ABSTRACT
Psoriasis is an immune-mediated chronic inflammatory skin disease. Keratinocyte hyperproliferation has been regarded as a significant event in psoriasis pathogenesis. Considering the vital role of miRNA-mediated mRNA repression in psoriasis pathogenesis, in the present study, we attempted to investigate the mechanism of keratinocyte overproliferation from the point of miRNA-mRNA regulation. Both online microarray expression profiles and experimental results indicated that the expression of LXR-α and PPAR-γ was downregulated in psoriasis lesion skin. LXR-α or PPAR-γ overexpression alone was sufficient to inhibit keratinocyte proliferation, decrease KRT5 and KRT14 protein levels and increase KRT1 and KRT10 protein levels. miR-203 negatively regulated LXR-α and PPAR-γ expression through direct targeting. miR-203 inhibition exerted the opposite effects to LXR-α or PPAR-γ overexpression on HaCaT cells. More importantly, LXR-α or PPAR-γ overexpression could markedly remarkably attenuate the effects of miR-203 overexpression in keratinocytes, indicating that miR-203 promotes keratinocyte proliferation by targeting LXR-α and PPAR-γ. In conclusion, the miR-203-LXR-α/PPAR-γ axis modulates the proliferation of keratinocytes and might be a novel target for psoriasis treatment, which needs further in vivo investigation.
KEYWORDS: Psoriasis, keratinocyte hyperproliferation, miR-203, LXR-α, PPAR-γ
Introduction
Psoriasis is an immune-mediated chronic inflammatory skin disease. Epidermal hyperplasia, angiogenesis, and inflammatory cell infiltration are the major characteristic features of psoriasis [1–4]. Several studies have indicated that a large number of internal and external factors, including the immune system, keratinocytes, susceptibility genes, and environmental factors, might participate in psoriasis pathogenesis [5]. However, the cause of psoriasis is complicated and not fully understood. As reviewed by Boehncke et al. [5] in Lancet, psoriasis can be induced by nonspecific triggers such as mild trauma (scratching, piercings, and tattoos), sunburn, or chemical irritants. Systemic drugs such as β blockers, lithium, antimalarials, and nonsteroidal anti-inflammatory agents can exacerbate the disease. However, previous investigations have indicated that dysregulation of the interaction between epidermal keratinocytes and immune cells results in hyperplasia of the epidermis within psoriatic lesions [6–8]. Further investigation of the mechanisms of keratinocyte hyperproliferation might provide novel strategies for psoriasis treatment.
Dozens of genes and signaling pathways related to psoriasis, especially related to keratinocyte hyperproliferation, have been found in genetic studies [9]. In terms of immunity, the cutaneous and systemic overexpression of several proinflammatory cytokines, particularly type-1 cytokines such as IL-2, IL-6, IL-8, IL-12, IFN-γ and TNF-α, has been demonstrated. The overexpression of these proinflammatory cytokines is considered to be responsible for the initiation, maintenance and recurrence of skin lesions. The cellular composition of the inflammatory infiltrate within the plaques as well as keratinocyte hyperproliferation appears to be directed by cytokines [10]. In terms of genetics, HLA-Cw6 is regarded as the primary genetic determinant of psoriatic lesions [11,12]. The function of the LXR-α (liver X receptor-α) gene is to modulate genes coding for inflammatory cytokines and genes involved in the cell cycle, immune regulation, and reactive oxygen species clearance in human keratinocytes. Furthermore, LXR-α gene silencing in healthy keratinocytes simulated the genomic profile shown within psoriasis lesions, and after knocking down LXR-α, the genes coding for PPAR-γ (peroxisome proliferator-activated receptor-γ), catalase and VDR (vitamin D receptor) were be reduced [13]. In contrast, thegenes coding for IFN-γ (interferon-γ), c-myc, IL-6, and IL-8 were dramatically increased [13]. By analyzing ten sets of online microarray profiles (EG-EOD-54456, GSE114286, GSE121212, GSE13355, GSE14905, GSE30999, GSE34248, GSE41662, GSE50790, GSE6710; Table 1), we determined that the expression of LXR-α was significantly reduced in psoriatic lesion tissues and that LXR-α and PPAR-γ expression in psoriasis lesion tissues had an apparent positive correlation (Table 2). Thus, LXR-α and PPAR-γ could be involved in regulating keratinocyte hyperproliferation.
Table 1.
Expression of NR1H3 in ten sets of online expression profiles.
Table 2.
The correlation between PPARG and LXR-a expression in psoriasis based on online data.
Recently, miRNAs have been reported to play a vital role in normal biological and disease pathological processes by targeting mRNA 3ʹUTRs (untranslated regions), subsequently inducing mRNA transcriptional inhibition or mRNA degradation [14]. Based on the miRNA and mRNA profiles, Zibert et al. [15] found that miR-21, −205, −221 and −222 might target the relevant mRNA targets in psoriasis lesions, including PDCD4, TPM1, P57, C-KIT, RTN4, SHIP2, TIMP3, RECK, and NFIB, factors that contribute to extracellular matrix cell growth, proliferation, apoptosis, and degradation [15], thus, these miRNAs also play a role in psoriasis pathogenesis. Herein, we also identified miRNAs that might target LXR-α and PPAR-γ with the use of online tools and examined their dynamic effects on keratinocyte hyperproliferation.
Herein, we examined LXR-α and PPAR-γ mRNA and protein expression in psoriasis lesions and noninvasive tissue samples and investigated the specific effects of LXR-α and PPAR-γ on keratinocyte hyperproliferation. Next, the predicted binding of miR-203 to LXR-α and PPAR-γ, as well as miR-203 negative regulation of LXR-α and PPAR-γ were investigated. Finally, we determined the dynamic effects of miR-203 and LXR-α or PPAR-γ on keratinocyte hyperproliferation. Our findings provide a rationale for further studies on the potential therapeutic benefits of miRNA-mRNA interactions regulating keratinocyte hyperproliferation.
Materials and methods
Tissue samples and pathological analysis
A total of 12 of psoriasis lesion tissues and 12 nonlesion tissues were obtained from psoriasis patients who received treatment in the Second Affiliated Hospital of Hunan University of Chinese Medicine. Before performing surgeries, informed consent was obtained from every patient with psoriasis, and this study was approved by the Research Ethics Committee of the Second Affiliated Hospital of Hunan University of Chinese Medicine. Tissue samples were stored at −80°C until further study.
