the streptavidin poly-HRP was prepared by diluting the stock solution 1500 times with the poly-HRP dilution buffer. See Fig. S1(D) for the incubation time for each step.
The protocol for the conventional plate-based ELISA can be found in the Supplementary Material.
2.4. ELISA measurements
The signal intensities of the microfluidic ELISA were measured with a chemiluminescent imaging method using a CMOS camera. The blue channel of the photos was extracted for signal quantification (with ImageJ). To enhance the dynamic ranges of the ELISA, multiple exposures with adjustable exposure time were applied (see Fig. S2). All chemiluminescent intensities (CL intensity) were normalized to 3 s of exposure time (an intermediate value between 0.5 s and 15 s). The Details of the chemiluminescent imaging and the multiple exposure approaches can be found in our previous publication (Tan et al., 2017).
2.5. Patient sample collection
Serum samples were obtained from subjects enrolled in Hackensack University Medical Center's prospective Phase IIa clinical trial assessing the safety and efficacy of convalescent serum from recovered COVID-19 donors (ClinicalTrials.gov study # NTC04343755, FDA IND approval 4/4/20). The subjects were aged 18–60 years and consisted of patients with documented infection with SARS-CoV-2 or donors who were recovered with least 14 days from resolution of symptoms and have high titers of neutralizing antibodies and swab negative.
3. Results
3.1. Affinity evaluation of monoclonal antibodies against SARS-CoV-2 S1
A good calibration standard of anti-SARS-CoV-2 S1-IgG should be a human-originated or humanized IgG with a high affinity and specificity towards SARS-CoV-2 S1. It should also have a large linear dynamic range against SARS-CoV-2 S1. However, due to nascence of SARS-CoV-2 there is no “gold standard” antibody that can be used in IgG tests. Consequently, it is urgent to find humanized antibodies with high binding affinities. Here we present a simple, rapid and effective approach for the affinity assessment of monoclonal antibodies, especially for ELISA applications.
An overview of the assay mechanism and the corresponding protocol are illustrated in Fig. 1(A) and S1(B), respectively. In order to avoid potential sources of error, our assay follows a single-step ELISA format shown in Fig. 1(B) rather than the more complicated sandwich ELISA format. Recombinant S1 protein (2 μg/mL) was first immobilized on the supporting surface via anti-polyhistidine-tag antibody (see Fig. S3) for better access to the epitopes (especially the RBD) on the S1 antibody. Then six different concentrations of HRP-conjugated IgG (molar ratio IgG: HRP = 1 : 4) IgG in 2.5% BSA buffer were withdrawn into the capillary reactors. The nature of microfluidics (high surface-to-volume ratio) expedites the process of IgG binding to S1 protein. Consequently, the entire assay can be completed in 8 min, including 6 min of IgG incubation, 4 times of rinsing, addition of substrate, and optical reading.
To compare the antibody's affinity towards the S1 protein of SARS-CoV-2 and SARS-CoV, we performed a side-by-side experiment for antibodies CR3022, D001, D003, and D006. The first antibody, CR3022, is a therapeutic human IgG originally found in recovered SARS patients. It was recently reported to have cross-reactivity towards the S1 protein of SARS-CoV-2 (Joyce et al., 2020; Tian et al., 2020; Yuan et al., 2020). It was also used as a positive control antibody in several COVID-19 related researches (Amanat et al., 2020; Dingens et al., 2020; McDade et al., 2020). The remaining three antibodies, D001, D003, and D006, are humanized chimeric IgGs (the precursors of D001 and D003 were originally raised in mouse and D006 was originally raised in rabbit) that were developed against the S1 protein of SARS-CoV. They were also believed to have cross-reactivities with the S1 protein of SARS-CoV-2.
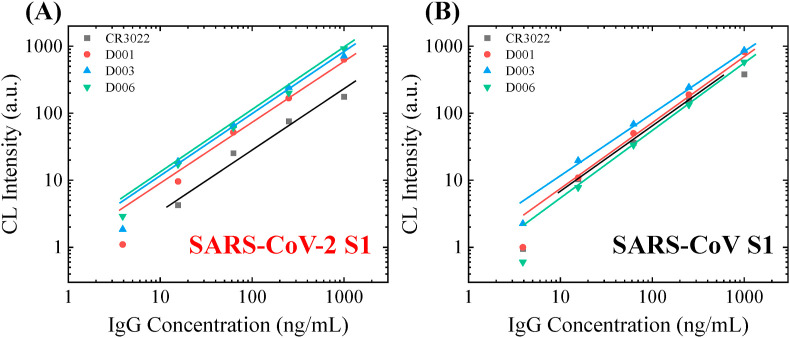
Due to the nature of the single-incubation protocol, our assay exhibits very good (generally <5%) intra-assay consistency (see, for example, Fig. S4), thus ensuring highly reliable measurements. As shown in Fig. 2 (A), these four antibodies have very different affinities towards the S1 protein of SARS-CoV-2. Note that the data for 1 ng/mL are not presented because the signal is not detectable for all four antibodies. We also performed a group of negative control experiments by directly applying one of the antibody candidates (D006) to a properly blocked capillary array (without the immobilization of anti-His antibody and recombinant S1 protein). No signal was detected at all concentrations.
