Abstract
The alveolar epithelium is comprised of two cell types, alveolar epithelial type 1 (AT1) and type 2 (AT2) cells, the latter being capable of self-renewal and transdifferentiation into AT1 cells for normal maintenance and restoration of epithelial integrity following injury. MicroRNAs (miRNAs) are critical regulators of several biological processes, including cell differentiation; however, their role in establishment/maintenance of cellular identity in adult alveolar epithelium is not well understood. To investigate this question, we performed genome-wide analysis of sequential changes in miRNA and gene expression profiles using a well-established model in which human AT2 (hAT2) cells transdifferentiate into AT1-like cells over time in culture that recapitulates many aspects of transdifferentiation in vivo. We defined three phases of miRNA expression during the transdifferentiation process as “early,” “late,” and “consistently” changed, which were further subclassified as up- or downregulated. miRNAs with altered expression at all time points during transdifferentiation were the largest subgroup, suggesting the need for consistent regulation of signaling pathways to mediate this process. Target prediction analysis and integration with previously published gene expression data identified glucocorticoid signaling as the top pathway regulated by miRNAs. Serum/glucocorticoid–regulated kinase 1 (SGK1) emerged as a central regulatory factor, whose downregulation correlated temporally with gain of hsa-miR-424 and hsa-miR-503 expression. Functional validation demonstrated specific targeting of these miRNAs to the 3′-untranslated region of SGK1. These data demonstrate the time-related contribution of miRNAs to the alveolar transdifferentiation process and suggest that inhibition of glucocorticoid signaling is necessary to achieve the AT1-like cell phenotype.
Keywords: glucocorticoid signaling, human alveolar epithelial cells (hAEC), human lung, microRNA, transdifferentiation
INTRODUCTION
The adult mammalian lung is comprised of the conducting airways that extend from the trachea to the small bronchioles and terminate in the alveolar airspaces where gas exchange occurs. The epithelium lining the internal surface of the lung is comprised of specialized cell types organized in a regio-specific manner along a proximo-distal axis (55). The alveolar epithelium which lines the most distal compartment of the lung is comprised of two cell types, cuboidal surfactant-producing alveolar epithelial type 2 (AT2) cells and large, flattened type 1 (AT1) cells (15, 33). AT2 cells constitute ~60% of all alveolar epithelial cells (AEC) and are located at the corners of the alveoli, whereas AT1 cells cover ~95% of the internal surface (15, 33) and play a critical role in gas exchange (68). AT2 cells serve as progenitors for the alveolar epithelium (1, 24), being capable of both self-renewal and differentiation to AT1 cells during normal homeostasis and following injury (5).
Re-epithelialization of the alveolar epithelial surface following injury is critical for restoration of normal barrier function of the lung, making it important to understand mechanisms that underlie AT2 to AT1 cell phenotypic transitions. The process of AT2-to-AT1 cell transition that occurs in vivo has been recapitulated in an in vitro model in which isolated AT2 cells grown on inflexible substrata transdifferentiate to an AT1 cell-like phenotype over time in culture (12, 18, 22, 23). We and others have used this model to investigate mechanisms regulating transdifferentiation of AT2 cells to an AT1-like cell phenotype (9, 10, 21, 28, 53, 75, 78). By integrating transcriptomic and epigenetic profiles of human AEC undergoing transdifferentiation in vitro, we identified transcription factors and validated novel signaling pathways involved in the phenotypic transition, such as the retinoid X receptor (RXR) pathway (44).
In addition to regulation at the transcriptional level during differentiation and regeneration following injury, gene expression can also be regulated at a posttranscriptional level. One example of such posttranscriptional regulation is via microRNAs (miRNAs). MiRNAs are a class of small noncoding RNAs that regulate their target gene expression by either inducing RNA degradation or inhibiting protein translation (6). MiRNAs are transcribed as double strands of nucleotides, namely pri-miRNAs, which are cleaved to pre-miRNAs and then processed to generate a 21–22 nucleotide single-strand mature miRNA. Canonical recognition occurs through base pairing of the 3′-untranslated region (UTR) of the target gene with the miRNA SEED sequence, a heptameric sequence at positions 2–7 from the miRNA 5′-end. One miRNA can have multiple targets (7) thereby regulating an entire signaling pathway by affecting several of its components. In addition, one gene can be the target of multiple miRNAs that cooperate to synergistically regulate gene expression (50).
MiRNAs have been implicated at various stages of lung development. For example, knockout of miR-26 promotes lung development and pulmonary surfactant synthesis (62), while miR-142, miR-17–92, and members of the miR-29 family play crucial roles in AT1 and AT2 cell maturation in the developing alveolar epithelium (31, 59, 64). Although some miRNAs have been found to be involved in alveolarization and in diseases such as bronchopulmonary dysplasia (BPD), their functional role in the sequence of events that leads to alveolarization has not yet been fully clarified (48). A few in vitro studies have explored the role of miRNAs in the transdifferentiation of AT2 to AT1-like cells in the adult lung. In rat AT2 cells, miR-375 was found to be downregulated during the transdifferentiation process and to inhibit the Wnt/β-catenin pathway (65). A pilot study where one of the enzymes involved in miRNA biogenesis (Drosha) was knocked down suggested that miRNAs can regulate the expression of surfactant protein A (SP-A) in human AT2 (hAT2) cells cultured in vitro for 5 days (60). The same study suggested the expression of specific miRNAs in hAT2 and intermediate/AT1 cells (60). However, global analysis of miRNA expression and integration with gene expression data to more fully elucidate involvement of miRNAs in the transition from AT2 to AT1 cells in the adult lung has not yet been performed.
MiRNAs have been implicated in the pathogenesis of several lung diseases (61). Idiopathic pulmonary fibrosis (IPF) is characterized by disruption of the alveolar epithelium that fails to regenerate and becomes replaced by fibrotic tissue. A number of miRNAs [e.g., miR-326 (19), let-7d and miR-200 (49, 72), miR-153 (38), and miR-17–92 (17)] have been shown to have an antifibrotic role in IPF. In contrast, miR-21 has been found to be upregulated in IPF and play a profibrotic role by enhancing epithelial-mesenchymal transition (EMT) and fibrosis (40, 79). Similar to IPF, anti- and profibrotic miRNAs have been identified in mouse lungs injured with bleomycin, the in vivo model most commonly used to study pulmonary fibrosis (16, 46, 69). More recently, the downregulation of miR-29c and its antiapoptotic role specifically in AT2 cells have been reported in both the bleomycin model and IPF (71). In contrast, at least two miRNAs have been reported to have a proapoptotic function in the bleomycin model, miR-155 (52) and miR-145, of which knockout of the latter is protected from fibrosis following bleomycin injury in vivo (73). While the analysis of IPF lungs sheds light on the role of miRNAs at the end stages of abnormal wound repairing, it does not inform as to the molecular mechanisms underlying initial failure of alveolar epithelial cell regeneration. The use of simplified in vitro models that recapitulate aspects of hAT2-to-AT1 cell differentiation are therefore useful to investigate regulation of this process and identify key players implicated in the pathogenesis of IPF.
To investigate the role of miRNAs and identify pathways targeted by miRNAs involved in the phenotypic transition of hAT2 to AT1 cells, we conducted a global analysis of miRNA changes during hAT2-to-AT1-like cell transdifferentiation in vitro and integrated these with transcriptomic changes occurring over the same period. Bioinformatic analysis identified subsets of miRNAs that changed early, late, or that were consistently changed throughout transdifferentiation, indicating time-dependent regulation of gene expression. Interestingly, the largest group of miRNAs was represented by those consistently changing, suggesting the need for continuous regulation of specific signaling pathways to achieve complete phenotypic transition. Target prediction and integration of miRNA and mRNA expression profiles, focusing on miRNAs consistently changing over the course of differentiation, identified glucocorticoid signaling as the top pathway regulated by miRNAs during AEC transdifferentiation. Furthermore, we identified serum/glucocorticoid-regulated kinase 1 (SGK1) as a direct target for two of the most upregulated miRNAs in our study, hsa-miR-424 and hsa-miR-503. Our study reveals the existence of a finely tuned process of miRNA regulation of gene expression during hAT2-to-AT1-like cell transdifferentiation, providing insights into potential molecular mechanisms underlying alveolar epithelial regeneration, and identifying miRNAs as a potential tool for promoting transition to the AT1 cell phenotype.
MATERIALS AND METHODS
Ethics statement.
Human donor lungs were obtained in compliance with Institutional Review Board-approved protocols for the use of human source material in research (HS-07-00660) and processed within 3 days of death.
Sample selection, isolation, and culture of human alveolar epithelial cells.
Human lung tissue was processed as previously described (4, 25) with inclusion of anti-EpCAM-conjugated beads to select for epithelial cells (44). Human AT2 (hAT2) cells were plated in 50:50 DMEM high-glucose media (catalog no. 21063, GIBCO, Gaithersburg, MD):DMEM-F12 (catalog no. D6421, Sigma, Burbank, CA) with 10% fetal bovine serum (FBS). Differentiation into AT1-like cells was verified by measuring pro-SFTPC and Nkx2.1 expression by immunostaining, and aquaporin-5 (AQP5) expression by qRT-PCR and Western blotting as previously reported (44).