Pathological analysis was generated using Hematoxylin-eosin (H&E) staining. After fixation for 24 h in 4% paraformaldehyde, the blocks were dehydrated, embedded in paraffin, and cut into 4-μm-thick slices. Slices were heated overnight at 37°C, dewaxed, and stained with H&E using standard procedures [16,17].
Cell line and cell transfection
Human immortalized keratinocytes, HaCaT cells, were obtained from ProCell (CL-0090, Wuhan, China) and cultured in MEM (PM150410, ProCell) supplemented with 15% FBS (164,210–500, ProCell) and 1% P/S (PB180120, ProCell). Cells were cultured at 37°C in 5% CO2.
The expression of miRNA was manipulated by the transfection of miRNA-overexpressing or anti-miRNA vectors (Genepharma, Shanghai, China). The overexpression of LXR-α or PPAR-γ was achieved by transfection of an LXR-α or PPAR-γ-overexpressing vector (Genepharma). For transfection, HaCaT cells were seeded into a 12-well plate at a density of 2 × 105/well before transfection. At 80% confluence, cells were transfected with the above-indicated vectors. After 6 h of incubation in serum-free medium, cells were cultured in DMEM with serum for 48 h. To assess transfection efficiency, cells were harvested, and the levels of target factors were confirmed by real-time qPCR. All transfections were conducted with Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA).
IL-22-induced psoriasis-like dermatitis in an in vitro model of HaCaT cells
HaCaT cells were seeded onto a 6-well plate at a density of 4 × 105 cells/well until reaching 80% to 90% confluence. Cells were stimulated with IL-22 (100 ng/ml; Pepro Tech, Rocky Hill, NJ, USA) for 24 h to establish anIL-22-induced psoriasis-like dermatitis in vitro model.
Immunohistochemical (IHC) staining
Immunohistochemistry was performed to examine the distribution and protein levels of LXR-α and PPAR-γ in tissue samples. Tissue sections were incubated with anti-LXR-α (ab41902, Abcam, Cambridge, UK) or anti-PPAR-γ (ab45036, Abcam). Image Pro-plus 6.0 software (NIH, USA) was used to assess the immunohistochemical staining of sections by measuring the integrated optical density.
PCR-based analyses
Total RNA was extracted from target tissues or cells using TRIzol reagent (Invitrogen), and then miRNA and mRNA expression was determined using a SYBR Green qPCR kit (Takara, Dalian, China) according to the manufacturer’s instructions and a previously described method [18]. The expression of RNU6B or GAPDH was used as an endogenous control for miRNA or mRNA expression determination, respectively. The 2−ΔΔCT method was applied for data processing. The sequences used here are provided in Table S1.
Immunoblotting
The protein levels of LXR-α, PPAR-γ, ki67, KRT1, KRT10, KRT5, and KRT14 were examined by immunoblotting following methods described before [18] with antibodies against LXR-α (ab41902; 50 kDa), PPAR-γ (ab45036; 57 kDa), ki67 (ab16667; 359 kDa), KRT1 (ab93652; 66 kDa), KRT10 (ab9025; 55 kDa), KRT5 (ab64081; 62 kDa), and KRT14 (ab7800; 50 kDa) followed by another incubation with proper HRP-conjugated secondary antibodies. GAPDH (ab8245; 37 kDa) was used as an endogenous control. All the antibodies were obtained from Abcam unless otherwise noted. Signals were visualized using enhanced chemiluminescent (ECL) substrates (Millipore, MA, USA) normalizing to GAPDH.
Cell viability determination by MTT assay
A modified MTT assay was used to evaluate cell viability following previously described methods [18]. DMSO was added after the supernatant was discarded to dissolve the formazan. OD values were measured at 450 nm. The viability of the non-treated cells (control) was defined as 100%, and the viability of cells from all other groups was calculated separately from that of the control group.
DNA synthesis determined by EdU assays
The DNA synthesis assay is based on the method of incorporating thymidine analog 5-ethoxy 2-deoxyuridine (EdU) into genomic DNA by using the Click-IT EdU Alexa Fluor 488 kit following previously describedmethods [19,20]. Apollo staining and DAPI staining were performed, and EdU positive cells were observed with a fluorescence microscope. The blue fluorescence represents the nucleus stained by DAPI and the green fluorescence represents the newly synthesized DNA stained by EdU. The incorporation rate of EdU was calculated as the ratio of EdU-positive cells (green cells) to total DAPI-positive cells (blue cells).
Dual-luciferase reporter assay
To assess the binding between miR-203 and LXR-α or PPAR-γ, the dual-luciferase reporter assay was performed following previously described methods [21]. The3'UTRs of the LXR-α and PPAR-γ genes were amplified by PCR using genomic DNA of the HaCaT cell line and each was cloned downstream of the Renilla luciferase open reading frame in the Renilla psiCHECK2 vector (Promega, Madison, WI, USA) using XhoI and NotI restriction sites. Mutations were introduced in the seed region of the miR-203 binding site in the LXR-α and PPAR-γ genes and the constructs were named mut-LXR-α and PPAR-γ 3'UTR, respectively. Next, 293 T cells were seeded in 96-well plates and cotransfected with miR-203/anti-miR-203 together with psiCHECK-2 reporter vectors (wt-/mut-LXR-α or PPAR-γ 3'UTR). Forty-eight hours after transfection, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Values were double normalized to firefly luciferase activity and the luciferase activity of cells transfected with the empty psiCHECK-2 control vector.
Statistical analysis
All experiments were repeated at least three times. Data from at least three independent experiments were processed using SPSS17.0 (IBM, Armonk, NY, USA) and are presented as the mean ± S.D. Student’s t-test was used for statistical comparisons between means where applicable. Differences among more than two groups in the above assays were estimated using one-way ANOVA followed by Turkey’S post hoc test. *P < 0.05; **P < 0.01.