Fig. 2.
Affinity screening of the calibration antibodies. (A) Calibration curves of 4 different monoclonal humanized S1 specific IgG against the S1 protein from SARS-CoV-2. (B) Calibration curves of 4 different monoclonal humanized S1 specific IgG against the S1 protein from SARS-CoV (B). The solid lines are the linear fit of the data in the log-log scale. D006 is the only antibody that has a high affinity and high specificity towards SARS-CoV-2 S1. Illustration of the assay mechanism, which uses a single-step ELISA format, is shown in Fig. 1(A). The sample-to-answer time of this assay is 8 min.
This set of experiments provide valuable affinity information about the four candidate IgGs. D001, D003, and D006 have similar affinity towards SARS-CoV-2 S1 and much better than CR3022. For the points within the linear dynamic ranges, the signal intensities with the strongest antibody (D006) are 3–5 times higher than the weakest antibody (CR3022). For example, the signal intensities for these two antibodies at 1000 ng/mL are 916.2 and 175.9, respectively. These observations agree with several recently published results and the BLI (bio-layer interferometry) measurements at Sino Biological, which shows the equilibrium dissociation constant, KD, of 6–12 nM and <1 nM, for CR3022 and D001, respectively (Joyce et al., 2020; Tian et al., 2020; Yuan et al., 2020). Note that although our current method allows us to rapidly evaluate the relatively affinity among the antibody candidates in ELISA applications, it is not designed to extract the exact value of KD.
For comparison purposes, the antibodies’ affinities towards SARS-CoV S1 are shown in Fig. 2(B), which suggests that all four candidates have a similar binding affinity. According to the results, D006 is the only one that shows a higher affinity toward SARS-CoV-2 S1 (see Fig. S4). Consequently, D006 shows a great potential as the antibody standard, which has not only the high affinity but also the relatively high specificity towards SARS-CoV-2.
3.2. Evaluation of anti-SARS-CoV-2 S1 IgG calibrators in realistic settings
Our next step was to evaluate SARS-CoV-2 S1 IgG calibrators in a clinically relevant setting, which is illustrated in Fig. 1(B). Same as in the previous section, recombinant S1 protein was first immobilized on the capillary inner surface through poly-histidine mediated immobilization. Then, spiked-in S1 specific IgG in the sample was attracted to the surface through immunosorbent reaction. Finally, the HRP-conjugated detection antibody was used to visualize the binding of the immobilized IgG. To ensure detection specificity, a monoclonal detection antibody that binds specifically to the Fc domain on human IgG was used. According to the protocol in Fig. S1(C), the entire assay was completed in 15 min.
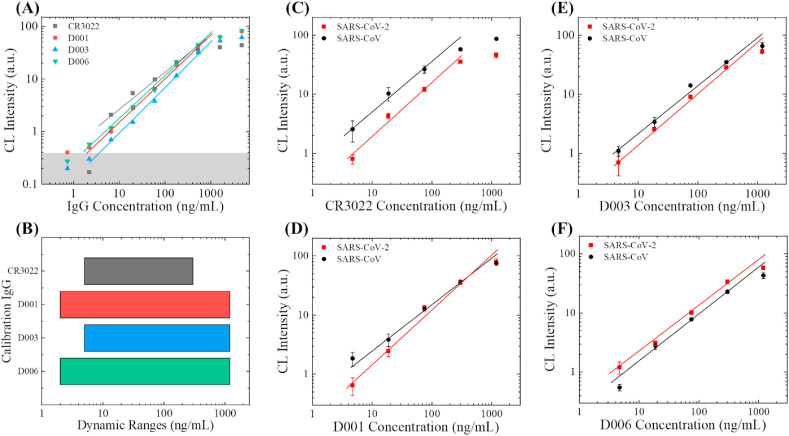
To validate the feasibility of our assay, we tested all four aforementioned humanized monoclonal anti-SARS-CoV-2 S1 antibodies. In order to mimic the actual clinical situation, we decided to use 50 times diluted human serum as the solvent of the IgG antibodies in all following IgG experiments (50-10000 is the typical dilution factor of serum in actual serological IgG analyses). The entire dynamic ranges of these three antibodies against SARS-CoV-2 S1 can be found in Fig. 3 (A). In general, the chemiluminescent intensities are proportional to the concentration of the spiked-in monoclonal antibodies. The linear dynamic range in the log-log scale for CR3022, D001, D003, and D006 is 5–300 ng/mL, 2-1200 ng/mL, 5-1200 ng/m, and 2-1200 ng/mL, respectively (as shown in Fig. 3(B)). The corresponding slopes in the linear range are 0.64, 0.83, 0.88, and 0.81 (on the log-log-scale). As a negative control, the human IgG isotype does not generate any detectable signal within the entire range of detection (0.7–4800 ng/mL).
