Abstract
Dysregulation of transcription is found in nearly every human disease, and as a result there has been intense interest in developing new therapeutics that target regulators of transcription. CREB binding protein (CBP) and its paralogue p300 are attractive targets due to their function as ‘master coactivators’. Although inhibitors of several CBP/p300 domains have been identified, the selectivity of many of these compounds has remained underexplored. Here, we review recent successes in the development of chemical tools targeting several CBP/p300 domains with selectivity acceptable for use as chemical probes. Additionally, we highlight recent studies which have used these probes to expand our understanding of interdomain interactions and differential coactivator usage.
Introduction
As the search for new therapeutic targets broadens, transcriptional coactivators have emerged as exciting opportunities. Nowhere is this more evident than with the socalled ‘master coactivators’ p300 and CBP [1]. Due to the involvement of CBP and p300 in many signaling pathways that contribute to diseases including cancer and neurodegeneration, a significant focus has been placed on the development of CBP and p300 probes [2–4]. In these endeavors, the domains of CBP/p300 are often considered in isolation or significantly shortened constructs; however, the domains are likely interrelated in their function and work together to modulate transcription (Figure 1). CBP and p300 diverged due to gene duplication 450 million years ago and were initially believed to be functionally redundant as they show high sequence similarity (75% sequence similarity and 58% sequence identity). This sequence conservation is particularly within the conserved activator-binding domains (93% sequence identity for TAZ1, 90% sequence identity for KIX) [5,6]. Nonetheless, increasing evidence now suggests that CBP and p300 serve distinct roles in some pathways [7–12].
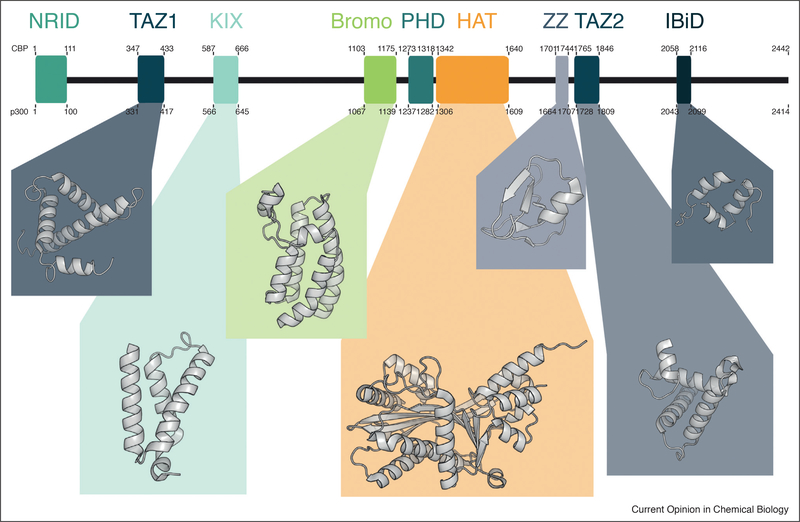
Figure 1.
Structure of CBP and p300. CBP and p300 have high sequence identity (58% overall), particularly within the conserved domains. These include an N-terminal nuclear receptor interaction domain (NRID), four zinc finger domains (TAZ1, PHD, ZZ, and TAZ2), a KIX domain, a bromodomain, a histone acetyltranferase (HAT), and the interferon binding domain (IBiD). Using these domains CBP and p300 can perform a variety of tasks, including modifying chromatin and transcription factors, reading epigenetic signals, and acting as a bridge between DNA bound transcription factors and the general transcriptional machinery (PDB IDs 1U2N, 4I9O, 4OUF, 5KJ2, 1TOT, 1F81, and 1JJS).
A better understanding of the roles CBP and p300 play in signaling pathways and how the domains within CBP and p300 work together to regulate transcription will be essential for the development of new therapeutic approaches. To accomplish this, it will be critical for chemical biologists to have access to a complete toolbox of selective probes [13–15]. However, the selectivity of CBP and p300 ligands has been largely underexplored. In this review, we will high- light some of the recent successes in CBP and p300 probe development in the context of each domain. Additionally, we will emphasize areas of concern regarding selectivity and remaining challenges that should be addressed.