Results
Distribution and protein levels of LXR-α and PPAR-γ in tissue samples
Based on online microarray profiles, the expression of LXR-α and PPAR-γ was found to be downregulated in psoriasis (EG-EOD-54,456, GSE114286, GSE121212, GSE13355, GSE14905, GSE30999, GSE34248, GSE41662, GSE50790, GSE6710) (Table 1); in the present study, we first examined LXR-α and PPAR-γ mRNA and protein expression in psoriasis lesion and nonlesion tissue samples. Pathological analysis of psoriasis lesion and nonlesion tissues was performed using H&E staining and shown in Figure 1(a). Compared to the healthy skin tissues, there were apparent erythema, plaque, infiltration, and scales in psoriasis lesions. IHC staining showed that, in psoriasis lesions, the protein levels of LXR-α and PPAR-γ were lower than those in nonlesion tissue samples (Figure 1(b-c)). As a further confirmation, LXR-α and PPAR-γ mRNA expression was remarkably downregulated in psoriasis lesions (n = 12) with normal tissue samples (Figure 1(d-e)). Moreover, LXR-α and PPAR-γ mRNA expression levels were positively correlated with each other as determined using Pearson’s correlation analysis (figure 1((f)). In summary, the expression of LXR-α and PPAR-γ is downregulated in psoriasis and may be involved in the development of psoriasis.
Figure 1.
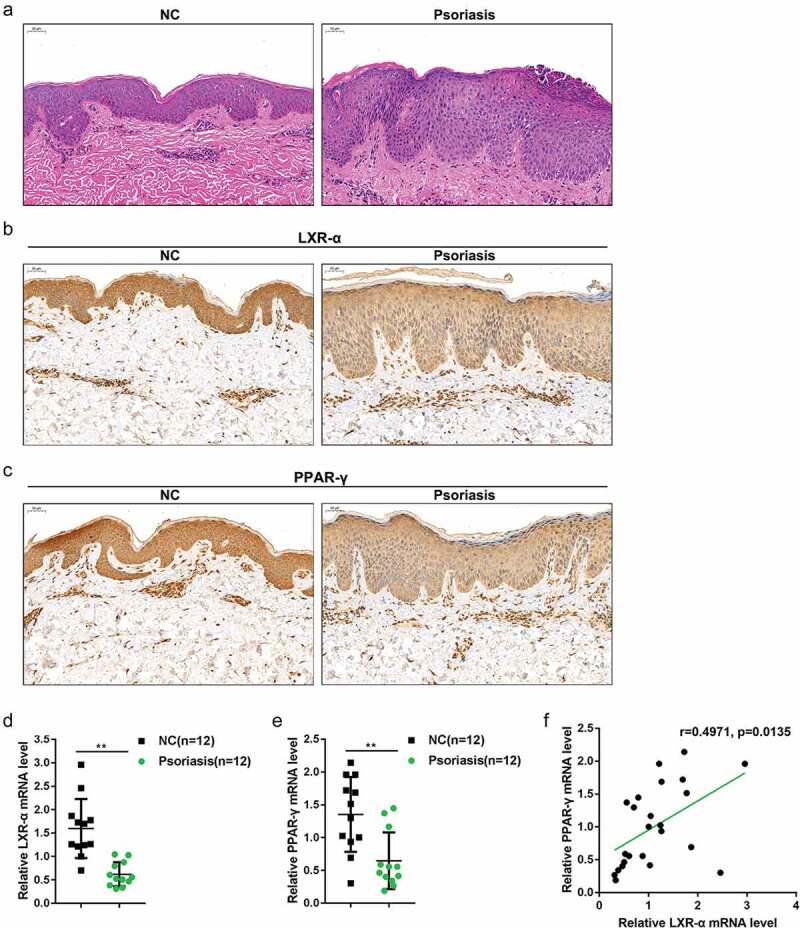
Distribution and protein levels of LXR-α and PPAR-γ in tissue samples (a) Pathological analysis of psoriasis lesion and nonlesion tissues using hematoxylin and eosin (H&E) staining. (b-c) The distribution and protein levels of LXR-α and PPAR-γ in psoriasis lesion and nonlesion tissues determined by immunohistochemical (IHC) staining. (d-e) The mRNA expression of LXR-α and PPAR-γ in psoriasis lesion (n = 12) and nonlesion tissues (n = 12) was determined by real-time PCR. (f) The correlation of LXR-α and PPAR-γ expression. **P < 0.01.
Effects of LXR-α and PPAR-γ on keratinocytes
The above-described findings indicate the abnormal downregulation of LXR-α and PPAR-γ in psoriasis, suggesting their potential roles in psoriasis pathogenesis. Thus, we continued to examine the specific effects of LXR-α and PPAR-γ on keratinocyte HaCaT cells. A psoriasis-like dermatitis in vitro model was generated by treating HaCaT cells with IL-22; in response to IL-22 stimulation, LXR-α and PPAR-γ proteins were both significantly decreased (Figure 2(a)). We transfected an LXR-α or PPAR-γ-overexpressing vector to achieve LXR-α or PPAR-γ overexpression HaCaT cells and performed western blotting to verify the transfection efficiency (Figure 2(b)).
Figure 2.
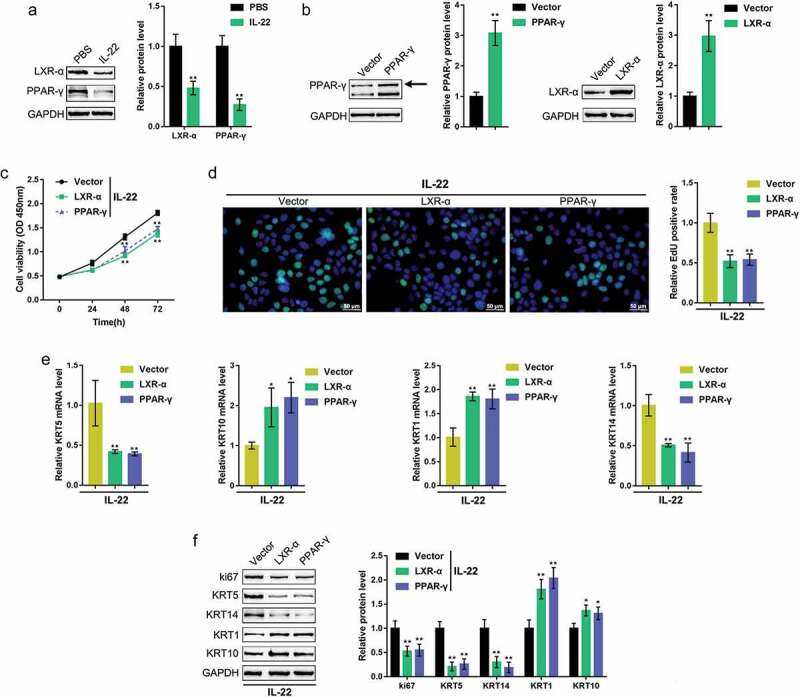
Effects of LXR-α and PPAR-γ on keratinocytes HaCaT cells were treated with IL-22 to generate a psoriasis-like dermatitis model in vitro and examined for (a) the protein levels of LXR-α and PPAR-γ using immunoblotting. (b) an LXR-α or PPAR-γ-overexpressing vector was transfected into HaCaT cells, and the transfection efficiency was verified by western blot (n = 3). Next, HaCaT cells were transfected with the LXR-α or PPAR-γ-overexpressing vector, treated with IL-22, and examined for (c) cell viability using the MTT assay (n = 5); (d) DNA synthesis capacity by the EdU assay (n = 3); (e) the mRNA expression of KRT5, KRT10, KRT1, and KRT14 using real-time PCR (n = 5); and (f) the protein levels of ki67, KRT1, KRT10, KRT5, and KRT14 using immunoblotting (n = 3). *P < 0.05, **P < 0.01.