Fig. 3.
Evaluation of anti-S1 calibration antibodies. (A) Entire dynamic ranges for the detection of the four humanized monoclonal antibodies (against SARS-CoV-2 S1). The concentrations were prepared from 3 times of serial dilution (starting from 4800 ng/mL). The averaged background is subtracted from all data points. The solid lines are the linear fit of the data in the log-log scale. The grey shaded area marks 3 × standard deviation of the background. (B) Comparison of the linear dynamic ranges. (C)–(F) Detection of the calibration antibodies in 50 times diluted serum, against the S1 protein from SARS-CoV-2 (red squares) and SARS-CoV (black circles). The calibration curves are generated with three different monoclonal humanized antibodies (CR3022 in (C), D001 in (D), D003 in (E), and D006 in (D)). The solid lines are the linear fit for the data in the log-log scale. Error bars are generated from duplicate measurements. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We also examined the intra-assay consistency for all four candidates. The corresponding results are shown in Fig. 3(C)–(F). To evaluate the differences in antibody's specificity towards SARS-CoV-2 S1 and SARS-CoV S1, we performed a side-by-side study with these two types of S1 proteins for all four clones of antibodies. Apparently, all these four antibodies are still detectable at 4.7 ng/mL with both types of S1 proteins.
For CR3022, the signal for SARS-CoV-2 S1 is systematically lower than that for SARS-CoV S1, indicative of a significantly weakened affinity of CR3022 towards SARS-CoV-2 S1 than SARS-CoV S1. Due to the narrow linear dynamic range, poor specificity and relatively low binding affinity, CR3022 should not be used as the calibration standard of anti-SARS-CoV-2 S1 IgG. For D001, D003, and D006, signal for both types of S1 proteins is similar, indicating that the antibodies’ binding affinity towards SARS-CoV-2 S1 and SARS-CoV S1 should also be similar. Out of these three chimeric antibodies, D001 and D003 may have slightly higher affinities toward SARS-CoV S1 than SARS-CoV-2 S1. D006 is the only one that has a higher affinity towards SARS-CoV-2 S1 than SARS-CoV S1. These results are consistent with our observations in Fig. 2. Since D006 shows superior performance in both affinity and specificity, it is the best candidate as a calibration antibody. We will use it to calculate the effective concentration of anti-SARS-CoV-2 S1 IgG in the serum of convalescent COVID-19 patients.
3.3. Quantifying the concentration of anti-SARS-CoV-2 S1 IgG in convalescent sera
We performed microfluidic chemiluminescent ELISA measurements with serum samples collected from 16 convalescent COVID-19 patients and 3 healthy donors. For assay validation purposes, the serum samples were first analyzed with conventional plate-based ELISA, which took >3 h to complete (see the Supplementary Material for the assay protocol). Based on conventional ELISA measurements shown in Fig. S5 and Table S1, three patients’ samples are marked as negative (NS) and the remaining 13 samples are marked as positive (PS).
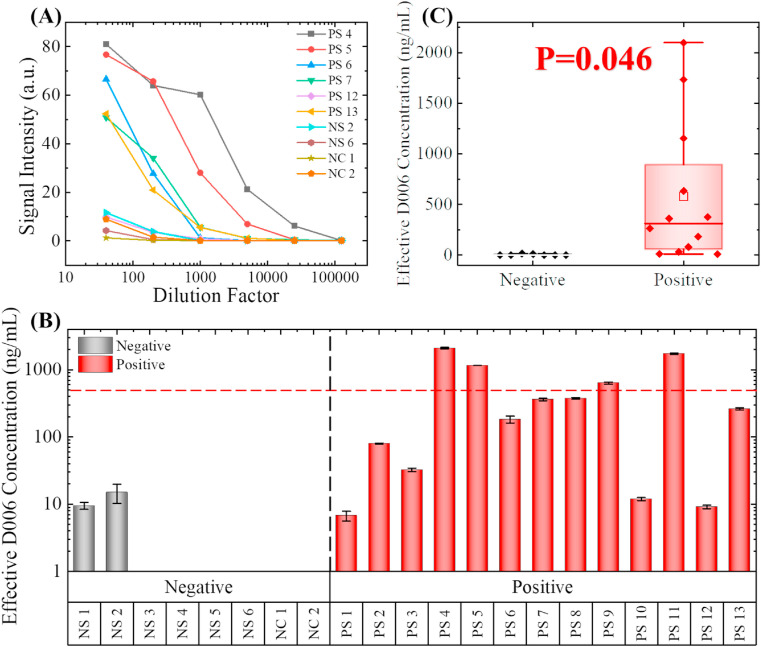
To identify the best dilution factor for our measurements, we performed a group of serial serum dilution tests, which is a classical ELISA test for the semi-quantitative measurement of the specific IgG in serum samples. An exemplary group of the serial dilution tests (including six positive samples, two negative samples, and two negative controls) can be found in Fig. 4 (A). All samples were diluted with 2.5% BSA in PBS. The dilution factors were 40 × , 200 × , 1000 × , 5000 × , 25,000 × , and 100,000 × , respectively. The titration curves for these six positive samples appear to be very different from each other. The maximum titer for PS 4 was 2,5000 × ; in contrast, the maximum titer for PS 12 was only 200 ×, indicating that the abundance of anti-SARS-CoV-2 S1 IgG differs significantly within the convalescent serum donors.