HAT
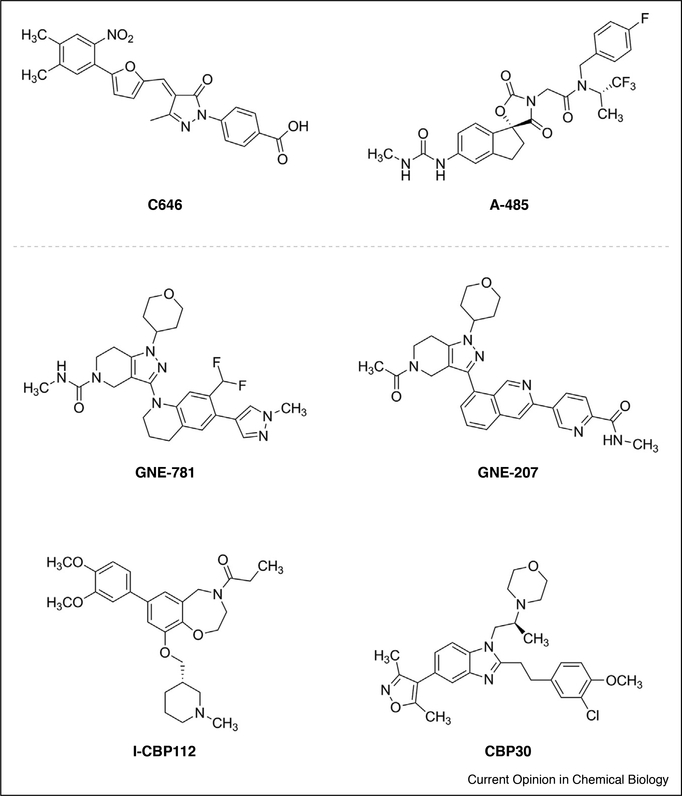
A central challenge for targeting HATs has been obtaining ligands with high affinity and specific inhibition [16,17]. Most reported HAT inhibitors contain electrophiles or motifs common in pan assay interference com-pounds (PAINs) [18]. For example, curcumin was reported to selectively inhibit p300 HAT over other related HATs, but curcumin is a well-known PAIN with a broad activity profile [19]. Another small molecule inhibitor, C646, was among the first small molecules to demonstrate submicromolar affinity for a HAT and is now commonly used as a CBP/p300 HAT inhibitor. C646 was identified through a virtual screen and found to be selective for p300 against six other HATs, and although it contains a potentially reactive conjugated pyrazolonefuran it was demonstrated to be a reversible inhibitor of the p300 HAT [20]. In 2016, Shrimp and coworkers used a chemoproteomics approach to identify possible covalent targets of C646 and found through LC–MS/MS analysis that C646 labeled tubulin proteins as well as highly abundant cellular proteins known to have reactive cysteines [21]. It was further demonstrated that C646 inhibits tubulin polymerization in vitro at concentration typically used in cellular assays. The problems facing HAT inhibitor development have been further expanded upon in a recent report by Dahlin and coworkers [22••]. Using a suite of screens including ALARM NMR, glutathione reactivity, and detergent sensitivity assays, it was demonstrated that out of 23 reported HAT inhibitors tested half showed non-specific thiol reactivity or compound aggregation. Furthermore, evaluation against unrelated targets and in cellular assays showed many of these inhibitors have significant off target effects.
The most recently reported CBP/p300 HAT inhibitor, A-485, represents a significant step forward in the development of HAT chemical probes [23•,24]. A-485 is an acetyl-CoA competitive inhibitor with low nanomolar potency for both the CBP and p300 HATs. Upon selectivity testing against 6 other HATs as well as 3 bromodomains and over 150 non-epigenetic targets, A-485 was found to be highly selective for the CBP/p300 HAT. When tested with 124cancer cell lines, A-485 showed broad, potent inhibition against hematological cancers as well as androgen receptor (AR) positive prostate cancers. In a mouse xenograft model of AR-positive castration-resistant prostate cancer, treatment with A-485 resulted in a 54% reduction in tumor growth. Finally, it should be noted that a structure of A-485 in the initial publication contained an error, and the correct structure is provided in Figure 2.
Figure 2.
Structures of CBP/p300 HAT inhibitors (top) and bromodomain inhibitors (bottom).
Bromodomain
The CBP and p300 bromodomains are the second most common family of bromodomain targets after the BET family bromodomains [25–27]. Of late, the bromodomain has also been the most targeted domain within CBP and p300, and over 50% of small molecule CBP/p300 ligands published since 2016 are bromodomain inhibitors. Although potent bromodomain inhibitors have been reported, achieving selectivity for the CBP/p300 bromodomain relative to the other 59 bromodomains has posed a significant challenge [28].