Samples derived from three donors, of which two were the same as those previously used for gene expression analysis (44), were used for the miRNA microarray and qRT-PCR for miRNA expression validation. Cells isolated from three additional donors were used for RNA-seq to validate SGK1 expression.
RNA isolation and miRNA expression analysis by microarray.
Purified human AEC (hAEC) were harvested at multiple time points [day (D) 0, 2, 4, 6, and 8] and total RNA and miRNA were extracted for genome-wide profiling. RNA and miRNA were extracted with the Illustra TriplePrep Kit (catalog no. 28-9425-44, GE Healthcare LifeSciences, Pittsburgh, PA) (44) and the MirVana miRNA isolation kit (catalog no. AM1560, Thermo Fisher Scientific, Waltham, MA), respectively. MiRNA expression was assessed on the Agilent MicroRNA Array v2.0 platform. This array contains 2446 unique probes, targeting 887 unique RNAs. MicroRNA array data were processed using AgiMicroRNA package in R (version 3.4.3) including quality control, quantile normalization, and filtering of the data (43). Differential miRNA expression was determined using the Limma package (67). Trend analysis was performed in R using the geom_smooth function.
RNA-seq analysis.
RNA was isolated from ~1 × 107 freshly isolated hAT2 cells or in vitro differentiated AT1-like cells from three donors using the Illustra Triple Prep Kit (catalog no. 28-9425-44, GE Healthcare LifeSciences). RNA (2 μg) was made into libraries using the Ribo-Zero Gold Magnetic Kit (catalog no. MRZG 12324, Illumina, Madison, WI) and sequenced using the Illumina HiSeq2000 at the University of Southern California Epigenome Center Core. Base calls were converted into FASTQ files and aligned to the hg19 genome using tophat2–2.0.8b and assembled into transcripts and RPKMs using Gencode Version 96 GTF files as the reference transcriptome.
Quantitative RT-PCR.
MiRNA was reverse transcribed using the miRNA cDNA synthesis kit (Applied Biological Materials, Richmond, BC), and quantitative (q) RT-PCR was performed using the EvaGreen miRNA qRT-PCR Mastermix ROX (Applied Biological Materials) following the manufacturer’s directions and RNA U6 as endogenous control for normalization. U6 primers were 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse). The hsa-miR-424 (CAGCAGCAATTCATGTTTTGAA), hsa-miR-503 (TAGCAGCGGGAACAGTTCTG), and hsa-miR-223 (TGTCAGTTTGTCAAATACCCCA) primers were used in combination with the universal 3′ miRNA Reverse Primer (Applied Biological Materials).
Integrated analysis of miRNA and gene expression.
mRNA gene expression (GEO DataSet GSE38571) was analyzed as previously described (44). The MicroCosm database (using the miRanda algorithm) was utilized to determine miRNA/mRNA pairing (30). Ingenuity Pathways Analysis (IPA) was used to assess for significantly enriched signaling pathways.
Luciferase assay.
A portion (37 nucleotides) of the 3′-UTR of the SGK1 gene containing the native or mutated SEED sequence for hsa-miR-424 and hsa-miR-503 was synthetized by oligo annealing and further cloned downstream of the firefly luciferase gene in the pmirGLO Dual-Luciferase miRNA Target Expression Vector expressing the Renilla luciferase gene for normalization (catalog no. E1330, Promega, San Luis Obispo, CA). The native SEED sequence was GCT and the mutated sequence AAG CTA. HEK293 cells (3 × 104/well) were plated in 96-well plates and after 24 h transfected using Lipofectamine LTX Reagent with PLUS Reagent (catalog no. 15338030, Thermo Scientific) with 30 ng of SGK1 construct in the presence or absence of 6 μmol of either hsa-miR-424 or hsa-miR-503 (miScript miRNA mimic, Qiagen, Valencia, CA). Cells were harvested 48 h posttransfection, and luciferase activity was tested using the Dual-Glo Luciferase Reporter Assay System (catalog no. E2920, Promega).
Statistics.
Significantly changed miRNAs over the course of AEC transdifferentiation were identified on the microarray using two-way ANOVA with Benjamini-Hochburg (BH) False Discovery Rate (FDR) correction. Significance of the qRT-PCR, microarray, and RNA-seq data comparisons over time was calculated using the y~I[sqrt(x)] polynomical regression with BH-FDR correction. Significance of the luciferase assay was determined with two-way ANOVA followed by Tukey's multiple comparisons test. The significance threshold was P < 0.05 for all tests performed.
All statistical analyses were performed using GraphPad Prism for Windows (GraphPad Software, San Diego, CA, https://www.graphpad.com/).
Data access.
All data have been deposited in GEO (SuperSeries number for the materials and methods section: GSE140073).
RESULTS
Identification of genome-wide changes in miRNA expression during hAT2-to-AT1-like cell transdifferentiation.
We and others (9, 10, 21, 28, 53, 75, 78) have previously reported that AT2 cells cultured on inflexible substrata in vitro shift their gene expression signatures and characteristics from the AT2 cell phenotype by day 3 (D3) in culture, and fully acquire an AT1-like cell phenotype between D6 and D8. To determine whether miRNAs play a regulatory role in AEC transdifferentiation, we evaluated global changes in miRNA expression during transdifferentiation of hAT2 cells cultured in vitro for 8 days. Two of the samples used in the present manuscript were reported in our previous study (44), and purity was verified by immunostaining as described previously (44), with purity of hAT2 cells averaging 89.1 ± 2.9 and 91.5 ± 1.5% as determined by pro-surfactant protein C (SPC) and Nkx2.1 expression, respectively. Furthermore, genes known to become activated during AT2-to-AT1-like cell transdifferentiation (PDPN, AQP5, CAV1) were upregulated over time while AT2 cell-specific genes (SFTPC and SFTPA2) were downregulated (44), indicating that transdifferentiation occurred.
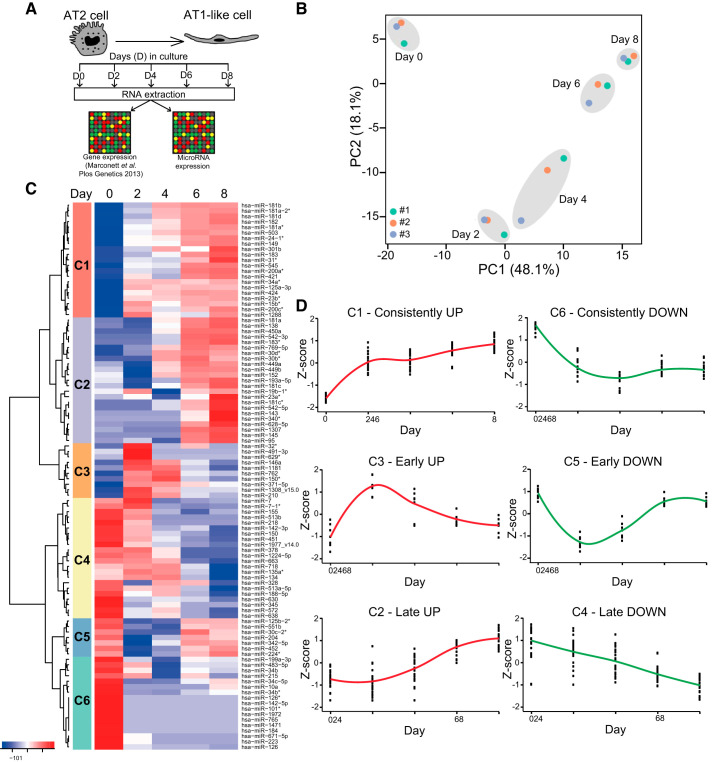
Genome-wide miRNA expression was determined using the experimental approach described schematically in Fig. 1A. The majority of the probes in the miRNA array were not expressed in hAEC at detectable levels, as shown by the low signal distribution (boxplot) for each sample (Supplemental Fig. S1A; see https://doi.org/10.6084/m9.figshare.11414766). After normalization and filtering, 299 miRNAs were found to be expressed at detectable levels at one or more time points. The density distribution for all samples was similar (Supplemental Fig. S1B), indicating that technical variation on the array was negligible. The coefficient of variation for miRNA probe sets was less than 1, indicating a high degree of reproducibility (Supplemental Fig. S1C). Principal component analysis (PCA) showed that samples clustered closely according to time point during differentiation, rather than to individual sample origin, suggesting that the largest contributor to variation in the data set is differentiation (Fig. 1B). Some variation was observed among samples at D4, which may have been due to differences in rates of transdifferentiation among individual cell preparations (Fig. 1B). Samples from D6 and D8 of differentiation clustered tightly together (Fig. 1B). This observation is consistent with previous reports of hAT2 cells becoming fully differentiated to an AT1 cell-like phenotype between D6 and D8 in vitro (44). Unsupervised hierarchical clustering of the top 100 most variant miRNAs across the data set similarly showed that sample clustering is driven by days in culture (Supplemental Fig. S2; see https://doi.org/10.6084/m9.figshare.12252233) and revealed that the major branch point differentiating samples occurred between D0 and D2 (Supplemental Fig. S2), consistent with initiation of the transdifferentiation process.