Fig. 4.
Detection of anti-S1 IgG in recovered COVID-19 patients' serum. (A) Serial dilution tests with 10 representative samples, including six positive samples (PS), two negative samples (NS), and two commercially available negative controls (NC). Note that the positive/negative was determined with traditional plate-based ELISA. 200 X dilution in 2.5% BSA was determined to be the optimum dilution factor for differentiating the strong positive samples from the weak positive and negative samples. (B). Effective D006 concentrations for all nineteen samples and the two negative controls. The concentrations are marked as 0 ng/mL if the calculated concentration was below 2 ng/mL (too close to LLOD). The error bars are generated from duplicate measurements. Only four samples have effective D006 concentrations higher than 500 ng/mL after 200 times of dilution. (Note that PS4 and PS11 exceeded the upper limit of detection). (C). Statistical comparison between the negative samples and the positive samples. Since p < 0.05, the difference between these two groups is statistically significant.
Although titration curves can be used to select the strongest donors for convalescent serum therapy, the requirement of multiple dilution factors still greatly limits the throughput of the assay. In order to perform the serum assessment in a single-point test, an appropriate dilution factor must be selected. Based on the data shown in Fig. 4(A), at 40 × dilution, the false positive rate is too high, and it is hard to differentiate the strongly positive samples (e.g., PS 4 and PS 5) from the intermediate positive samples (e.g., PS 6). On the other hand, if we select higher dilution factors (>1000), the false negative rate will be too high. For these reasons, the most appropriate dilution factor for COVID-19 convalescent serum evaluation by our microfluidic chemiluminescent ELISA platform should be 200 ×. At this dilution factor, the signal intensities for most of the positive samples are within the central part of the linear dynamic range, thus the reliability of the measurements can be ensured. Note that in the conventional ELISA (see results in Fig. S5 and Table. S1 in the Supplementary Material), four different dilution factors ranging from 100 × to 10,000 × are needed for IgG evaluation due to the limited dynamic range (OD = 0.2 to OD = 1.2).
The original signal intensity measurements for all 19 patient samples and the two negative controls can be found in Fig. S6; all error bars were generated from duplicate measurements. Although some overlap exists (e.g., PS 1 vs. NS 2), the signal intensities for the positive samples appear to be significantly higher than the negative samples (p = 0.003, see Fig. S7), which means that our measurements generally agree with the conventional plate-based ELISA measurements. Using the plate-based ELISA as the gold standard, our results will correlate to a 0% false-negative rate and a 25% false-positive rate. For the two “false positive” measurements, NS 1 was collected from a COVID-19 patient and should have probably been classified as a “weak positive” rather than a “negative”. NS 2 was collected from a healthy donor; the signal was possibly caused by the cross-reactivity of IgG that might have been induced by other types of coronaviruses.
Owing to the employment of the calibration standard, we were also able to convert our measurements results into effective calibrator IgG (D006) concentration using the calibration curve in Fig. 3(F). The effective D006 concentrations of the 21 samples can be found in Fig. 4(B). Note that the effective concentrations for six of the negative samples were below our lower limit of detection (2 ng/mL) and they were marked as 0 ng/mL; two of the calculated effective concentrations exceeded our upper limit of detection (1200 ng/mL) and they were marked as the calculated D006 concentration. Due to the variation in antibody affinity, the effective D006 concentrations are not necessarily equal to the actual concentration of anti-S1 IgG. Nevertheless, using this internal calibrator provides us a way to better quantify and compare anti-S1 IgG concentrations among patients.
The statistical analysis of the effective concentrations is shown in Fig. 4(C). The difference between the positive samples and the negative samples are still statistically significant (p = 0.046) in the two-tailed t-test. As shown in Fig. 3(B) and (C), the calculated effective anti-SARS-CoV-2 S1 IgG concentration in the positive samples distributed in a broad range between 7 and 2100 ng/mL, corresponding to the original concentration of the effective IgG of 1.4–4200 μg/mL (before the 200 × dilution).