Until recently, most CBP/p300 bromodomain inhibitors showed appreciable binding to at least one off-target bromodomain. Groups from Genentech have reported a series of inhibitors based on the initial compound GNE-272 that have cleared this selectivity hurdle (Figure 2) [29–31,32•,33] Two optimized, orally-available inhibitors, GNE-207 and GNE-781, have subnanomolar binding affinities for the CBP/p300 bromodomains [32•,33]. When evaluated against a diverse panel of bromodomains, GNE-207 was found to be >4700-fold selective for CBP/p300 and GNE-781 was >7200-fold selective. Additional analysis of both inhibitors at 10 μM in a Cerep off-target screening panel found that at none of the 43 receptors tested were inhibited at >40%.
CellCentric recently disclosed the development of CCS1477, an orally-available small molecule bromodomain inhibitor with single digit nanomolar affinity for CBP/p300 [34–36]. In a mouse xenograft model of AR-positive castration resistant prostate cancer, treatment with CCS1477 resulted in complete inhibition of tumor growth and reduction of tumor biomarkers. CellCentric has announced plans to begin phase I clinical trials of CCS1477 in late stage prostate cancer this year.
In addition to serving as probes of direct bromodomain functions, inhibitors of the CBP and p300 bromodomains are contributing to the increasing evidence that these motifs can modulate HAT activity [37]. Zucconi and coworkers have shown that treatment of recombinant full length p300 with the inhibitor I-CBP112 increased acetylation of reconstituted nucleosomes up to 3-fold; importantly, this effect was not observed with the isolated HAT domain [38]. These effects were further observed in acute leukemia and prostate cancer cells, where treatment with I-CBP112 resulted in increased acetylation of the known CBP/p300 target H3K18. In contrast, Shrimp and colleagues found that I-CBP112, as well as the CBP/p300 bromodomain inhibitor CBP30, inhibited transcriptional activation by a fusion protein consisting of catalytically dead Cas9 (dCas9) and p300. Interestingly, activation was also inhibited by mutations in the bromodomain that blocked acetyllysine binding [39]. It should be noted that while Zucconi and coworkers used full length p300, the dCas9–p300 fusion used by Shrimp et al. only contained the p300 bromodomain and HAT. The differing effects on HAT activity observed in these two studies could stem from additional interdomain effects that are present in full length p300. While the mechanisms by which the CBP/p300 bromodomain, and potential additional domains, influences HAT activity are still not understood the availability of potent and specific chemical probes for these motifs will aid in dissecting how interdomain interactions modulate function.
KIX
Unlike the HAT and bromodomain, the KIX domain lacks well-defined binding pockets and instead uses two broad surfaces on opposite faces of the protein to interact with a diverse group of transcriptional activators [40]. Consequently, the development of highly potent CBP and p300 KIX inhibitors has remained challenging. Additionally, determining the probe binding site can be difficult and time consuming due to allosteric communication between the two transcription factor interaction sites and the overall conformationally lability of the domain [41]. There has thus been a need for new ligand discovery and characterization strategies for this domain.
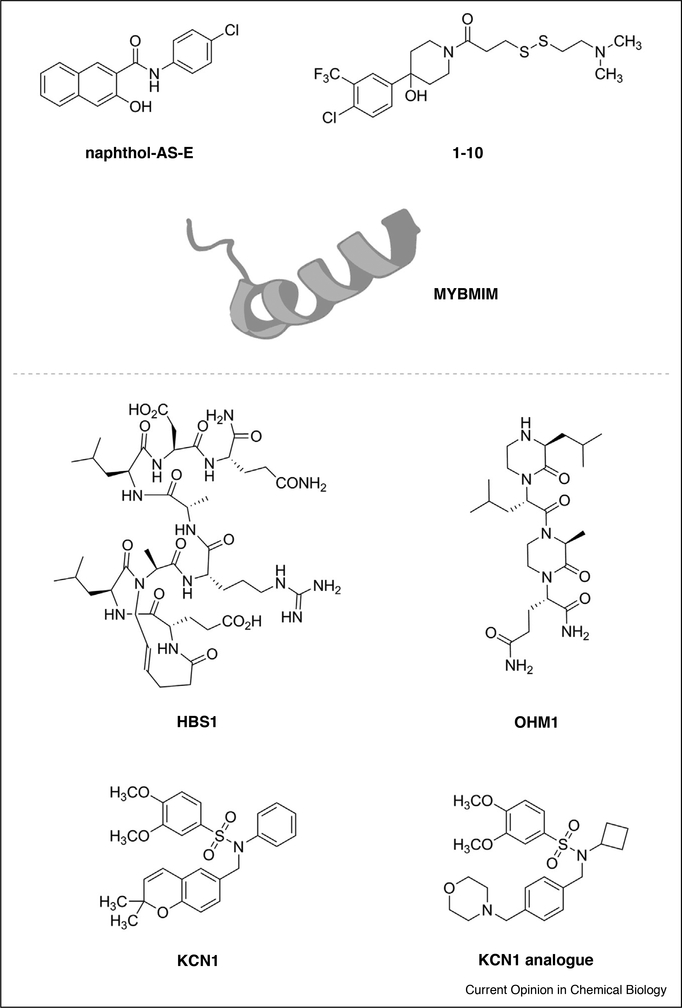