Fig. 1.
Genome-wide microRNA (miRNA) expression changes during AT2-to-AT1-like cell transdifferentiation. A: schematic of experimental design. Human AT2 (hAT2) cells were plated at day 0 (D0) and harvested at D0, 2, 4, 6, and 8 for RNA extraction and gene and miRNA expression analysis by microarray. B: principal component (PC) analysis of human alveolar epithelial cell samples from the miRNA microarray analysis following quality control and quantile normalization in R. #1, #2, and #3 indicate cells isolated from 3 different human donors (#1 = female, 61 yr old; #2 = female, 49 yr old; #3 = male, 47 yr old). C: cluster (C) analysis of the top 100 altered miRNAs based on z-score value. D: trend of miRNA expression over time for each cluster. Each dot represents a single miRNA. Lines are plotted along the means. Red, upregulated; green, downregulated.
To reveal miRNA expression patterns over time during hAT2-to-AT1-like cell transdifferentiation, we performed cluster analysis on the top 100 altered miRNAs across the data set using the z-score value, which indicates how many SDs each miRNA moves from the mean of its expression across time (Fig. 1C). This analysis identified six clusters (Fig. 1C). Trend analysis, which evaluated the tendency toward changes in miRNA expression over time, suggested the existence of three main categories of miRNAs, with further classification based on the direction of expression changes (up or down) for a total of six clusters (designated C) (Fig. 1D). We defined miRNAs in C1 and C6 as “consistently” up- and downregulated, respectively, because changes in z-score value occurred at D2 and were retained during the following days. Conversely, C3 and C5 were defined as “early” up- and downregulated, respectively, as they showed their maximal expression changes at D2 which tended to revert to baseline levels at subsequent time points. Finally, we identified miRNAs in C2 and C4 as “late” up- and downregulated, respectively, because they displayed the maximal perturbation of their expression at D8. This initial analysis demonstrated that the miRNA expression signature of AEC differentiating in culture changes dramatically during the hAT2-to-AT1-like cell transdifferentiation process.
MiRNA expression correlates with specific stages in the process of alveolar transdifferentiation.
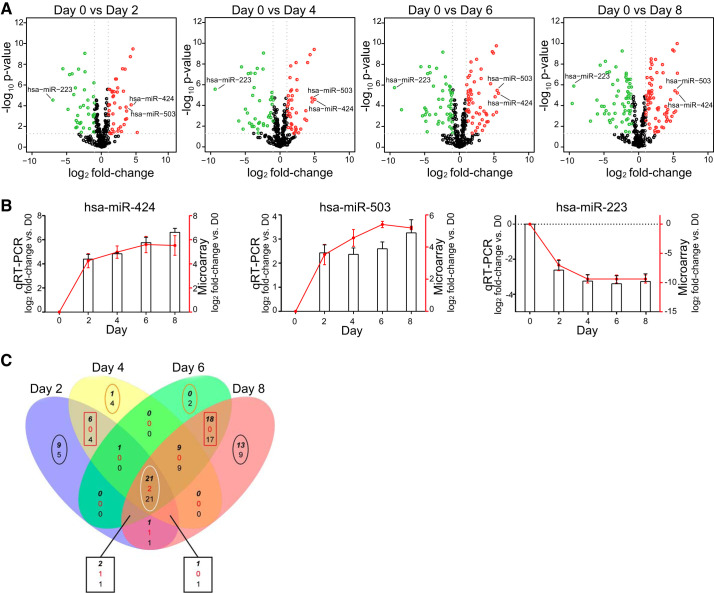
To identify specific miRNAs playing a role at different stages of transdifferentiation, we compared the expression of miRNAs at each day (D2, D4, D6, and D8) with D0. Significant changes in miRNA expression were observed across time points as shown by Volcano plots (Fig. 2A). At D2 and D4, the expression of 41 and 40 miRNAs was increased while 36 and 39 miRNAs were decreased, respectively (Supplemental Table S1; see https://doi.org/10.6084/m9.figshare.12151680). At later time points, 53 and 67 miRNAs were upregulated and 50 and 60 downregulated at D6 and D8, respectively (Supplemental Table S1). Two miRNAs, hsa-miR-424 and hsa-miR-503, were among the most upregulated miRNAs while hsa-miR-223 was the most downregulated miRNA over time. Microarray results for these three miRNAs were validated by qRT-PCR on the same samples used for the microarray study (Fig. 2B).
Fig. 2.
MicroRNAs (miRNAs) are differentially expressed during transdifferentiation. A: volcano plots showing significantly changing miRNAs over the course of alveolar epithelial cell (AEC) differentiation. Red dots are significantly upregulated; green dots are significantly downregulated. x-Axis shows log2 fold-change in miRNA expression; y-axis shows negative log10 of the Benjamini-Hochberg-corrected P values (−10logBH-pvalues). B: quantitative (q) RT-PCR and microarray quantification of 2 of the most upregulated (miR-424 and miR-503) and 1 of the most downregulated (miR-223) microRNAs. Columns, expression levels assessed by qRT-PCR; red line, expression level assessed by microarray; n = 3. Statistical analysis was performed with polynomial regression. hsa-mR-424: qRT-PCR P < 0.01; microarray P < 0.01. hsa-mR-503: qRT-PCR P < 0.05; microarray P < 0.01. hsa-mR-223: qRT-PCR P < 0.05; microarray P < 0.05. C: Venn diagram for microRNAs altered at day 2 (D2), D4, D6, and D8 compared with D0. At intersections (3 numbers): top bold italic number represents number of microRNAs with increased expression; middle red number indicates number of microRNAs with increased expression in 1 comparison and decreased expression in the other comparison; and bottom number indicates number of microRNAs that are decreased in expression. For miRNAs altered only at specific time points (uniquely altered), the top number indicates upregulated and bottom number downregulated. Black ovals, miRNAs uniquely altered at D2 and D8; orange oval, miRNAs uniquely altered at D4 and D6; red rectangles, miRNAs shared by D2 with D4, and D6 and D8; white oval, consistently altered microRNAs. One of the miRNAs upregulated at D2 (hsa-miR-886-3p_v15.0) and 1 of the consistently altered (hsa-miR-1977_v14.0) that were included in our analyses have since been withdrawn from miRBase.
To determine whether miRNA changes over time were distinctive for specific stages in the process of alveolar transdifferentiation, we parsed the data into a Venn diagram among days of in vitro culture (Fig. 2C). This analysis allowed us to further characterize patterns of expression previously identified by trends revealing subsets of miRNAs altered only at specific time points (uniquely altered) or shared among time points. Overall, 160 miRNAs were significantly altered with more than or equal to twofold changes in expression at any given time point. As shown in Fig. 2C (black ovals), a number of miRNAs were altered only at D2 (13 miRNAs, Table 1) and at D8 (22 miRNAs, Table 2). In contrast, very few miRNAs were uniquely altered at D4 (5 miRNAs) and D6 (2 miRNAs) (Fig. 2C, orange ovals). The difference in the extent of the number of miRNAs uniquely altered at different days might reflect the fact that more intermediate stages, such as D4 and D6, are characterized by a transitioning and not yet well-defined cell identity (39). Furthermore, a number of miRNAs were shared by some but not all time points. In particular, D2 and D4 shared 10 altered miRNAs, while D6 and D8 shared 35 miRNAs (Fig. 2C, red rectangle). A total of 43 miRNAs was altered at all days compared with D0 (D2, 4, 6, and 8 vs. D0) and designated as “consistently changing” (Fig. 2C, white oval), of which 21 were upregulated, with hsa-miR-424 and hsa-miR-503 being among the top 10, and 20 downregulated with hsa-miR-223 being the most downregulated (Fig. 2A, Table 3). For almost all of the upregulated miRNAs, the maximal increase was achieved between D6 and D8. In contrast, for most of the downregulated miRNAs, the minimum expression level was already achieved at an early stage and maintained throughout the transdifferentiation process. These results demonstrate that time-related changes in miRNA expression pattern occur during the hAT2-to-AT1-like cell transition.
Table 1.
MicroRNAs uniquely altered at day 2 vs. day 0 during AEC differentiation
|
D2 vs. D0 | |
|---|---|
| Upregulated | Downregulated |
| hsa‐miR‐32‐3p | hsa‐miR‐30b‐5p |
| hsa‐miR‐629‐3p | hsa‐miR‐30d‐5p |
| hsa‐miR‐491‐3p | hsa‐miR‐125b‐2‐3p |
| hsa‐miR‐18b‐5p | hsa‐miR‐224‐3p |
| hsa‐miR‐574‐5p | hsa‐miR‐342‐5p |
| hsa‐miR‐18a‐5p | |
| hsa‐miR‐574‐3p | |
| hsa‐miR‐100‐5p | |
MicroRNAs with ≥2-fold change (Benjamini-Hochberg-corrected P ≤ 0.05) in expression between day 2 (D2) and day 0 (D0) of alveolar epithelial cell (AEC) differentiation. Upregulated, boldface; downregulated, underlined. MiRNAs are listed from the most to the least altered.