Next, we transfected HaCaT cells with the LXR-α or PPAR-γ-overexpressing vector, treated them with IL-22, and examined the effects of LXR-α and PPAR-γ on HaCaT cells. As shown in Figure 2(c,d), overexpression of either LXR-α or PPAR-γwas sufficient to significantly inhibit HaCaT cell viability and DNA synthesis. To further confirm the effects of these two factors on keratinocyte hyperplasia, we monitored the changes in the mRNA expression of KRT1/10, the keratin pair mainly expressed in differentiated epidermal cells on the basal layer, and KRT5/14, the keratin pair mainly expressed in proliferative epidermal basal cells. Upon IL-22 stimulation, KRT1 and KRT10 mRNA expression was dramatically increased, whereas KRT5 and KRT14 mRNA expression was reduced by either overexpression of LXR-α or PPAR-γ (Figure 2(e)). Similarly, LXR-α overexpression enhanced KRT1, and KRT10 proteins, whereas decreased ki67, KRT5, and KRT14; PPAR-γ overexpression dramatically enhanced KRT1, and KRT10 proteins but decreased ki67, KRT5, and KRT14 (figure 2(f)). These data indicate that LXR-α or PPAR-γ overexpression significantly inhibits the proliferation and hyperplasia of keratinocytes.
miR-203 targets LXR-α and PPAR-γ to negatively regulate their expression negatively
As mentioned earlier, miRNAs exert an essential effect on psoriasis pathogenesis. As predicted by online tool LncTar (http://www.cuilab.cn/lnctar), miR-203 could simultaneously target LXR-α and PPAR-γ; Moreover, It has been reported that miR-203 was higher expressed in psoriatic lesion and regulated keratinocytes differentiation [22,23]. Here, we first examined miR-203 expression in psoriasis lesion and nonlesion tissues. Figure 3 shows that the expression of miR-203 was significantly upregulated in psoriasis lesions. Moreover, upon IL-22 stimulation, the expression of miR-203 was increased within HaCaT cells (Figure 3(b)). To validate the miR-203 regulation of LXR-α and PPAR-γ, we transfected the miR-203/anti-miR-203 vector to express miR-203 in HaCaT cells and performed real-time PCR to verify the transfection efficiency (Figure 3(c)). Figure 3(d) shows that LXR-α and PPAR-γ proteins were both significantly decreased upon the overexpression of miR-203 but increased upon the inhibition of miR-203. To further verify the putative binding between miR-203 and LXR-α or PPAR-γ, we performed a luciferase reporter assay. We constructed two different types of luciferase reporter vectors, wild-type and mutant, of LXR-α and PPAR-γ; within the putative miR-203 binding site of the mutant reporter vectors, 5 bases were mutated (Figure 3(e)). We cotransfected these vectors into 293 T cells with miR-203/anti-miR-203 vectors and examined luciferase activity. Figure 3(e) shows that the overexpression of miR-203 dramatically reduced, whereas the inhibition of miR-203 increased, the luciferase activity of both wild-type vectors, while mutating the predicted binding site could abolish miR-203 overexpression or inhibition-induced alterations of luciferase activity. In summary, miR-203 negatively regulates the expression of LXR-α and PPAR-γ by directly targeting their 3ʹUTRs.
Figure 3.
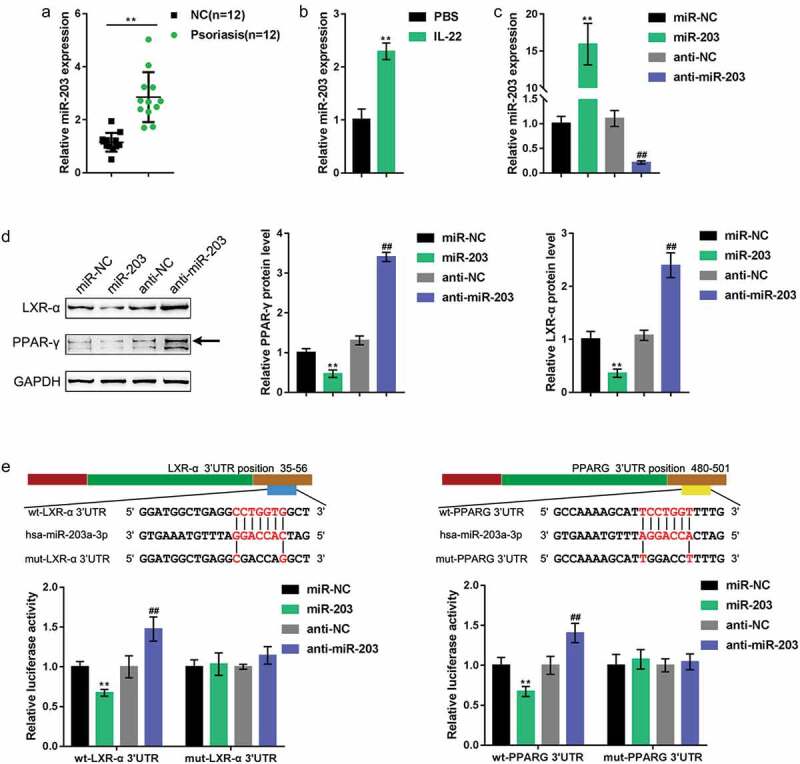
miR-203 targets LXR-α and PPAR-γ to negatively regulate their expression. (a) miR-203 expression in psoriasis lesion and nonlesion tissues examined by real-time PCR (n = 12). (b) miR-203 expression determined in HaCaT cells in the presence or absence of IL-22 (n = 5). (c) miR-203 expression in HaCaT cells after transfection of miR-203 or anti-miR-203 vector, as confirmed by real-time PCR. miR-NC and anti-NC were transfected as negative control for miR-203 and anti-miR-203, respectively. (d) The protein levels of LXR-α and PPAR-γ in response to miR-203 overexpression or inhibition determined in HaCaT cells using Immunoblotting (n = 3). (e) A luciferase reporter assay was performed by constructing wild-type and mutant LXR-α and PPAR-γ luciferase reporter vectors. These vectors were cotransfected in 293 T cells with miR-203 or anti-miR-203 and the luciferase activity was determined (n = 3). **P < 0.01, ##P < 0.01, compared to miR-NC (negative control) or anti-NC group (negative control).