The Pomerantz group has advanced protein-observed 19F (PrOF) NMR as a ligand discovery strategy for bromodomains and the CBP KIX domain that enables preliminary selectivity to be determined in the course of the primary screen and for determination of the binding mode of hit molecules [42–44]. Most recently, they have expanded this work by using PrOF NMR with dually 3-fluorotyrosine and 4-fluorophenylalanine labeled CBP KIX. Dual labeling facilitated identification of distinct signatures for native ligands of each CBP KIX binding site as well as determination of the binding sites of previously identified small molecule ligands [45••]. Interestingly, the CBP/p300 KIX inhibitor naphthol-AS-E (Figure 3), for which there is conflicting data concerning the binding site, produced a PrOF signature that is inconsistent with binding at the Myb interface as has been proposed [44,46,47]. The PrOF NMR signature combined with 1H–15N HSCQ NMR data and docking studies instead suggested naphthol-AS-E binds at a previously unidentified site between the α1 and α2 helices. This is an exciting finding as it indicates that purely allosteric modulation of CBP/p300 KIX with small molecules is achievable.
Figure 3.
Structures of CBP/p300 KIX inhibitors (top) and TAZ1 inhibitors (bottom).
The Mapp laboratory has used the site-directed ligand discovery strategy Tethering to identify fragments with affinity for CBP KIX at defined binding sites [48–50]. These chemical co-chaperones such as 1–10 enable distinct conformations of KIX to be characterized biophysically and functionally. These probes have enabled us to dissect the kinetics of allosteric effects between the two activator binding interfaces and to obtain the first crystal structure of a KIX domain [49,51]. Most relevant for future probe discovery, these studies show that KIX can be regulated by a dual orthosteric/allosteric modulator through targeting the most conformationally mobile region of the motif, the flexible loop that connects α2 and α3 and comprises a portion of the MLL binding site. Although these probes have been powerful tools in vitro, the requirement of the engineered cognate cysteine has thus far limited use of these probes in vivo.
Ramaswamy and coworkers recently developed MYB-MIM, a retro-inverso peptidomimetic of the Myb transcriptional activation domain with low micromolar affinity for CBP/p300 KIX [52•]. MYBMIM inhibits co-immunoprecipitation of CBP/p300 by Myb, and in various acute myeloid leukemia cell lines it was found that MYBMIM accumulated in the nuclei and had robust effects on the transcription of Myb-dependent genes including MYC and BCL2. Additionally, MYBMIM treatment of immunodeficient mice grafted with patient-derived leukemias resulted in decreased disease progression and extended survival. The in vivo efficacy of MYBMIM should make it a powerful tool for studying Myb-dependent transcription.
It should be noted that KIX domains are also found in the Med15 subunit of the Mediator complex and the DNA helicase RECQL5 [53]. However, selectivity evaluation for KIX domain ligands is often incomplete. To the best of our knowledge, no ligands have been evaluated for binding to the RECQL5 KIX domain, and extensive off-target profiling also has not been completed. As we continue to move forward using CBP/p300 KIX domain probes in vivo, complete selectivity profiles will be critical for accurate evaluation of data.
TAZ1
Despite significant differences in their primary sequences, the TAZ1 and TAZ2 domains (also referred to as the CH1 and CH3 regions, respectively) adopt a similar fold, and this fold has not been observed in other zinc finger proteins [54]. The unique fold makes the TAZ domains attractive drug targets, and several groups have targeted the TAZ1/HIF-1α interaction using natural pro-ducts, small molecules, and peptidomimetics (Figure 3) [55]. Early inhibitors such as chetomin and chaetocin induced ejection of zinc from the TAZ1 protein, and this mechanism of action also lead to inhibition of other proteins such as thioredoxin reductase and the histone lysine methyltransferase G9a [56–59]. Later inhibitors such as KCN1, OHM1, and HBS1 have been demonstrated to inhibit the TAZ1/HIF-1α interaction through direct binding to TAZ1 without affecting zinc binding [60–63]. Most recently, Ferguson and coworkers have reported an analogue of KCN1 with a >50-fold increase in aqueous solubility and similar activity in a reporter assay for HIF transcriptional activity [64•].