According to recent studies about COVID-19 neutralization antibodies, to ensure an effective protection (>50% protection) the concentration of the neutralization antibodies should be higher than 1 μg/mL, depending on the binding epitope and the affinity of the antibodies (Brouwer et al., 2020; Cao et al., 2020; Ju et al., 2020; Pinto et al., 2020; Wu et al., 2020). In a typical convalescent serum therapy, 100–200 mL of donor serum is injected into the receptor's bloodstream of ~5000 mL, meaning that the convalescent serum is diluted 25–50 times. Based on this calculation, it is reasonable to use 500 ng/mL as the cut-off value of the effective anti-SARS-CoV-2 S1 IgG concentration in selecting qualified convalescent serum donors in order to produce 2 μg/mL antibody. As shown by the data and the red dashed line in Fig. 4(B), only four positive samples (PS 4, PS 5, PS 9, and PS 11) can be classified as qualified convalescent serum samples. On the other hand, traditional plate-based ELISA has identified six potentially qualified convalescent serum samples (PS 4, PS 5, PS 6, PS 7, PS 9, and PS 11, see Fig. S5) based on the criterion given in the Supplementary Material (i.e., OD > 0.4 at 10000x dilution), which included all of the four qualified samples identified using our method. The difference in the qualified samples between the two methods is simply due to the different criteria used. From the results above, we can conclude that our technology, along with the single-point absolute quantification approach, can facilitate the quantitative evaluation of the convalescent serum.
3.4. Detection of SARS-CoV-2 antigens
In addition to the evaluation of convalescent serum, our technology may also be able to facilitate the diagnosis of COVID-19 by sensitively detecting the viral antigens. Several publications have suggested that some of the viral antigen (such as S1 protein or N protein) may appear in blood for the patients who develop coronavirus viremia (Che et al., 2004; Diao et al., 2020). These viral proteins may also exist in mucus samples, such as saliva (Li et al., 2005; Vinayachandran and Saravanakarthikeyan, 2020).
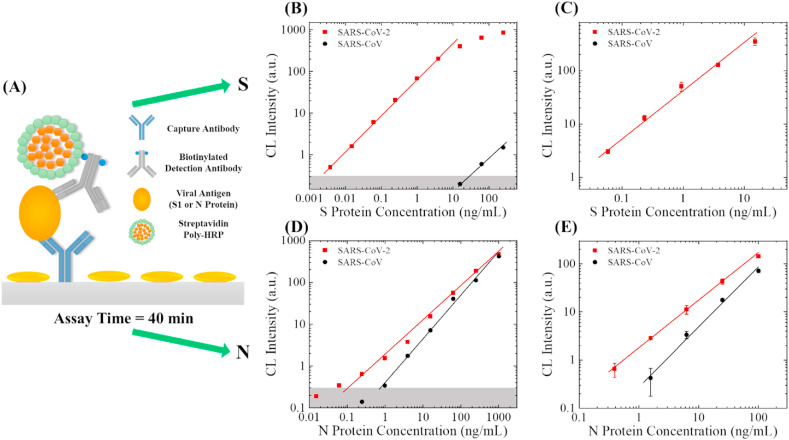
To sensitively detect the viral antigens, we employed the standard sandwich ELISA, along with the poly-HRP signal amplification technique, as illustrated in Fig. 5 (A). For S1 protein detection, SARS-CoV-2/SARS-CoV S1 RBD-specific antibody (D006) was used as the capture antibody that has previously shown a high affinity and high specificity towards SARS-CoV-2 S1 protein. SARS-CoV-2 S1-specific antibody (MM43) was used as the detection antibody. For N protein detection, SARS-CoV-2/SARS-CoV N-specific antibody (R001) was used as the capture antibody, and another SARS-CoV-2/SARS-CoV N-specific antibody (MM05) was used as the detection antibody. In order to enhance the detection sensitivity, we biotinylated the detection antibodies and employed streptavidin poly-HRP to improve the limit of detection by 5–10 times. In this set of experiments, the sample-to-answer time for both analytes were 40 min (see Fig. S1 for protocol).
Fig. 5.