Effects of miR-203 on keratinocytes
Next, we determined the specific effects of miR-203 on the proliferation and hyperplasia of HaCaT cells. We treated HaCaT cells with IL-22 to generate a psoriasis-like dermatitis model in vitro, transfected these cells with anti-miR-203, and examined the related indexes. Figure 4(a-b) shows that the inhibition of miR-203 suppressed cell viability and DNA synthesis. Similarly, LXR-α, PPAR-γ, KRT1, and KRT10 mRNA and protein expression was increased, whereas ki67, KRT5, and KRT14 expression levels were decreased upon the inhibition of miR-203 (Figure 4(c-d)). In summary, miR-203 inhibition suppressed the proliferation and hyperplasia of keratinocytes.
Figure 4.
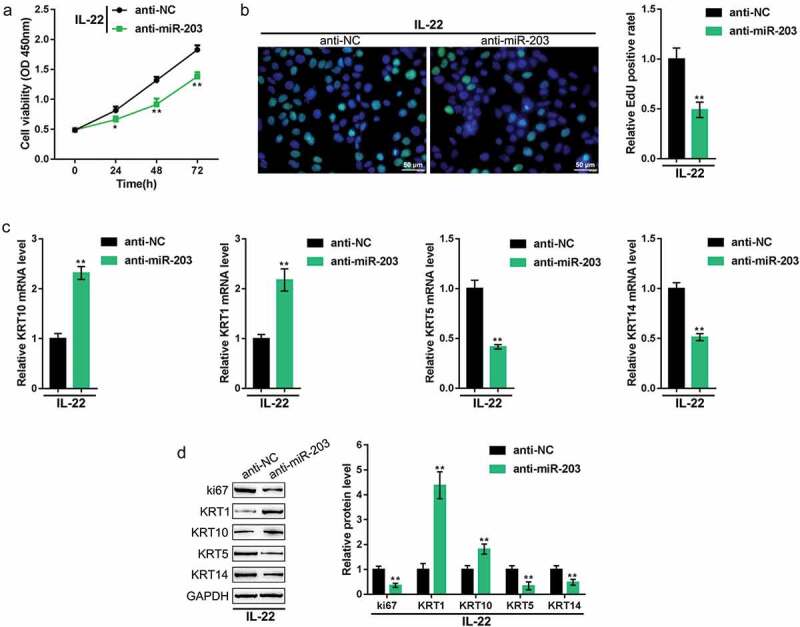
Effects of miR-203 on keratinocytes HaCaT cells were treated with IL-22 to generate a psoriasis-like dermatitis model in vitro, transfected with anti-miR-203 (anti-NC was used as a negative control), and examined for (a) cell viability using the MTT assay; (b) DNA synthesis capacity by the EdU assay; (c) the mRNA expression of KRT5, KRT10, KRT1, and KRT14 using real-time PCR; and (d) the protein levels of LXR-α, PPAR-γ, ki67, KRT1, KRT10, KRT5, and KRT14 using immunoblotting. *P < 0.05, **P < 0.01.
Dynamic effects of miR-203 and LXR-α or PPAR-γ on keratinocytes
Finally, we examined the dynamic effects of miR-203 and LXR-α or PPAR-γ on keratinocytes to investigate whether miR-203 exerts its functions through LXR-α or PPAR-γ. HaCaT cells were treated with IL-22 to generate a psoriasis-like dermatitis model in vitro. The cells were then co-transfected with miR-203 and the LXR-α-overexpressing vector or PPAR-γ-overexpressing vector, and examined for related indexes. miR-203 overexpression significantly promoted HaCaT cell proliferation, decreased LXR-α, PPAR-γ, KRT1, and KRT10 mRNA and protein expression and increased ki67, KRT5, and KRT14 mRNA and protein expression (Figures 5, 6). Overexpression of either LXR-α or PPAR-γexerted the opposite effects on these indexes (Figures 5,6). Moreover, the overexpression of LXR-α or PPAR-γ might remarkably attenuate the effects of miR-203 overexpression (Figures 5,6), indicating that miR-203 exerts its functions on keratinocytes through LXR-α and PPAR-γ.
Figure 5.