Transcription activation domains are often highly specific for TAZ1 or TAZ2, and although this may hold true for TAZ domain probes, the selectivity of TAZ1/HIF-1α inhibitors for TAZ1 over TAZ2 has not yet been tested [65]. Preliminary evaluation of off-target effects for HBS1 and OHM1 has been completed through monitoring changes in global gene expression, but beyond this the selectivity of TAZ1 inhibitors remains largely unexplored [62,63].
NRID
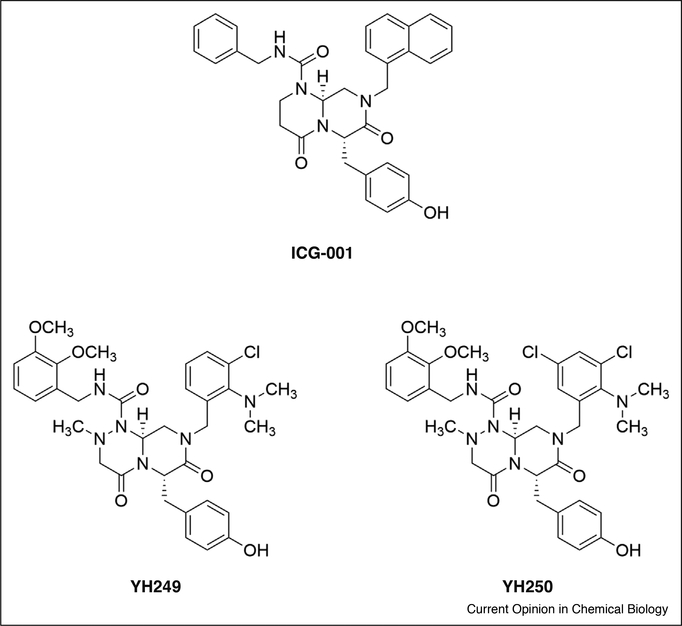
The NRID domain shows the least sequence identity (63%) between the CBP and p300 domains and is the only domain for which compounds that can discriminate between CBP and p300 have been developed (Figure 4). Previously, the Kahn lab identified ICG-001 as a selective inhibitor of the β-catenin/CBP NRID interaction, and an analogue of ICG-001 has completed phase I clinical trials [66–68]. More recently, they also reported the first direct inhibitors of the p300/β-catenin interaction [69••]. Using a differential cell-based reporter gene assay to detect compounds that were selective for p300 over CBP, the closely related compounds YH249 and YH250 were identified from a focused library of ICG-001 analogues. YH249 and YH250 inhibited p300-dependent transcription at nanomolar concentrations and showed >2400-fold selectivity for p300 over CBP. Co-immunoprecipitation and pulldown experiments in the presence of YH249 confirmed that it selectively engaged p300 and antagonized the p300/β-catenin interaction. Although not demonstrated, the binding location is assumed to be the p300 NRID based on the structural similarity to ICG-001.
Figure 4.
Structures of CBP and p300 NRID inhibitors. ICG-001 is selective for the CBP NRID domain, and YH249 and YH250 are selective for the p300 NRID. These are the only small molecule inhibitors reported that discriminate between CBP and p300.
The development of selective inhibitors of the CBP and p300 NRID/beta-catenin interactions has enabled the study of the differential coactivator usage by β-catenin. A combination of genetic methods and treatment with ICG-001 were used to demonstrate that β-catenin/TCF mediated expression of the survivin gene requires CBP [70]. Additionally, these probes have been used to generate a model of coactivator usage by β-catenin in stem cells in which interaction with CBP results in maintenance of pluripotency while interaction with p300 leads to differentiation [69••,71–73].
Outlook
To fully interrogate the roles of CBP and p300 in various pathways, it will be vital to have access to a complete toolbox of probes, including inhibitors as well as artificial activators. While we assemble this toolbox, the community will need to pay greater attention to probe selectivity. This should include testing against related domains as well as common off targets. Furthermore, target occupancy in cells has been underexplored for many CBP/p300 probes. As probes with improved potency and selectivity continue to be reported, the variety of available approaches for determining target engagement in cells should enable a thorough evaluation of target occupancy [74]. The establishment of a full toolbox of vetted probes will enable the community to robustly establish causal relationship between target engagement and phenotype [15].