Table 2.
MicroRNAs uniquely altered at day 8 vs. day 0 during AEC differentiation
|
D8 vs. D0 | |
|---|---|
| Upregulated | Downregulated |
| hsa‐miR‐542‐5p | hsa‐miR‐29a‐3p |
| hsa‐miR‐143‐3p | hsa‐miR‐26b‐5p |
| hsa‐miR‐181c‐3p | hsa‐miR‐29c‐5p |
| hsa‐miR‐340‐3p | hsa‐miR‐877‐3p |
| hsa‐miR‐301b | hsa‐miR‐195‐5p |
| hsa‐miR‐95 | hsa‐miR‐1246 |
| hsa‐miR‐193a‐3p | hsa‐miR‐188‐5p |
| hsa‐miR‐31‐5p | hsa‐miR‐718 |
| hsa‐miR‐28‐5p | hsa‐miR‐134‐5p |
| hsa‐miR‐331‐3p | |
| hsa‐miR‐224‐5p | |
| hsa‐miR‐21‐3p | |
| hsa‐miR‐200b‐5p | |
MicroRNAs with ≥2-fold change (Benjamini-Hochberg-corrected P ≤ 0.05) in expression between day 8 (D8) and D0 of alveolar epithelial cell (AEC) differentiation. Upregulated, boldface; downregulated, underlined. MiRNAs are listed from the most to the least altered.
Table 3.
MicroRNAs consistently altered at days 2, 4, 6, and 8 vs. day 0 during AEC differentiation
|
D2, 4, 6, and 8 vs. D0 | |
|---|---|
| Upregulated | Downregulated |
| hsa‐miR‐424‐5p | hsa‐miR‐223‐3p |
| hsa‐miR‐181a‐2‐3p | hsa‐miR‐142‐5p |
| hsa‐miR‐503‐5p | hsa‐miR‐218‐5p |
| hsa‐miR‐30d‐3p | hsa‐miR‐671‐5p |
| hsa‐miR‐181a‐3p | hsa‐miR‐126‐5p |
| hsa‐miR‐23b‐5p | hsa‐miR‐1471 |
| hsa‐miR‐545‐3p | hsa‐miR‐765 |
| hsa‐miR‐200c‐5p | hsa‐miR‐34c‐5p |
| hsa‐miR‐34a‐3p | hsa‐miR‐184 |
| hsa‐miR‐371a‐5p | hsa‐miR‐345‐5p |
| hsa‐miR‐24‐1‐5p | hsa‐miR‐1972 |
| hsa‐miR‐182‐5p | hsa‐miR‐126‐3p |
| hsa‐miR‐15b‐3p | hsa‐miR‐572 |
| hsa‐miR‐149‐5p | hsa‐miR‐10a‐5p |
| hsa‐miR‐181b‐5p | hsa‐miR‐638 |
| hsa‐miR‐125a‐3p | hsa‐miR‐34b‐5p |
| hsa‐miR‐24‐3p | hsa‐miR‐101‐5p |
| hsa‐miR‐27b‐3p | hsa‐miR‐630 |
| hsa‐miR‐34a‐5p | hsa‐miR‐135a‐5p |
| hsa‐miR‐93‐5p | hsa‐miR‐34c‐3p |
| hsa‐miR‐744‐5p | |
MicroRNAs that consistently showed changes in expression ≥2-fold (Benjamini-Hochberg- corrected P ≤ 0.05) during alveolar epithelial cell (AEC) transdifferentiation. Upregulated, boldface; downregulated, underlined. MiRNAs are listed from the most to the least altered. D8, day 8; D0, day 0.
MicroRNAs shared among all days are predicted to regulate pathways functionally involved in hAT2-to-AT1-like transdifferentiation.
The observation that the majority of miRNA changes were sustained throughout the entire cell transdifferentiation process (Fig. 2C, white oval) suggested the need for a consistent regulation of targets/pathways crucial to complete and maintain the full differentiation program. In Table 3, we report the changed miRNAs on D2, 4, 6, and 8, ranked by largest fold-change. Consistent with a role in the transdifferentiation process, the three most upregulated miRNAs (hsa-miR-424, hsa-miR-181a, and hsa-miR-503) have been previously implicated in multiple differentiation processes (42, 58, 70, 77), while some of the downregulated miRNAs (e.g., hsa-miR-223, hsa-miR-126) have been shown to promote cell proliferation (11, 74), suggesting that differentiation is favored over proliferation in hAT2 plated in the 2D in vitro system.
The in vitro transdifferentiation process is a dynamic continuous process, where cells lose the expression of AT2 cell markers while concurrently acquiring AT1 cell markers. We therefore investigated whether any of the known ”classic” AT2 (SFTPA, SFTPB, SFTPC, ABCA3) or AT1 (AQP5, HOPX, PDPN, AGER, GRAMD2) cell-specific markers was a predicted target of consistently changed miRNAs. While SFTPC and ABCA3 were predicted to be regulated by upregulated miRNAs (miR-181a-2p, miR-23b-5p, and miR-27b; Supplemental Table S2, see https://doi.org/10.6084/m9.figshare.12151683) and AQP5 expression was inversely correlated to the one of the miRNAs predicted to target it (miR-345–5p; Supplemental Table S2), other AEC markers were not. These data suggest the existence of both direct effects as well more indirect effects of miRNAs on pathways that in turn regulate gene expression.
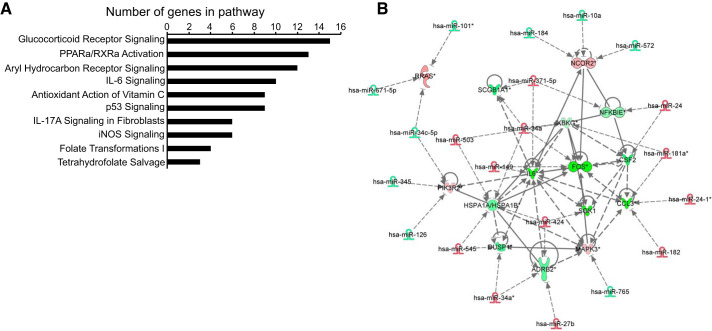
To identify potential new targets and signaling pathways regulated by miRNAs consistently altered at D2, 4, 6, and 8, we first utilized the MicroCosm database (using the miRanda algorithm) to determine predicted miRNA/mRNA pairs. Subsequently, we integrated the genome-wide miRNA microarray data with our previously published RNA microarray data across all days (44). Integration analysis was performed using Ingenuity Pathway Analysis (IPA) software. Putative target genes were identified based on the expression profile being inversely correlated to that of the miRNA. The top 10 most significant pathways were sorted according to the number of genes potentially regulated in each pathway (Fig. 3A). Glucocorticoid receptor signaling emerged as the top pathway (P < 0.05) and included more potentially regulated genes than any other signaling pathway (Fig. 3A). PPAR-α/RXR-α activation, which we previously showed to be important for AEC transdifferentiation (44), had the second largest number of potentially regulated genes (Fig. 3A) and shared some overlap in gene signature with glucocorticoid signaling. Of the 41 miRNAs altered consistently across time points, 21 were predicted to regulate the glucocorticoid pathway (Fig. 3B). We therefore focused on this pathway for further analyses.
Fig. 3.
MicroRNAs (miRNAs) shared among all days regulate pathways involved in hAT2-to-AT1-like cell transdifferentiation. A: Ingenuity Pathways Analysis (IPA) analysis of genes potentially regulated by the miRNAs altered at day 2 (D2), 4, 6, and 8 vs. D0. The top 10 most significant pathways were sorted according to the number of genes potentially regulated in each pathway. B: glucocorticoid pathway, relationship of miRNAs and their putative target genes. miRNAs shown here are shared by D2, 4, 6, and 8 vs. D0. Gene expression changes are D8 vs. D0. Red, increased; green, decreased expression vs. D0. Dotted lines indicate predicted relationships; solid lines indicate validated relationships.
Glucocorticoid pathway component SGK1 is a target of hsa-mir-424 and hsa-miR-503.