SARS-CoV-2 antigen detection. (A) Illustration of the assay mechanism. The sample-to-answer time of this assay is 40 min. (B) Entire dynamic ranges of SARS-CoV-2 S1 protein (red squares) and SARS-CoV S1 protein (black circles) in 10 times diluted human serum. The averaged background is subtracted from all data points. The solid lines are the linear fit of the data in the log-log scale. The grey shaded area marks 3 × standard deviation of the background. The lower limit of detection (LLOD) for SARS-CoV-2 S1 protein is 0.004 ng/mL <0.01% cross reactivity was observed with SARS-CoV S1 protein. (C) Calibration curves for S1 proteins between 0.06 and 15 ng/mL. The error bars are generated from duplicate measurements. (D) Entire dynamic ranges of SARS-CoV-2 N protein (red squares) and SARS-CoV N protein (black circles) in 10 times diluted human serum. The averaged background is subtracted from all data points. The solid lines are the linear fit of the data in the log-log scale. The grey shaded area marks 3 × standard deviation of the background. The lower limit of detection (LLOD) for SARS-CoV-2 N protein and SARS-CoV S1 are 0.06 ng/mL and 1 ng/mL, respectively. (E) Calibration curves for N proteins between 0.39 and 100 ng/mL. The error bars are generated from duplicate measurements. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Same as in the calibration antibody experiments, we performed a side-by-side study with the recombinant viral antigens from SARS-CoV-2 and SARS-CoV. To mimic actual clinical setting, we used 10 times diluted human serum as the solvent of the viral antigen, as we do not expect to see a high concentration of viral S1 protein in serum (or saliva). The entire dynamic range of the S1 detection assay is presented in Fig. 5(B). The linear dynamic range for SARS-CoV-2 is 0.004–15.6 ng/mL with a slope of 0.88 in the log-log scale. Very weak (<0.01%) cross-reactivity towards SARS-CoV was observed at high concentrations (>62.5 ng/mL), indicating that this antibody pair has high specificity. Intra-assay variance for most of the concentrations were smaller than 10% (except 16.3% for the 15 ng/mL data point), as shown in Fig. 5(C). The entire dynamic range of the N protein assay can be found in Fig. 5(D). The linear dynamic range for SARS-CoV-2 N and SARS-CoV N is 0.062–1000 ng/mL and 0.98–1000 ng/mL with a slope of 0.78 and 1.00 in the log-log scale, respectively. According to Fig. 5(D) and (E), the detection of SARS-CoV-2 N protein appears to have a better limit of detection than the detection of SARS-CoV N. However, the specificity of this assay is still not as good as in the S1 detection assay. Due to the relatively weak signal, in intra-assay variance for the lower concentrations were around 15–25% (see Fig. 5(E)). Compared to the dynamic range obtained with the conventional ELISA and provided on Sino Biological's product datasheets (0.156–10 ng/mL for S1 and 0.094–6 ng/mL for N), our technology is able to extend the dynamic range on both ends of the calibration curves for S1 and N. A summary of the parameters in the antigen tests can be found in Table. S2.
4. Discussion
Although the results appear to be promising, there are still a few points worthy of further discussion. For the calibration antibody, since D006 was originally developed against SARS-CoV S1, its specificity towards SARS-CoV-2 S1 is still not perfect. Since our methodology is highly flexible, if we are able to identify a better calibration antibody with a better specificity (e.g., the humanized version of MM43 or one of the potential therapeutic antibodies), we can easily adapt the entire protocol by simply conducting a few groups of calibration experiments as shown in Fig. 2, Fig. 3.
For convalescent serum evaluation, the serial dilution results generated with our technology are not exactly comparable with the titration curves generated with conventional plate-based ELISA. This was caused mainly by the short sample incubation time in our assay (8 min vs. 1–2 h in conventional ELISA). At this incubation time, the high-affinity antibodies (such as the calibrator antibody) dominate the binding process and the low-affinity antibodies cannot bind sufficiently with the immobilized SARS-CoV-2 S1. As a screening technology, our assay cannot be used for examining the neutralization efficacy of the convalescent serum. A separate virus neutralization assay may be required in high-standard applications (such as the clinical trials). Besides, in this study, the antibody measurement protocol was intentionally designed for selecting the strongest donor candidate for convalescent serum therapy. If we aim to detect and quantify the concentration of anti-SARS-CoV-2 S1 IgG in low-abundance samples (such as the serum collected shortly after infection), we will need to re-optimize our assay protocol. For example, the sample incubation time may be extended to 15–20 min. We may also need to replace the HRP-conjugated detection antibody with a biotinylated detection antibody and use it along with the streptavidin poly-HRP for signal enhancement. Note that changing the immobilized antigen from S1 to S1-RBD cannot improve the LLOD, see Fig. S8.
Our approach also opens a door for other COVID-19 related clinical or laboratory researches. For example, the diagnostic value of the COVID-19 related biomarkers in serum and saliva (especially S1 specific IgA and IgM) is currently under intensive evaluation (Li et al., 2020; Sabino-Silva et al., 2020; To et al., 2020). The IgG detection method described in this work can be easily adapted for other types of SARS-CoV-2-specific antibodies such as IgM and IgA (Gai et al., 2008, 2011; Sabino-Silva et al., 2020). The concept of microfluidic chemiluminescent ELISA can also be used to study the neutralization efficacy of therapeutic antibodies (Jiang et al., 2020) (see Fig. S9), as well as for the recognition, evaluation, and phenotyping of natural SARS-CoV-2 particles and COVID-19 pseudovirus particles(Arnold et al., 2008).