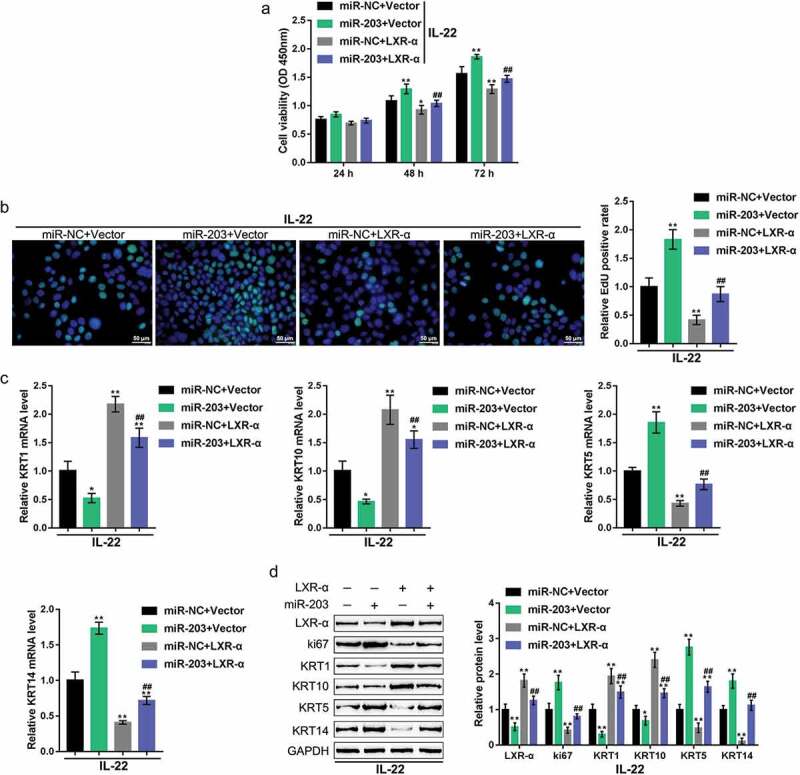
Dynamic effects of miR-203 and LXR-α on keratinocytes HaCaT cells were treated with IL-22 to generate a psoriasis-like dermatitis model in vitro. The cells were cotransfected with miR-203 and the LXR-α-overexpressing vector (miR-NC and Vector were used as negative controls, respectively), and examined for (a) cell viability using the MTT assay; (b) DNA synthesis capacity by EdU assay; (c) the mRNA expression of KRT5, KRT10, KRT1, and KRT14 using real-time PCR; and (d) the protein levels of LXR-α, ki67, KRT1, KRT10, KRT5, and KRT14 using immunoblotting. *P < 0.05, **P < 0.01, compared to control group; ##P < 0.01, compared to the miR-NC (negative control) + LXR-α group.
Figure 6.
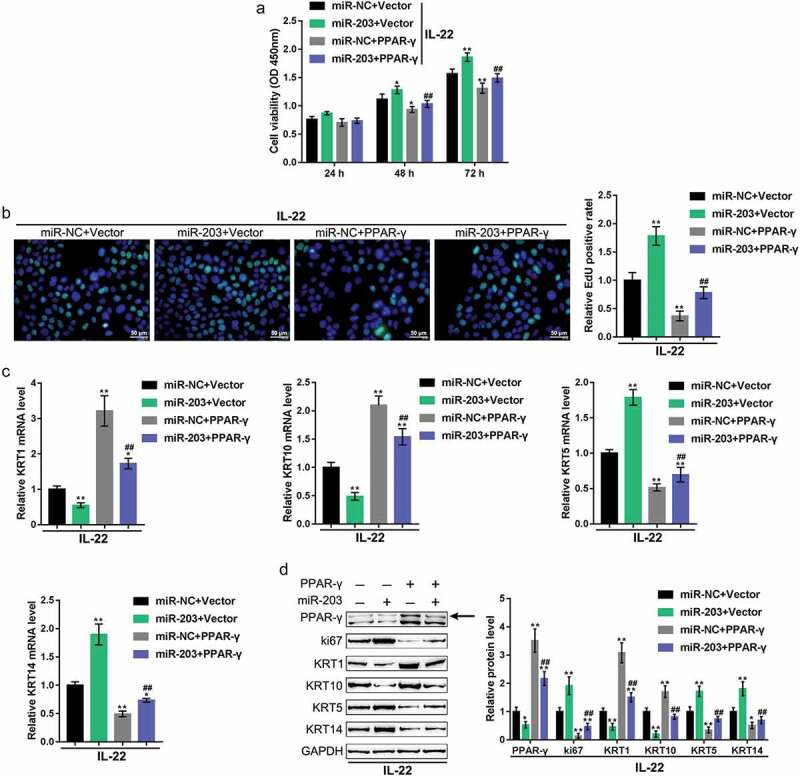
Dynamic effects of miR-203 and PPAR-γ on keratinocytes HaCaT cells were treated with IL-22 to generate a psoriasis-like dermatitis model in vitro and cotransfected with miR-203 and PPAR-γ-overexpressing vector (miR-NC and empty vector were used as negative controls, respectively), and examined for (a) cell viability using MTT assay; (b) DNA synthesis capacity by EdU assay; (c) the mRNA expression of KRT5, KRT10, KRT1, and KRT14 using real-time PCR; (d) the protein levels of PPAR-γ, ki67, KRT1, KRT10, KRT5, and KRT14 using immunoblotting. *P < 0.05, **P < 0.01, compared to control group; ##P < 0.01, compared to miR-NC (negative control) + PPAR-γ group.
Discussion
Herein, we confirmed that the mRNA expression and protein levels of LXR-α and PPAR-γ were downregulated in psoriasis lesion tissues. LXR-α or PPAR-γ overexpression alone was sufficient to inhibit keratinocyte proliferation, increase KRT1 and KRT10 proteins, whereas decrease KRT5 and KRT14 protein levels. miR-203 negatively regulated LXR-α and PPAR-γ expression through direct targeting. miR-203 inhibition exerted the opposite effects on HaCaT cells to those of the LXR-α or PPAR-γ overexpression. More importantly, LXR-α or PPAR-γ overexpression could markedly attenuate the effects of miR-203 overexpression on keratinocytes, indicating that miR-203 promotes keratinocyte proliferation by targeting LXR-α and PPAR-γ.
Psoriasis is a prevalent, chronic inflammatory disease of the skin. The increased cell proliferation of keratinocytes, dermal angiogenesis changes and the increase in proinflammatory cytokine production are the major characteristic features of psoriasis [24,25]. The nuclear receptor LXR-α exerts a well-documented effect on cholesterol homeostasis and inflammation [26]. As recently reported, the LXR-α gene exerts a significant effect on the proliferation and differentiation of keratinocytes [27–29]. In keratinocytes, the LXR-α gene modulates the expression of proinflammatory cytokines, VDR, PPAR-γ, and catalase. Catalase is considered a major antioxidant enzyme for the clearance of ROS (reactive oxygen species) produced in primary epidermal keratinocytes [13]. In the present study, LXR-α and PPAR-γ mRNA and protein expression levels were remarkably downregulated in psoriasis lesions, consistent with the results reported by ten datasets of online microarray profiles.