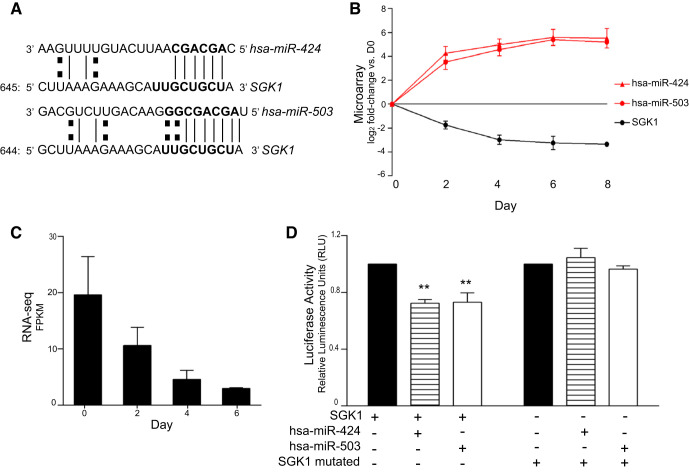
Glucocorticoids have been shown to regulate human fetal AT2-to-AT1-like cell differentiation in vitro (63), but the underlying molecular mechanisms have not been fully elucidated. Our IPA analyses predicted, among others, regulation of SGK1 by hsa-miR-424 (Fig. 3B), which is transcribed as one polycistron with hsa-miR-503. Given that both miRNAs were highly upregulated across days of transdifferentiation with a 40- to 50-fold increase by D6 compared with D0, we proceeded with target validation analyses.
The 3′-UTR of SGK1 contains predicted binding sites for both hsa-miR-424 and hsa-miR-503 (Fig. 4A). Inverse correlation between the two miRNAs and SGK1 was assessed by microarray analysis of their expression profiles over time, with SGK1 being consistently downregulated across days (Fig. 4B). Furthermore, we confirmed downregulation of SGK1 during transdifferentiation in in vitro culture by RNA-seq analysis of AEC from three additional human donors (Fig. 4C).
Fig. 4.
Glucocorticoid pathway component serum/glucocorticoid–regulated kinase 1 (SGK1) is a target of microRNAs miR-424 and miR-503. A: target prediction and sequence alignment of SGK1 with hsa-miR-424 and hsa-miR-503. B: expression profile of hsa-miR-424, hsa-miR-503, and SGK1 over time during hAT2-to-AT1-like cell transdifferentiation based on integrated analysis of microarray data; n = 3. Statistical analysis was performed with polynomial regression. hsa-mR-424 P < 0.01; hsa-miR-503 P < 0.01; SGK1 P < 0.01. C: expression profile of SGK1 over days in culture by RNA-seq analysis. Statistical analysis was performed by polynomial regression; P < 0.01. D: luciferase assay of miR-424 and 503 with SGK1. A luciferase plasmid carrying the native or mutated forms of SGK1 was transfected separately or together with synthetic hsa-miR-424 or hsa-miR-503 into 293T cells. Results are shown as relative luciferase units (RLU); n = 3. Statistical analysis was performed by 2-way ANOVA. **P < 0.01.
To confirm that SGK1 was a direct target of hsa-miR-424 and hsa-miR-503, we inserted a fragment of the 3′-UTR of SGK1 downstream of the firefly luciferase gene and transfected HEK-293T cells with the construct in the presence and absence of either hsa-miR-424 or hsa-miR-503. In the presence of each miRNA, a decrease of 27.61% (SD 4.47) for has-miR-424 and 26.93% (SD 11.45) for hsa-miR-503 (P < 0.05) in luciferase activity was observed (Fig. 4D), demonstrating that each of the miRNAs can target SGK1. To verify that the binding of hsa-miR-424 and hsa-miR-503 to the SGK1 3′-UTR was specific, we mutated the predicted SEED sequence within the SGK1 3′-UTR. Neither hsa-miR-424 nor hsa-miR5-03 affected the mutated SGK1-3′-UTR luciferase activity, confirming direct targeting of SGK1 by hsa-miR-424 and hsa-miR-503 (Fig. 4D). These data functionally validate SGK1 as a direct target for hsa-miR-424 and hsa-miR-503, and more generally support a role for miRNAs in the regulation of the glucocorticoid pathway during hAT2-to-AT1-like cell transdifferentiation. SGK1 targeting is part of the global miRNA/target gene expression changes that dictate alterations in the expression of several genes, that together lead to phenotypic cell changes during the AT2-to-AT1-like cell transdifferentiation.
DISCUSSION
In the more than 20 years since their discovery, the role of miRNAs in the regulation of biological processes has been extensively documented. MiRNAs have been shown to induce differentiation and regulate cell fate decisions to direct among other processes, organ development (34, 35). More recently, miRNAs have been employed to promote differentiation of induced pluripotent stem cells (iPSC) (20, 54, 66). There is thus an increasing interest in the identification of miRNAs that regulate differentiation and their potential application to regulation of biological processes.
A number of in vitro (26, 47, 56) and in vivo (5, 36) studies have focused on elucidating the signaling pathways regulated during the transdifferentiation of AT2 into AT1 cells, but the potential role of miRNAs in regulating this process has not been fully investigated. In the current study, we assessed genome-wide changes in miRNA expression occurring over the entire hAT2 to AT1-like cell transdifferentiation process in vitro and integrated these with the corresponding mRNA expression data (44). This in vitro system is well suited for such a study because AT2 cells undergo a relatively synchronous transdifferentiation process, as underscored by the fact that cultured AEC isolated from three human lungs clustered together based on the expression patterns of miRNAs at different days during transdifferentiation. This indicates a well-orchestrated miRNA expression pattern during the process. The in vitro model offered us the unique opportunity of identifying candidate pathways regulated by miRNAs at specific stages of hAT2-to-AT1-like cell transdifferentiation that can subsequently be validated in vivo.
Cluster and trend analyses allowed us to identify trends within the miRNA expression pattern showing alteration either at early/late stages or consistently altered throughout transdifferentiation. This indicated that while subsets of miRNAs are involved in initiation and completion of hAT2-to-AT1-like cell transdifferentiation, the majority of miRNAs were involved throughout the differentiation process. We further investigated which miRNAs were specifically involved in the process by differential expression analysis of each day vs. D0 and compared miRNA changes at each time point by Venn diagram analysis. This allowed us to identify miRNAs that were altered only at specific time points (unique) or shared among days. Interestingly, the majority of altered miRNAs were shared among all days and maintained their expression change over time compared with D0. Among these, the downregulated miRNAs were already at their minimal expression level at D2, while upregulated miRNAs reached maximal expression levels by D6 or D8. We speculate that the downregulated miRNAs shared by all time points, together with those uniquely altered at D2, regulate genes involved in initiation and progression of the process while the upregulated miRNAs shared by all time points, together with those uniquely altered at D8, are crucial to ensure completion of hAT2-to-AT1-like cell transdifferentiation and maintenance of the AT1 cell phenotype. Thus we hypothesize that constant regulation of specific pathways is needed to fully effect hAT2-to-AT1-like transdifferentiation.
To identify which signaling pathways might be regulated by the consistently changed miRNAs relative to D0, we performed target prediction analysis and then combined it with the integration analysis for miRNA versus gene expression microarrays. The glucocorticoid pathway was predicted to be the most regulated pathway by miRNAs altered at D2, 4, 6, and 8 vs. D0. Glucocorticoids are routinely used clinically to induce maturation of the alveolar epithelium in premature infants. During development, administration of glucocorticoids to mice promotes alveolar maturation and differentiation, while deletion of the glucocorticoid receptor causes an additional round of bronchiolar branching, thus impairing alveolar differentiation (2). Dexamethasone promotes and/or accelerates alveolar epithelial gene expression although it is not absolutely required for alveolar differentiation in vitro (37). A study in mice reported that deletion of the glucocorticoid receptor led to a decrease in AT1 cells but an increase of AT2 cells, suggesting that receptor-mediated glucocorticoid signaling is not required for AT2 cell differentiation during murine embryonic development (14). However, a more recent study showed that activation of glucocorticoid signaling in vitro in murine cells led to an increase in the AT2 cell marker SFTPC and a decrease in miR-142, suggesting the existence of a glucocorticoid-miR-142-p300 signaling axis controlling pneumocyte maturation (59). In humans, dexamethasone has been shown to promote hAT2 cell differentiation and maintain hAT2 cell phenotypic characteristics and gene expression of epithelial cells isolated from fetal lungs (4, 29, 63). To the best of our knowledge, the function of glucocorticoid signaling in the transdifferentiation from hAT2 to AT1-like cells in vitro in the adult has not been investigated, and our data support the hypothesis that inhibition of this pathway is important to allow the process of transdifferentiation. In fact, two of the most upregulated miRNAs shared by D2, 4, 6, and 8 vs. D0 in our study, namely hsa-miR-424 and hsa-miR-503, regulate the expression of the glucocorticoid pathway component SGK1, indicating a role for these miRNAs in promoting the AT1-like cell phenotype through inhibition of the glucocorticoid pathway.
Hsa-miR-424 (mmu-miR-322 in mouse) and hsa-miR-503 have been previously shown to regulate cell fate in other cell types, such as cardiac progenitor cells, inducing their differentiation (58). These two miRNAs are transcribed on the same polycistron and belong to the miR-16 family together with miR-16/15/195/497, which can act synergistically to regulate a number of cell cycle genes and determine cell cycle arrest (27, 41, 51). In our study, another member of the family, hsa-miR-15b-3p, was found to be consistently upregulated (Table 3), suggesting its involvement in promoting the AT1 cell-like phenotype in synergy with hsa-miR-424/503. Furthermore, consistent with a function for hsa-miR-424/503 in promoting differentiation over proliferation, these miRNAs have been found to exert a tumor suppressor function and be downregulated in a variety of human tumors, including lung cancer (32, 76).