5. Conclusion
In this study, we present a portable chemiluminescent microfluidic ELISA platform, which provides sensitive detection and quantification of anti-SARS-CoV-2 S1 IgG for COVID-19 convalescent serum evaluation, in only 15 min. In order to perform a fully quantitative assessment of the convalescent serum, we first assessed four potential calibration antibodies in terms of their affinity, specificity, and dynamic range against SARS-CoV-2 S1. Based on the assessment results, we have successfully identified a recombinant humanized antibody, D006 (out of four candidates), which can be used as the calibration antibody for the quantitative evaluation of anti-SARS-CoV-2 S1 IgG. The LLOD of 2 ng/mL for IgG in serum was achieved using the D006 as the model system.
Then, we have successfully quantified the abundance of anti-SARS-CoV-2 S1 IgG in 16 COVID-19 convalescent serum with our technology platform. Based on our results, 200 × was an optimal dilution factor for performing IgG serological evaluation. With the employment of the calibration antibody, we calculated the effective D006 concentration in these samples. With a cut-off concentration at 500 ng/mL (100 μg/mL in the undiluted serum), only four out of the 16 samples were identified as “qualified donors” for convalescent serum therapy. Since our system is portable and fully integrated, it can be deployed at front-line medical clinics, regional blood banks or other blood-draw stations. Attractive features such as high sensitivity and short assay time will make it suitable for performing rapid quantitative point-of-care pre-evaluation for potential convalescent serum donors.
In addition to the quantification of anti-SARS-CoV-2 S1 IgG, we also demonstrated that our microfluidic ELISA technology can sensitively detect SARS-CoV-2 antigens (S1 and N proteins) with LLODs at 4 pg/mL and 62 pg/mL (for S1 and N, respectively) in 40 min (in the model system). The linear dynamic ranges for both antigens were close to 4 orders of magnitudes. With these results, our technology may serve as one of the new approaches for the rapid diagnosis of COVID-19 (Nachtigall et al., 2020).
CRediT authorship contribution statement
Xiaotian Tan: Conceptualization, Methodology, Investigation, Formal analysis, Writing. Mila Krel: Investigation. Enriko Dolgov: Investigation. Steven Park: Resources, Supervision, Writing. Xuzhou Li: Investigation. Weishu Wu: Investigation. Yun-Lu Sun: Investigation. Jie Zhang: Formal analysis, Resources, Writing. Maung Kyaw Khaing Oo: Methodology, Investigation, Resources, Formal analysis, Writing. David S. Perlin: Methodology, Resources, Supervision, Writing. Xudong Fan: Conceptualization, Formal analysis, Resources, Supervision, Writing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: The authors declare the following competing financial interest): M. K. K. O. and X. F. are co-founders of and have an equity interest in Optofluidic Bioassay, LLC.
Acknowledgement
This work was supported by NSF, ECCS-2029484. The authors thank the financial support from the University of Michigan, Department of Biomedical Engineering. The authors also thank the help from Dr. C. Lin in Sino Biological.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2020.112572.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold S., Ramjit R., Keng D., Kolchenko V., Teraoka I. Faraday Discuss. 2008;137:65–83. doi: 10.1039/b702920a. [DOI] [PubMed] [Google Scholar]
- Bloch E.M., Shoham S., Casadevall A., Sachais B.S., Shaz B., Winters J.L., van Buskirk C., Grossman B.J., Joyner M., Henderson J.P. J. Clin. Invest. 2020;130(6):2757–2765. doi: 10.1172/JCI138745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., Bentlage A.E.H., van Haaren M.M., Guerra D., Burger J.A., Schermer E.E., Verheul K.D., van der Velde N., van der Kooi A., van Schooten J., van Breemen M.J., Bijl T.P.L., Sliepen K., Aartse A., Derking R., Bontjer I., Kootstra N.A., Wiersinga W.J., Vidarsson G., Haagmans B.L., Ward A.B., de Bree G.J., Sanders R.W., van Gils M.J. Science. 2020;369(6504):643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C. Cell. 2020;182(1):73–84. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Pirofski L.-a. J. Clin. Invest. 2020;130(4):1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che X.-Y., Hao W., Wang Y., Di B., Yin K., Xu Y.-C., Feng C.-S., Wan Z.-Y., Cheng V.C., Yuen K.-Y. Emerg. Infect. Dis. 2004;10(11):1947. doi: 10.3201/eid1011.040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xiong J., Bao L., Shi Y. Lancet Infect. Dis. 2020;20(4):398–400. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Wen K., Chen J., Liu Y., Yuan Z., Han C., Chen J., Pan Y., Chen L., Dan Y. 2020. medRxiv. [Google Scholar]