As evidence continues to mount that the functions of CBP and p300 are not always redundant, the next frontier will likely be the development of probes that are selective for CBP or p300. The successful development of CBP and p300 selective NRID inhibitors demonstrates that in some cases selective probes may be identified through counter screening efforts. For domains that show higher sequence identity between CBP and p300, such as the TAZ1 and KIX domains, the CRISPR/Cas9 system may facilitate engineering probe selectivity. Mutations to CBP or p300 could facilitate bump and hole strategies to convert ligands into selective probes, or engineered cysteine residues could enable the use of selective tethering fragments in vivo.
Probes of CBP and p300 will continue to be valuable tools as the field works towards a more complete understanding of transcriptional regulation. A complete toolbox of well-characterized, potent, and selective probes will be essential for this task. Insight gained from the development and use of these probes will refine our model of CBP and p300 function and may also guide the development of new therapeutic approaches.
Acknowledgements
MEB is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation. We thank Dr. Rachel Pricer, Dr. Stephen Joy, and Dr. Steven Sturlis for assistance with manuscript preparation.
Footnotes
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as
• of special interest
•• of outstanding interest
- 1.Dyson HJ, Wright PE: Role of intrinsic protein disorder in the function and interactions of the transcriptional coactivators CREB-binding protein (CBP) and p300. J Biol Chem 2016, 291:6714–6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang F, Marshall CB, Ikura M: Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition. Cell Mol Life Sci 2013, 70:3989–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dutta R, Tiu B, Sakamoto KM: CBP/p300 acetyltransferase activity in hematologic malignancies. Mol Gen Metab 2016, 119:37–43. [DOI] [PubMed] [Google Scholar]
- 4.Valor LM, Viosca J, Lopez-Atalaya JP, Barco A: Lysine acetyltransferases CBP and p300 as therapeutic targets in cognitive and neurodegenerative disorders. Curr Pharm Des 2013, 19:5051–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giles RH, Dauwerse HG, van Ommen GJ, Breuning MH: Do human chromosomal bands 16p13 and 22q11–13 share ancestral origins? Am J Hum Gen 1998, 63:1240–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arany Z, Sellers WR, Livingston DM, Eckner R: E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell 1994, 77:799–800. [DOI] [PubMed] [Google Scholar]
- 7.Kalkhoven E: CBP and p300: HATs for different occasions. Biochem Pharmacol 2004, 68:1145–1155. [DOI] [PubMed] [Google Scholar]
- 8.Kasper LH, Fukuyama T, Biesen MA, Boussouar F, Tong C, de Pauw A, Murray PJ, van Deursen JMA, Brindle PK: Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol Cell Biol 2006, 26:789–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramos YFM, Hestand MS, Verlaan M, Krabbendam E, Ariyurek Y, van Galen M, van Dam H, van Ommen G-JB, den Dunnen JT, Zantema A et al. : Genome-wide assessment of differential roles for p300 and CBP in transcription regulation. Nucleic Acids Res 2010, 38:5396–5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santer FR, Höschele PPS, Oh SJ, Erb HHH, Bouchal J, Cavarretta IT, Parson W, Meyers DJ, Cole PA, Culig Z: Inhibition of the acetyltransferases p300 and CBP reveals a targetable function for p300 in the survival and invasion pathways of prostate cancer cell lines. Mol Cancer Ther 2011, 10:1644–1655. [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Jia W, Wang K, Si X, Zhu S, Duan T, Kang J: Distinct roles for CBP and p300 on the RA-mediated expression of the meiosis commitment gene Stra8 in mouse embryonic stem cells. PLoS ONE 2013, 8:e66076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas PD, Kahn M: Kat3 coactivators in somatic stem cells and cancer stem cells: biological roles, evolution, and pharmacologic manipulation. Cell Biol Toxicol 2016, 32:61–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arrowsmith CH, Audia JE, Austin C, Baell J, Bennett J, Blagg J, Bountra C, Brennan PE, Brown PJ, Bunnage ME et al. : The promise and peril of chemical probes. Nat Chem Biol 2015, 11:536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blagg J, Workman P: Choose and use your chemical probe wisely to explore cancer biology. Cancer Cell 2017, 32:9–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Copeland RA, Boriack-Sjodin PA: The elements of translational chemical biology. Cell Chem Biol 2018, 25:128–134. [DOI] [PubMed] [Google Scholar]
- 16.Fiorentino F, Mai A, Rotili D: Lysine acetyltransferase inhibitors: structure–activity relationships and potential therapeutic implications. Fut Med Chem 2018, 10:1067–1091. [DOI] [PubMed] [Google Scholar]
- 17.Andre R, Angela NK: Epigenetic modulation using small molecules — targeting histone acetyltransferases in disease. Curr Med Chem 2017, 24:4121–4150. [DOI] [PubMed] [Google Scholar]
- 18.Baell JB, Holloway GA: New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 2010, 53:2719–2740. [DOI] [PubMed] [Google Scholar]
- 19.Nelson KM, Dahlin JL, Bisson J, Graham J, Pauli GF, Walters MA: The essential medicinal chemistry of curcumin: miniperspective. J Med Chem 2017, 60:1620–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H et al. : Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem Biol 2010, 17:471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shrimp JH, Sorum AW, Garlick JM, Guasch L, Nicklaus MC, Meier JL: Characterizing the covalent targets of a small molecule inhibitor of the lysine acetyltransferase P300. ACS Med Chem Lett 2016, 7:151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahlin JL, Nelson KM, Strasser JM, Barsyte-Lovejoy D, Szewczyk MM, Organ S, Cuellar M, Singh G, Shrimp JH, Nguyen N et al. : Assay interference and off-target liabilities of reported histone acetyltransferase inhibitors. Nat Commun 2017, 8:1527.•• Using a suite of assays, the authors demonstrate that most commonly used HAT inhibitors have features that should exclude their use as probes. This includes demonstrating compound reactivity, aggregation, and binding to off targets at concentrations commonly used cellular studies.