In the present study, SGK1, a component of the glucocorticoid signaling pathway, was first predicted and then proven to be a direct target of hsa-miR-424/503 and its expression during the hAT2-to-AT1-like transdifferentiation was inversely correlated with each of the two miRNAs. Interestingly, SGK1 overexpression has been previously shown to promote cell cycle progression in tumors (3), suggesting that its inhibition by hsa-miR-424/503 may account, at least in part, for cell cycle arrest in hAT2 cells and promotion of transdifferentiation into AT1-like cells.
In the epithelium of the mammary gland, the expression of both hsa-miR-424 and hsa-miR-503 is regulated by transforming growth factor (TGF)-β (42). In the alveolar epithelium, AT2 cell autocrine production of TGF-β is required for AT2-to-AT1 cell transdifferentiation (8), where the rate of AT2 cell transdifferentiation is regulated by the opposing effects of bone morphogenetic protein (BMP) and TGF-β signaling (78). A more recent study showed that TGF-β is necessary for AT2 cell cycle arrest following injury-induced proliferation but must be inactivated to allow transdifferentiation to AT1 cells (57). Interestingly, SGK1 has also been identified as a downstream component of the TGF-β signaling pathway in epithelial cells (45). Thus initial activation of TGF-β likely leads to expression of both miR-424/503 and SGK1. Since miRNA expression has a delayed effect on target gene expression, we speculate that SGK1 levels are initially maintained by TGF-β induction and later downregulated by the concomitant inactivation of TGF-β and targeting by miR-424/503. The rather complex effects of TGF-β signaling on miR/target regulation will be pursued in future studies.
In summary, we demonstrate that the miRNA expression profile is dramatically altered during hAT2-to-AT1-like cell transdifferentiation. MiRNAs that change significantly across all days are predicted to regulate genes in several pathways, including glucocorticoid signaling and RXR activation, that have been previously implicated in regulating AEC differentiation. Given that miRNAs are predicted to simultaneously regulate large numbers of genes, it is likely that they play major roles in regulation of the AEC differentiation process. Modulation of miRNA expression could offer a useful tool with which to promote AEC differentiation in in vitro models (e.g., induced pluripotent stem cells) and in vivo where hAT2-to-AT1 cell differentiation may be impaired, leading to failure of normal re-epithelialization (13).
GRANTS
This study was supported by National Heart, Lung, and Blood Institute Grants R35HL135747, HL112638, and HL126877 (Z.B.), R01 HL114094 (I.A.O. and Z.B.) and the Hastings Foundation. Z.B. is Ralph Edgington Chair in Medicine. C.N.M. was supported by the Baxter Foundation. M.E.R. was supported in part by National Institutes of Health Fellowship T32 CA009320. This work was supported in part by the Norris Comprehensive Cancer Center core grant, award number P30CA009320 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
M.E.R. and Z.B. conceived and designed research; A.C., M.E.R., M.D., M.S., K.P., and C.N.M. performed experiments; A.C., M.H., M.E.R., M.D., M.S., K.P., and C.N.M. analyzed data; A.C., M.H., M.E.R., B.Z., I.A.O., C.N.M., and Z.B. interpreted results of experiments; A.C., M.H., and M.E.R. prepared figures; A.C. and M.E.R. drafted manuscript; A.C., M.H., M.E.R., M.D., M.S., B.Z., I.A.O., C.N.M., and Z.B. edited and revised manuscript; Z.B. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Juan Ramon Alvarez for assistance with isolation of alveolar epithelial cells.
REFERENCES
- 1.Adamson IY, Bowden DH. Derivation of type 1 epithelium from type 2 cells in the developing rat lung. Lab Invest 32: 736–745, 1975. [PubMed] [Google Scholar]
- 2.Alanis DM, Chang DR, Akiyama H, Krasnow MA, Chen J. Two nested developmental waves demarcate a compartment boundary in the mouse lung. Nat Commun 5: 3923, 2014. doi: 10.1038/ncomms4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amato R, D’Antona L, Porciatti G, Agosti V, Menniti M, Rinaldo C, Costa N, Bellacchio E, Mattarocci S, Fuiano G, Soddu S, Paggi MG, Lang F, Perrotti N. Sgk1 activates MDM2-dependent p53 degradation and affects cell proliferation, survival, and differentiation. J Mol Med (Berl) 87: 1221–1239, 2009. doi: 10.1007/s00109-009-0525-5. [DOI] [PubMed] [Google Scholar]
- 4.Ballard PL, Lee JW, Fang X, Chapin C, Allen L, Segal MR, Fischer H, Illek B, Gonzales LW, Kolla V, Matthay MA. Regulated gene expression in cultured type II cells of adult human lung. Am J Physiol Lung Cell Mol Physiol 299: L36–L50, 2010. doi: 10.1152/ajplung.00427.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, Hogan BL. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 123: 3025–3036, 2013. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297, 2004. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233, 2009. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhaskaran M, Kolliputi N, Wang Y, Gou D, Chintagari NR, Liu L. Trans-differentiation of alveolar epithelial type II cells to type I cells involves autocrine signaling by transforming growth factor β1 through the Smad pathway. J Biol Chem 282: 3968–3976, 2007. doi: 10.1074/jbc.M609060200. [DOI] [PubMed] [Google Scholar]
- 9.Borok Z, Danto SI, Lubman RL, Cao Y, Williams MC, Crandall ED. Modulation of t1α expression with alveolar epithelial cell phenotype in vitro. Am J Physiol Lung Cell Mol Physiol 275: L155–L164, 1998. [DOI] [PubMed] [Google Scholar]
- 10.Borok Z, Lubman RL, Danto SI, Zhang XL, Zabski SM, King LS, Lee DM, Agre P, Crandall ED. Keratinocyte growth factor modulates alveolar epithelial cell phenotype in vitro: expression of aquaporin 5. Am J Respir Cell Mol Biol 18: 554–561, 1998. doi: 10.1165/ajrcmb.18.4.2838. [DOI] [PubMed] [Google Scholar]
- 11.Chai B, Guo Y, Cui X, Liu J, Suo Y, Dou Z, Li N. MiR-223-3p promotes the proliferation, invasion and migration of colon cancer cells by negative regulating PRDM1. Am J Transl Res 11: 4516–4523, 2019. [PMC free article] [PubMed] [Google Scholar]
- 12.Cheek JM, Evans MJ, Crandall ED. Type I cell-like morphology in tight alveolar epithelial monolayers. Exp Cell Res 184: 375–387, 1989. doi: 10.1016/0014-4827(89)90337-6. [DOI] [PubMed] [Google Scholar]
- 13.Chilosi M, Poletti V, Murer B, Lestani M, Cancellieri A, Montagna L, Piccoli P, Cangi G, Semenzato G, Doglioni C. Abnormal re-epithelialization and lung remodeling in idiopathic pulmonary fibrosis: the role of ΔN-p63. Lab Invest 82: 1335–1345, 2002. doi: 10.1097/01.LAB.0000032380.82232.67. [DOI] [PubMed] [Google Scholar]
- 14.Cole TJ, Solomon NM, Van Driel R, Monk JA, Bird D, Richardson SJ, Dilley RJ, Hooper SB. Altered epithelial cell proportions in the fetal lung of glucocorticoid receptor null mice. Am J Respir Cell Mol Biol 30: 613–619, 2004. doi: 10.1165/rcmb.2003-0236OC. [DOI] [PubMed] [Google Scholar]
- 15.Crapo JD, Barry BE, Gehr P, Bachofen M, Weibel ER. Cell number and cell characteristics of the normal human lung. Am Rev Respir Dis 126: 332–337, 1982. [DOI] [PubMed] [Google Scholar]
- 16.Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, Thannickal VJ, Cardoso WV, Lü J. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol 45: 287–294, 2011. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dakhlallah D, Batte K, Wang Y, Cantemir-Stone CZ, Yan P, Nuovo G, Mikhail A, Hitchcock CL, Wright VP, Nana-Sinkam SP, Piper MG, Marsh CB. Epigenetic regulation of miR-17~92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med 187: 397–405, 2013. doi: 10.1164/rccm.201205-0888OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danto SI, Shannon JM, Borok Z, Zabski SM, Crandall ED. Reversible transdifferentiation of alveolar epithelial cells. Am J Respir Cell Mol Biol 12: 497–502, 1995. doi: 10.1165/ajrcmb.12.5.7742013. [DOI] [PubMed] [Google Scholar]