Regarding the molecular effects, we overexpressed LXR-α or PPAR-γin keratinocytes (HaCaT cells) and found that the expression of either LXR-α or PPAR-γ was sufficient to inhibit cell proliferation, which was manifested by inhibited cell viability and DNA synthesis. Besides, overexpression of either LXR-α or PPAR-γwas sufficient to decrease the protein level of ki67. Ki67 is a nuclear protein that is strictly associated with and necessary for cellular proliferation. The fact that the ki67 protein is present during all active phases of the cell cycle (G(1), S, G(2), and mitosis, but is absent in resting cells (G(0)) makes it an excellent marker for determining cell proliferation [30]. Moreover, KRT1 and KRT10 were significantly increased, whereas KRT5 and KRT14 were decreased by LXR-α or PPAR-γ overexpression. In the interfollicular epidermis, KRT14-KRT5 is the major type I-type II keratin pair expressed in proliferative basal keratinocytes, whereas KRT10-KRT1 is the major type keratin pair expressed in differentiated keratinocytes in the suprabasal layers [31]. In the process of skin damage or psoriatic lesions, suprabasal keratinocytes reduce KRT10-KRT1 and increase KRT14-KRT5, strongly inducing KRT16/KRT17-KRT6 expression [31]. These data indicate that LXR-α or PPAR-γ overexpression can suppress the proliferation of keratinocytes. However, LXR-α and PPAR-γ mRNA and protein expression levels were dramatically downregulated in psoriasis lesions; therefore, we further investigated the mechanism of LXR-α and PPAR-γ downregulation in psoriasis.
As predicted by online tools, LXR-α and PPAR-γ might be direct downstream targets of miR-203. The link between miRNAs and psoriasis was first described in 2007 [32]. To date, it has been reported that more than 250 miRNAs are abnormally expressed in psoriasis lesions, most of which are present in peripheral blood or related psoriatic skin [33]. Some investigations have compared the miRNA profiles of unrelated psoriatic skin with those of normal and healthy skin [22,34]. Many miRNAs, including miR-203, have been reported to be strongly implicated in the immunopathogenesis of psoriasis [15,32,35]. miR-203 was found to be specifically altered in psoriatic lesions [36]. miR-203 upregulation in psoriasis lesions is significantly correlated with the downregulation of SOCS3 (suppressor of cytokine signaling 3) and the subsequent increase in STAT3 (signal transducer and activator of transcription-3), which is well-known for its fundamental role in transcription in keratinocytes during psoriasis progression [37]. In the present study, we confirmed that the expression of miR-203 was upregulated in psoriasis lesions. By targeting the 3ʹUTRs of LXR-α and PPAR-γ, miR-203 negatively regulated their expression. As earlier indicated, LXR-α or PPAR-γ overexpression significantly suppressed the proliferation of keratinocytes; similarly, miR-203 inhibition in HaCaT cells also significantly suppressed cell proliferation and altered the protein levels of related factors. More importantly, miR-203 overexpression promoted HaCaT cell proliferation. In contrast, the overexpression of LXR-α or PPAR-γsignificantly attenuated its effects, suggesting that miR-203 promotes HaCaT cell proliferation by targeting LXR-α and PPAR-γ.
In conclusion, the miR-203-LXR-α/PPAR-γ axis modulates the proliferation of keratinocytes and might be a novel target for psoriasis treatment, which needs further in vivo and clinical investigations.
As further confirmation enhancement of the present findings, an RNA-Seq experiment, which has been performed by several previous studies [38,39], could be a promising tool. Moreover, the role of glyoxalase I in the proliferation and EMT of human keratinized epithelial cells [40,41] might be an extension of the current research.
Supplementary Material
Funding Statement
This study was supported by the National Natural Science Foundation [81573985].
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Beek CH, van Reede EC.. The nature and frequency of the histological changes found in psoriasis vulgaris. Arch Dermatol Res. 1977;257(3):255–264. [DOI] [PubMed] [Google Scholar]
- [2].Zhang J, Lin Y, Li C, et al. IL-35 decelerates the inflammatory process by regulating inflammatory cytokine secretion and M1/M2 macrophage ratio in psoriasis. J Immunol. 2016;197(6):2131–2144. [DOI] [PubMed] [Google Scholar]
- [3].Teng X, Hu Z, Wei X, et al. IL-37 ameliorates the inflammatory process in psoriasis by suppressing proinflammatory cytokine production. J Immunol. 2014;192(4):1815–1823. [DOI] [PubMed] [Google Scholar]
- [4].Nold MF, Nold-Petry CA, Zepp JA, et al. IL-37 is a fundamental inhibitor of innate immunity. Nat Immunol. 2010;11(11):1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Boehncke WH, Schon MP.. Psoriasis. Lancet. 2015;386(9997):983–994. [DOI] [PubMed] [Google Scholar]
- [6].Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445(7130):866–873. [DOI] [PubMed] [Google Scholar]
- [7].Monteleone G, Pallone F, MacDonald T, et al. Psoriasis: from pathogenesis to novel therapeutic approaches. Clin Sci (Lond). 2011;120(1):1–11. [DOI] [PubMed] [Google Scholar]
- [8].Cabrijan L, Lipozenčić J, Batinac T, et al. Psoriasis vulgaris - an inflammatory skin disease and/or benign epidermal hyperplasia. Acta Dermatovenerol Croat. 2011;19(2):117–119. [PubMed] [Google Scholar]
- [9].Perera GK, Di Meglio P, Nestle FO. Psoriasis. Annu Rev Pathol. 2012;7:385–422. [DOI] [PubMed] [Google Scholar]
- [10].Asadullah K, Döcke WD, Volk HD, et al. The pathophysiological role of cytokines in psoriasis. Drugs Today (Barc). 1999;35(12):913–924. [PubMed] [Google Scholar]
- [11].Henseler T, Christophers E. Psoriasis of early and late onset: characterization of two types of psoriasis vulgaris. J Am Acad Dermatol. 1985;13(3):450–456. [DOI] [PubMed] [Google Scholar]