- 23.Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, Hansen TM, Risi RM, Frey R, Manaves V et al. : Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 2017, 550:128.• The authors have developed a highly potent and drug-like inhibitor of the CBP/p300 HAT. A-485 is the first highly potent and specific inhibitor of the CBP/p300 HAT and is suitable for usein vivo.
- 24.Michaelides MR, Kluge A, Patane M, Van Drie JH, Wang C, Hansen TM, Risi RM, Mantei R, Hertel C, Karukurichi K et al. : Discovery of spiro oxazolidinediones as selective, orally bioavailable inhibitors of p300/CBP histone acetyltransferases. ACS Med Chem Lett 2018, 9:28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pérez-Salvia M, Esteller M: Bromodomain inhibitors and cancer therapy: from structures to applications. Epigenetics 2017, 12:323–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moustakim M, Clark PGK, Hay DA, Dixon DJ, Brennan PE: Chemical probes and inhibitors of bromodomains outside the BET family. Medchemcomm 2016, 7:2246–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero FA, Taylor AM, Crawford TD, Tsui V, Côté A, Magnuson S: Disrupting acetyl-lysine recognition: progress in the development of bromodomain inhibitors. J Med Chem 2016, 59:1271–1298. [DOI] [PubMed] [Google Scholar]
- 28.Andrieu G, Belkina AC, Denis GV: Clinical trials for BET inhibitors run ahead of the science. Drug Discov Today: Technol 2016, 19:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crawford TD, Romero FA, Lai KW, Tsui V, Taylor AM, de Leon Boenig G, Noland CL, Murray J, Ly J, Choo EF et al. : Discovery of a potent and selective in vivo probe (GNE-272) for the bromodomains of CBP/EP300. J Med Chem 2016, 59:10549–10563. [DOI] [PubMed] [Google Scholar]
- 30.Bronner SM, Murray J, Romero FA, Lai KW, Tsui V, Cyr P, Beresini MH, de leon Boenig G, Chen Z, Choo EF et al. : A unique approach to design potent and selective cyclic adenosine monophosphate response element binding protein, binding protein (CBP) inhibitors. J Med Chem 2017, 60:10151–10171. [DOI] [PubMed] [Google Scholar]
- 31.Jin L, Garcia J, Chan E, de la Cruz C, Segal E, Merchant M, Kharbanda S, Raisner R, Haverty PM, Modrusan Z et al. : Therapeutic targeting of the CBP/p300 bromodomain blocks the growth of castration-resistant prostate cancer. Cancer Res 2017, 77:5564–5575. [DOI] [PubMed] [Google Scholar]
- 32.Romero FA, Murray J, Lai KW, Tsui V, Albrecht BK, An L, Beresini MH, de Leon Boenig G, Bronner SM, Chan EW et al. : GNE-781, a highly advanced potent and selective bromodomain inhibitor of cyclic adenosine monophosphate response element binding protein, binding protein (CBP). J Med Chem 2017, 60:9162–9183.• Using structure-based design, the authors have improved the moderate potency and selectivity of a previously identified CBP/p300 bromodomain inhibitor. The optimized inhibitors have exceptional potency and are among the most selective bromodomain inhibitors reported.