- 19.Das S, Kumar M, Negi V, Pattnaik B, Prakash YS, Agrawal A, Ghosh B. MicroRNA-326 regulates profibrotic functions of transforming growth factor-β in pulmonary fibrosis. Am J Respir Cell Mol Biol 50: 882–892, 2014. doi: 10.1165/rcmb.2013-0195OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Souza Lima IM, Schiavinato JL, Paulino Leite SB, Sastre D, Bezerra HL, Sangiorgi B, Corveloni AC, Thomé CH, Faça VM, Covas DT, Zago MA, Giacca M, Mano M, Panepucci RA. High-content screen in human pluripotent cells identifies miRNA-regulated pathways controlling pluripotency and differentiation. Stem Cell Res Ther 10: 202, 2019. doi: 10.1186/s13287-019-1318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demaio L, Tseng W, Balverde Z, Alvarez JR, Kim KJ, Kelley DG, Senior RM, Crandall ED, Borok Z. Characterization of mouse alveolar epithelial cell monolayers. Am J Physiol Lung Cell Mol Physiol 296: L1051–L1058, 2009. doi: 10.1152/ajplung.00021.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diglio CA, Kikkawa Y. The type II epithelial cells of the lung. IV. Adaption and behavior of isolated type II cells in culture. Lab Invest 37: 622–631, 1977. [PubMed] [Google Scholar]
- 23.Dobbs LG, Williams MC, Brandt AE. Changes in biochemical characteristics and pattern of lectin binding of alveolar type II cells with time in culture. Biochim Biophys Acta 846: 155–166, 1985. doi: 10.1016/0167-4889(85)90121-1. [DOI] [PubMed] [Google Scholar]
- 24.Evans MJ, Cabral LJ, Stephens RJ, Freeman G. Transformation of alveolar type 2 cells to type 1 cells following exposure to NO2. Exp Mol Pathol 22: 142–150, 1975. doi: 10.1016/0014-4800(75)90059-3. [DOI] [PubMed] [Google Scholar]
- 25.Fang X, Song Y, Zemans R, Hirsch J, Matthay MA. Fluid transport across cultured rat alveolar epithelial cells: a novel in vitro system. Am J Physiol Lung Cell Mol Physiol 287: L104–L110, 2004. doi: 10.1152/ajplung.00176.2003. [DOI] [PubMed] [Google Scholar]
- 26.Flozak AS, Lam AP, Russell S, Jain M, Peled ON, Sheppard KA, Beri R, Mutlu GM, Budinger GR, Gottardi CJ. β-catenin/T-cell factor signaling is activated during lung injury and promotes the survival and migration of alveolar epithelial cells. J Biol Chem 285: 3157–3167, 2010. doi: 10.1074/jbc.M109.070326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forrest AR, Kanamori-Katayama M, Tomaru Y, Lassmann T, Ninomiya N, Takahashi Y, de Hoon MJ, Kubosaki A, Kaiho A, Suzuki M, Yasuda J, Kawai J, Hayashizaki Y, Hume DA, Suzuki H. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia 24: 460–466, 2010. doi: 10.1038/leu.2009.246. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh MC, Gorantla V, Makena PS, Luellen C, Sinclair SE, Schwingshackl A, Waters CM. Insulin-like growth factor-I stimulates differentiation of ATII cells to ATI-like cells through activation of Wnt5a. Am J Physiol Lung Cell Mol Physiol 305: L222–L228, 2013. doi: 10.1152/ajplung.00014.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzales LW, Guttentag SH, Wade KC, Postle AD, Ballard PL. Differentiation of human pulmonary type II cells in vitro by glucocorticoid plus cAMP. Am J Physiol Lung Cell Mol Physiol 283: L940–L951, 2002. doi: 10.1152/ajplung.00127.2002. [DOI] [PubMed] [Google Scholar]
- 30.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158, 2008. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo W, Benlhabib H, Mendelson CR. The microRNA 29 family promotes type II cell differentiation in developing lung. Mol Cell Biol 36: 2141, 2016. doi: 10.1128/MCB.00096-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta G, Chellappan DK, de Jesus Andreoli Pinto T, Hansbro PM, Bebawy M, Dua K. Tumor suppressor role of miR-503. Panminerva Med 60: 17–24, 2018. [DOI] [PubMed] [Google Scholar]
- 33.Haies DM, Gil J, Weibel ER. Morphometric study of rat lung cells. I. Numerical and dimensional characteristics of parenchymal cell population. Am Rev Respir Dis 123: 533–541, 1981. [DOI] [PubMed] [Google Scholar]
- 34.Ivey KN, Srivastava D. microRNAs as developmental regulators. Cold Spring Harb Perspect Biol 7: a008144, 2015. doi: 10.1101/cshperspect.a008144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell 7: 36–41, 2010. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 36.Jansing NL, McClendon J, Henson PM, Tuder RM, Hyde DM, Zemans RL. Unbiased quantitation of alveolar type II to alveolar type I cell transdifferentiation during repair after lung injury in mice. Am J Respir Cell Mol Biol 57: 519–526, 2017. doi: 10.1165/rcmb.2017-0037MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laresgoiti U, Nikolić MZ, Rao C, Brady JL, Richardson RV, Batchen EJ, Chapman KE, Rawlins EL. Lung epithelial tip progenitors integrate glucocorticoid- and STAT3-mediated signals to control progeny fate. Development 143: 3686–3699, 2016. doi: 10.1242/dev.134023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang C, Li X, Zhang L, Cui D, Quan X, Yang W. The anti-fibrotic effects of microRNA-153 by targeting TGFBR-2 in pulmonary fibrosis. Exp Mol Pathol 99: 279–285, 2015. doi: 10.1016/j.yexmp.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 39.Liebler JM, Marconett CN, Juul N, Wang H, Liu Y, Flodby P, Laird-Offringa IA, Minoo P, Zhou B. Combinations of differentiation markers distinguish subpopulations of alveolar epithelial cells in adult lung. Am J Physiol Lung Cell Mol Physiol 310: L114–L120, 2016. doi: 10.1152/ajplung.00337.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med 207: 1589–1597, 2010. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res 36: 5391–5404, 2008. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Llobet-Navas D, Rodríguez-Barrueco R, Castro V, Ugalde AP, Sumazin P, Jacob-Sendler D, Demircan B, Castillo-Martín M, Putcha P, Marshall N, Villagrasa P, Chan J, Sanchez-Garcia F, Pe’er D, Rabadán R, Iavarone A, Cordón-Cardó C, Califano A, López-Otín C, Ezhkova E, Silva JM. The miR-424(322)/503 cluster orchestrates remodeling of the epithelium in the involuting mammary gland. Genes Dev 28: 765–782, 2014. doi: 10.1101/gad.237404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.López-Romero P. Pre-processing and differential expression analysis of Agilent microRNA arrays using the AgiMicroRna Bioconductor library. BMC Genomics 12: 64, 2011. doi: 10.1186/1471-2164-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marconett CN, Zhou B, Rieger ME, Selamat SA, Dubourd M, Fang X, Lynch SK, Stueve TR, Siegmund KD, Berman BP, Borok Z, Laird-Offringa IA. Integrated transcriptomic and epigenomic analysis of primary human lung epithelial cell differentiation. PLoS Genet 9: e1003513, 2013. doi: 10.1371/journal.pgen.1003513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Massagué J, Gomis RR. The logic of TGFβ signaling. FEBS Lett 580: 2811–2820, 2006. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 46.Montgomery RL, Yu G, Latimer PA, Stack C, Robinson K, Dalby CM, Kaminski N, van Rooij E. MicroRNA mimicry blocks pulmonary fibrosis. EMBO Mol Med 6: 1347–1356, 2014. doi: 10.15252/emmm.201303604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mutze K, Vierkotten S, Milosevic J, Eickelberg O, Königshoff M. Enolase 1 (ENO1) and protein disulfide-isomerase associated 3 (PDIA3) regulate Wnt/β-catenin-driven trans-differentiation of murine alveolar epithelial cells. Dis Model Mech 8: 877–890, 2015. doi: 10.1242/dmm.019117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nardiello C, Morty RE. MicroRNA in late lung development and bronchopulmonary dysplasia: the need to demonstrate causality. Mol Cell Pediatr 3: 19, 2016. doi: 10.1186/s40348-016-0047-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, Richards T, Selman M, Watkins SC, Pardo A, Ben-Yehudah A, Bouros D, Eickelberg O, Ray P, Benos PV, Kaminski N. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 182: 220–229, 2010. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peter ME. Targeting of mRNAs by multiple miRNAs: the next step. Oncogene 29: 2161–2164, 2010. doi: 10.1038/onc.2010.59. [DOI] [PubMed] [Google Scholar]