- [12].Stuart P, Malick F, Nair RP, et al. Analysis of phenotypic variation in psoriasis as a function of age at onset and family history. Arch Dermatol Res. 2002;294(5):207–213. [DOI] [PubMed] [Google Scholar]
- [13].Gupta DS, Kaul D, Kanwar AJ, et al. Psoriasis: crucial role of LXR-alpha RNomics. Genes Immun. 2010;11(1):37–44. [DOI] [PubMed] [Google Scholar]
- [14].Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9(3):219–230. [DOI] [PubMed] [Google Scholar]
- [15].Zibert JR, Løvendorf MB, Litman T, et al. MicroRNAs and potential target interactions in psoriasis. J Dermatol Sci. 2010;58(3):177–185. [DOI] [PubMed] [Google Scholar]
- [16].Roberts WC, Siegel RJ, McManus BM. Idiopathic dilated cardiomyopathy: analysis of 152 necropsy patients. Am J Cardiol. 1987;60(16):1340–1355. [DOI] [PubMed] [Google Scholar]
- [17].Han D, Kim H-Y, Lee H-J, et al. Wound healing activity of gamma-aminobutyric Acid (GABA) in rats. J Microbiol Biotechnol. 2007;17(10):1661–1669. [PubMed] [Google Scholar]
- [18].Liu H, Deng H, Zhao Y, et al. LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J Exp Clin Cancer Res. 2018;37(1):279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liu Z, Dou C, Yao B, et al. Ftx non coding RNA-derived miR-545 promotes cell proliferation by targeting RIG-I in hepatocellular carcinoma. Oncotarget. 2016;7(18):25350–25365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tu K, LIU Z, YAO B, et al. MicroRNA-519a promotes tumor growth by targeting PTEN/PI3K/AKT signaling in hepatocellular carcinoma. Int J Oncol. 2016;48(3):965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Donato L, Scimone C, Rinaldi C, et al. Stargardt phenotype associated with two ELOVL4 promoter variants and ELOVL4 downregulation: new possible perspective to etiopathogenesis? Invest Ophthalmol Vis Sci. 2018;59(2):843–857. [DOI] [PubMed] [Google Scholar]
- [22].Joyce CE, Zhou X, Xia J, et al. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum Mol Genet. 2011;20(20):4025–4040Xu Y, Ji Y, Lan X, et al. miR‑203 contributes to IL‑17‑induced VEGF secretion by targeting SOCS3 in keratinocytes[J]. Mol Med Rep. 2017;16(6):8989.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].XU Y, Ji Y, Lan X, et al. miR‑203 contributes to IL‑17‑induced VEGF secretion by targeting SOCS3 in keratinocytes[J]. Mol Med Rep. 2017;16(6):8989. [DOI] [PubMed] [Google Scholar]
- [24].Liu Y, Krueger JG, Bowcock AM. Psoriasis: genetic associations and immune system changes. Genes Immun. 2007;8(1):1–12. [DOI] [PubMed] [Google Scholar]
- [25].Sabat R, Philipp S, Höflich C, et al. Immunopathogenesis of psoriasis. Exp Dermatol. 2007;16(10):779–798. [DOI] [PubMed] [Google Scholar]
- [26].Tontonoz P, Mangelsdorf DJ. Liver X receptor signaling pathways in cardiovascular disease. Mol Endocrinol. 2003;17(6):985–993. [DOI] [PubMed] [Google Scholar]
- [27].Hanley K, Ng DC, He S, et al. Oxysterols induce differentiation in human keratinocytes and increase Ap-1-dependent involucrin transcription. J Invest Dermatol. 2000;114(3):545–553. [DOI] [PubMed] [Google Scholar]
- [28].Komuves LG, Schmuth M, Fowler AJ, et al. Oxysterol stimulation of epidermal differentiation is mediated by liver X receptor-beta in murine epidermis. J Invest Dermatol. 2002;118(1):25–34. [DOI] [PubMed] [Google Scholar]
- [29].Schmuth M, Elias PM, Hanley K, et al. The effect of LXR activators on AP-1 proteins in keratinocytes. J Invest Dermatol. 2004;123(1):41–48. [DOI] [PubMed] [Google Scholar]
- [30].Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182(3):311–322. [DOI] [PubMed] [Google Scholar]
- [31].Zhang X, Yin M, Zhang LJ. Keratin 6, 16 and 17-critical barrier alarmin molecules in skin wounds and psoriasis. Cells. 2019;8(8):807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sonkoly E, Wei T, Janson PCJ, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2(7):e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hawkes JE, Nguyen GH, Fujita M, et al. microRNAs in psoriasis. J Invest Dermatol. 2016;136(2):365–371. [DOI] [PubMed] [Google Scholar]
- [34].Raaby L, Langkilde A, Kjellerup RB, et al. Changes in mRNA expression precede changes in micro RNA expression in lesional psoriatic skin during treatment with adalimumab. Br J Dermatol. 2015;173(2):436–447. [DOI] [PubMed] [Google Scholar]
- [35].Lerman G, Avivi C, Mardoukh C, et al. MiRNA expression in psoriatic skin: reciprocal regulation of hsa-miR-99a and IGF-1R. PLoS One. 2011;6(6):e20916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang M-J, Xu -Y-Y, Huang R-Y, et al. Role of an imbalanced miRNAs axis in pathogenesis of psoriasis: novel perspectives based on review of the literature. Oncotarget. 2017;8(3):5498–5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sonkoly E, Wei T, Pavez Loriè E, et al. Protein kinase C-dependent upregulation of miR-203 induces the differentiation of human keratinocytes. J Invest Dermatol. 2010;130(1):124–134. [DOI] [PubMed] [Google Scholar]
- [38].Donato L, Scimone C, Nicocia G, et al. Role of oxidative stress in Retinitis pigmentosa: new involved pathways by an RNA-Seq analysis. Cell Cycle. 2019;18(1):84–104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [39].Donato L, Scimone C, Rinaldi C, et al. Non-coding RNAome of RPE cells under oxidative stress suggests unknown regulative aspects of retinitis pigmentosa etiopathogenesis. Sci Rep. 2018;8(1):16638. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [40].Rinaldi C, Bramanti P, Famà A, et al. Glyoxalase I A111e, paraoxonase 1 Q192r and L55m polymorphisms in Italian patients with sporadic cerebral cavernous malformations: a pilot study. J Biol Regul Homeost Agents. 2015;29(2):493–500. [PubMed] [Google Scholar]
- [41].Donato L, Scimone C, Nicocia G, et al. GLO1 gene polymorphisms and their association with retinitis pigmentosa: a case-control study in a Sicilian population. Mol Biol Rep. 2018;45(5):1349–1355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