- Dingens A.S., Crawford K.H., Adler A., Steele S.L., Lacombe K., Eguia R., Amanat F., Walls A.C., Wolf C.R., Murphy M. 2020. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117(17):9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai W.-w., Zhang Y., Zhou D.-h., Chen Y.-q., Yang J.-y., Yan H.-m. Virol. Sin. 2011;26(2):81–94. doi: 10.1007/s12250-011-3183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai W., Zou W., Lei L., Luo J., Tu H., Zhang Y., Wang K., Tien P., Yan H. Viral Immunol. 2008;21(1):27–37. doi: 10.1089/vim.2007.0079. [DOI] [PubMed] [Google Scholar]
- Jiang S., Hillyer C., Du L. Trends Immunol. 2020;41(5):355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce M.G., Sankhala R.S., Chen W.-H., Choe M., Bai H., Hajduczki A., Yan L., Sterling S.L., Peterson C., Green E.C. 2020. bioRxiv. [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S. Nature. 2020;584(7819):115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Li X., Geng M., Peng Y., Meng L., Lu S. J. Pharm. Anal. 2020;10(2):102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.-H., Li J., Liu X.-E., Wang L., Li T., Zhou Y.-H., Zhuang H. J. Virol. Methods. 2005;130(1–2):45–50. doi: 10.1016/j.jviromet.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., Wang D.-Q., Hu Y., Ren J.-H., Tang N., Xu Y.-Y., Yu L.-H., Mo Z., Gong F., Zhang X.-L., Tian W.-G., Hu L., Zhang X.-X., Xiang J.-L., Du H.-X., Liu H.-W., Lang C.-H., Luo X.-H., Wu S.-B., Cui X.-P., Zhou Z., Zhu M.-M., Jing Wang6 C.-J.X., Li X.-F., Wang L., Li Z.-J., Wang K., Niu C.-C., Yang Q.-J., Tang X.-J., Zhang Y., Liu X.-M., Li J.-J., De-Chun Zhang10, Zhang F., Liu P., Yuan J., Li1 Q., Hu J.-L., Chen J., Huang A.-L. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- McDade T.W., McNally E., Zelikovich A., D'Aquila R., Mustanski B., Miller A., Vaught L., Reiser N., Bogdanovic E., Fallon K. 2020. PLoS One. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtigall F.M., Pereira A., Trofymchuk O.S., Santos L.S. Nat. Biotechnol. 2020:1–6. [Google Scholar]
- Padoan A., Cosma C., Sciacovelli L., Faggian D., Plebani M. Clin. Chem. Lab. Med. 2020;58(7):1081–1088. doi: 10.1515/cclm-2020-0443. [DOI] [PubMed] [Google Scholar]
- Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Sabino-Silva R., Jardim A.C.G., Siqueira W.L. Clin. Oral Invest. 2020;24(4):1619–1621. doi: 10.1007/s00784-020-03248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L. J. Am. Med. Assoc. 2020;323(16):1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Feng Y., Mo X., Zheng P., Wang Q., Li P., Peng P., Liu X., Chen Z., Huang H. Emerg. Microb. Infect. 2020;9(1):940–948. doi: 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Broses L., Zhou M., Day K., Liu W., Li Z., Weizer A.Z., Khaing Oo M.K., Day M., Fan X. Lab Chip. 2020;20:634–646. doi: 10.1039/c9lc01006h. [DOI] [PubMed] [Google Scholar]
- Tan X., David A., Day J., Tang H., Dixon E.R., Zhu H., Chen Y.-C., Khaing Oo M.K., Shikanov A., Fan X. ACS Sens. 2018;3(11):2327–2334. doi: 10.1021/acssensors.8b00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Khaing Oo M.K., Gong Y., Li Y., Zhu H., Fan X. Analyst. 2017;142(13):2378–2385. doi: 10.1039/c7an00523g. [DOI] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y. Emerg. Microb. Infect. 2020;9(1):382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., Yip C.C.-Y., Cai J.-P., Chan J.M.-C., Chik T.S.-H. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayachandran D., Saravanakarthikeyan B. J. Dent. Sci. 2020 doi: 10.1016/j.jds.2020.04.006. [DOI] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M., van Haperen R., Osterhaus A.D., van Kuppeveld F.J., Haagmans B.L., Grosveld F., Bosch B.-J. Nat. Commun. 2020;11(1):1–6. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200301-sitrep-41-covid-19.pdf
- WHO 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200401-sitrep-72-covid-19.pdf
- WHO 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y. Science. 2020;368(6496):1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T., Lv H., Mok C.K., Wilson I.A. Science. 2020;368(6491):630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.