- 51.Porrello ER, Johnson BA, Aurora AB, Simpson E, Nam YJ, Matkovich SJ, Dorn GW II, van Rooij E, Olson EN. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res 109: 670–679, 2011. doi: 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pottier N, Maurin T, Chevalier B, Puisségur MP, Lebrigand K, Robbe-Sermesant K, Bertero T, Lino Cardenas CL, Courcot E, Rios G, Fourre S, Lo-Guidice JM, Marcet B, Cardinaud B, Barbry P, Mari B. Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions. PLoS One 4: e6718, 2009. doi: 10.1371/journal.pone.0006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiao R, Zhou B, Liebler JM, Li X, Crandall ED, Borok Z. Identification of three genes of known function expressed by alveolar epithelial type I cells. Am J Respir Cell Mol Biol 29: 98–105, 2003. doi: 10.1165/rcmb.2002-0196OC. [DOI] [PubMed] [Google Scholar]
- 54.Qiao S, Deng Y, Li S, Yang X, Shi D, Li X. Partially reprogrammed induced pluripotent stem cells using microRNA cluster miR-302s in Guangxi Bama minipig fibroblasts. Cell Reprogram 21: 229–237, 2019. doi: 10.1089/cell.2019.0035. [DOI] [PubMed] [Google Scholar]
- 55.Rackley CR, Stripp BR. Building and maintaining the epithelium of the lung. J Clin Invest 122: 2724–2730, 2012. doi: 10.1172/JCI60519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rieger ME, Zhou B, Solomon N, Sunohara M, Li C, Nguyen C, Liu Y, Pan JH, Minoo P, Crandall ED, Brody SL, Kahn M, Borok Z. p300/β-catenin interactions regulate adult progenitor cell differentiation downstream of WNT5a/protein kinase C (PKC). J Biol Chem 291: 6569–6582, 2016. doi: 10.1074/jbc.M115.706416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riemondy KA, Jansing NL, Jiang P, Redente EF, Gillen AE, Fu R, Miller AJ, Spence JR, Gerber AN, Hesselberth JR, Zemans RL. Single cell RNA sequencing identifies TGFβ as a key regenerative cue following LPS-induced lung injury. JCI Insight 5: e123637, 2019. doi: 10.1172/jci.insight.123637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen X, Soibam B, Benham A, Xu X, Chopra M, Peng X, Yu W, Bao W, Liang R, Azares A, Liu P, Gunaratne PH, Mercola M, Cooney AJ, Schwartz RJ, Liu Y. miR-322/-503 cluster is expressed in the earliest cardiac progenitor cells and drives cardiomyocyte specification. Proc Natl Acad Sci USA 113: 9551–9556, 2016. doi: 10.1073/pnas.1608256113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shrestha A, Carraro G, Nottet N, Vazquez-Armendariz AI, Herold S, Cordero J, Singh I, Wilhelm J, Barreto G, Morty R, El Agha E, Mari B, Chen C, Zhang JS, Chao CM, Bellusci S. A critical role for miR-142 in alveolar epithelial lineage formation in mouse lung development. Cell Mol Life Sci 76: 2817–2832, 2019. doi: 10.1007/s00018-019-03067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silveyra P, Chroneos ZC, DiAngelo SL, Thomas NJ, Noutsios GT, Tsotakos N, Howrylak JA, Umstead TM, Floros J. Knockdown of Drosha in human alveolar type II cells alters expression of SP-A in culture: a pilot study. Exp Lung Res 40: 354–366, 2014. doi: 10.3109/01902148.2014.929757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stolzenburg LR, Harris A. The role of microRNAs in chronic respiratory disease: recent insights. Biol Chem 399: 219–234, 2018. doi: 10.1515/hsz-2017-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun YF, Kan Q, Yang Y, Zhang YH, Shen JX, Zhang C, Zhou XY. Knockout of microRNA-26a promotes lung development and pulmonary surfactant synthesis. Mol Med Rep 17: 5988–5995, 2018. doi: 10.3892/mmr.2018.8602. [DOI] [PubMed] [Google Scholar]
- 63.Wade KC, Guttentag SH, Gonzales LW, Maschhoff KL, Gonzales J, Kolla V, Singhal S, Ballard PL. Gene induction during differentiation of human pulmonary type II cells in vitro. Am J Respir Cell Mol Biol 34: 727–737, 2006. doi: 10.1165/rcmb.2004-0389OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y, Frank DB, Morley MP, Zhou S, Wang X, Lu MM, Lazar MA, Morrisey EE. HDAC3-dependent epigenetic pathway controls lung alveolar epithelial cell remodeling and spreading via miR-17-92 and TGF-β signaling regulation. Dev Cell 36: 303–315, 2016. doi: 10.1016/j.devcel.2015.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y, Huang C, Reddy Chintagari N, Bhaskaran M, Weng T, Guo Y, Xiao X, Liu L. miR-375 regulates rat alveolar epithelial cell trans-differentiation by inhibiting Wnt/β-catenin pathway. Nucleic Acids Res 41: 3833–3844, 2013. doi: 10.1093/nar/gks1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watanabe T, Yamazaki S, Yoneda N, Shinohara H, Tomioka I, Higuchi Y, Yagoto M, Ema M, Suemizu H, Kawai K, Sasaki E. Highly efficient induction of primate iPS cells by combining RNA transfection and chemical compounds. Genes Cells 24: 473–484, 2019. doi: 10.1111/gtc.12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics 20: 3705–3706, 2004. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 68.Williams MC. Alveolar type I cells: molecular phenotype and development. Annu Rev Physiol 65: 669–695, 2003. doi: 10.1146/annurev.physiol.65.092101.142446. [DOI] [PubMed] [Google Scholar]
- 69.Xiao J, Meng XM, Huang XR, Chung AC, Feng YL, Hui DS, Yu CM, Sung JJ, Lan HY. miR-29 inhibits bleomycin-induced pulmonary fibrosis in mice. Mol Ther 20: 1251–1260, 2012. doi: 10.1038/mt.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao X, Huang C, Zhao C, Gou X, Senavirathna LK, Hinsdale M, Lloyd P, Liu L. Regulation of myofibroblast differentiation by miR-424 during epithelial-to-mesenchymal transition. Arch Biochem Biophys 566: 49–57, 2015. doi: 10.1016/j.abb.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xie T, Liang J, Geng Y, Liu N, Kurkciyan A, Kulur V, Leng D, Deng N, Liu Z, Song J, Chen P, Noble PW, Jiang D. MicroRNA-29c prevents pulmonary fibrosis by regulating epithelial cell renewal and apoptosis. Am J Respir Cell Mol Biol 57: 721–732, 2017. doi: 10.1165/rcmb.2017-0133OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang S, Banerjee S, de Freitas A, Sanders YY, Ding Q, Matalon S, Thannickal VJ, Abraham E, Liu G. Participation of miR-200 in pulmonary fibrosis. Am J Pathol 180: 484–493, 2012. doi: 10.1016/j.ajpath.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang S, Cui H, Xie N, Icyuz M, Banerjee S, Antony VB, Abraham E, Thannickal VJ, Liu G. miR-145 regulates myofibroblast differentiation and lung fibrosis. FASEB J 27: 2382–2391, 2013. doi: 10.1096/fj.12-219493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yuan Y, Shen C, Zhao SL, Hu YJ, Song Y, Zhong QJ. MicroRNA-126 affects cell apoptosis, proliferation, cell cycle and modulates VEGF/TGF-β levels in pulmonary artery endothelial cells. Eur Rev Med Pharmacol Sci 23: 3058–3069, 2019. doi: 10.26355/eurrev_201904_17588. [DOI] [PubMed] [Google Scholar]
- 75.Zhang L, Zhao S, Yuan L, Wu H, Jiang H, Luo G. Hyperoxia-mediated LC3B activation contributes to the impaired transdifferentiation of type II alveolar epithelial cells (AECIIs) to type I cells (AECIs). Clin Exp Pharmacol Physiol 43: 834–843, 2016. doi: 10.1111/1440-1681.12592. [DOI] [PubMed] [Google Scholar]
- 76.Zhang M, Zeng J, Zhao Z, Liu Z. Loss of MiR-424-3p, not miR-424-5p, confers chemoresistance through targeting YAP1 in non-small cell lung cancer. Mol Carcinog 56: 821–832, 2017. doi: 10.1002/mc.22536. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Z, Gao Y, Xu MQ, Wang CJ, Fu XH, Liu JB, Han DX, Jiang H, Yuan B, Zhang JB. miR-181a regulate porcine preadipocyte differentiation by targeting TGFBR1. Gene 681: 45–51, 2019. doi: 10.1016/j.gene.2018.09.046. [DOI] [PubMed] [Google Scholar]
- 78.Zhao L, Yee M, O’Reilly MA. Transdifferentiation of alveolar epithelial type II to type I cells is controlled by opposing TGF-β and BMP signaling. Am J Physiol Lung Cell Mol Physiol 305: L409–L418, 2013. doi: 10.1152/ajplung.00032.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou J, Xu Q, Zhang Q, Wang Z, Guan S. A novel molecular mechanism of microRNA-21 inducing pulmonary fibrosis and human pulmonary fibroblast extracellular matrix through transforming growth factor β1-mediated SMADs activation. J Cell Biochem 119: 7834–7843, 2018. doi: 10.1002/jcb.27185. [DOI] [PubMed] [Google Scholar]