- 33.Lai KW, Romero FA, Tsui V, Beresini MH, de Leon Boenig G, Bronner SM, Chen K, Chen Z, Choo EF, Crawford TD et al. : Design and synthesis of a biaryl series as inhibitors for the bromodomains of CBP/P300. Bioorg Med Chem Lett 2018, 28:15–23. [DOI] [PubMed] [Google Scholar]
- 34.Pegg N, Brooks N, Worthington J, Young B, Prosser A, Lane J, Taddei D, Brown R, Harbottle G, Shannon J et al. : Characterisation of CCS1477: a novel small molecule inhibitor of p300/CBP for the treatment of castration resistant prostate cancer. J Clin Oncol 2017, 35 11590–11590. [Google Scholar]
- 35.Brooks N, Pegg N, Worthington J, Young B, Prosser A, Lane J, Taddei D, Schiewer MJ, Gordon N, Knudsen KE: A novel small molecule inhibitor of p300/CBP for the treatment of castration-resistant prostate cancer: preclinical evaluation. J Clin Oncol 2017, 35 168–168. [Google Scholar]
- 36.Brooks N, Pegg N, Worthington J, Young B, Prosser A, Lane J, Taddei D, Schiewer M, deLeeuw R, McCann J et al. : Abstract 1575: novel small molecule inhibitors of p300/CBP down-regulate AR and c-Myc for the treatment of castrate resistant prostate cancer. Cancer Res 2017, 77 1575–1575.28087598 [Google Scholar]
- 37.Zucconi BE, Cole PA: Allosteric regulation of epigenetic modifying enzymes. Curr Opin Chem Biol 2017, 39:109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zucconi BE, Luef B, Xu W, Henry RA, Nodelman IM, Bowman GD, Andrews AJ, Cole PA: Modulation of p300/CBP acetylation of nucleosomes by bromodomain ligand I-CBP112. Biochemistry 2016, 55:3727–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shrimp JH, Grose C, Widmeyer SRT, Thorpe AL, Jadhav A, Meier JL: Chemical control of a CRISPR–Cas9 acetyltransferase. ACS Chem Biol 2018, 13:455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thakur JK, Yadav A, Yadav G: Molecular recognition by the KIX domain and its role in gene regulation. Nucleic Acids Res 2014, 42:2112–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Law SM, Gagnon JK, Mapp AK, Brooks CL: Prepaying the entropic cost for allosteric regulation in KIX. Proc Natl Acad Sci 2014, 111:12067–12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gee CT, Arntson KE, Urick AK, Mishra NK, Hawk LML, Wisniewski AJ, Pomerantz WCK: Protein-observed 19F-NMR for fragment screening, affinity quantification and druggability assessment. Nat Protoc 2016, 11:1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arntson KE, Pomerantz WCK: Protein-observed fluorine NMR: a bioorthogonal approach for small molecule discovery. J Med Chem 2016, 59:5158–5171. [DOI] [PubMed] [Google Scholar]
- 44.Pomerantz WC, Wang N, Lipinski AK, Wang R, Cierpicki T, Mapp AK: Profiling the dynamic interfaces of fluorinated transcription complexes for ligand discovery and characterization. ACS Chem Biol 2012, 7:1345–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gee CT, Arntson KE, Koleski EJ, Staebell RL, Pomerantz WCK: Dual labeling of the CBP/p300 KIX domain for 19F NMR leads to identification of a new small molecule binding site. ChemBioChem 2018, 19:963–969.•• The authors have advanced the use of PrOF NMR for screening KIX domain inhibitors by using dually labeled protein. Using this technique, the authors identified a new small molecule binding site on the CBP KIX domain that can allosterically modulate transcription factor binding.
- 46.Best JL, Amezcua CA, Mayr B, Flechner L, Murawsky CM, Emerson B, Zor T, Gardner KH, Montminy M: Identification of small-molecule antagonists that inhibit an activator: coactivator interaction. Proc Nat Acad Sci U S A 2004, 101:17622–17627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uttarkar S, Dukare S, Bopp B, Goblirsch M, Jose J, Klempnauer K-H: Naphthol AS-E phosphate inhibits the activity of the transcription factor Myb by blocking the interaction with the KIX domain of the coactivator p300. Mol Cancer Ther 2015, 14:1276–1285. [DOI] [PubMed] [Google Scholar]
- 48.Erlanson DA, Wells JA, Braisted AC: Tethering: fragment-based drug discovery. Ann Rev Biophys Biomol Struct 2004, 33:199–223. [DOI] [PubMed] [Google Scholar]
- 49.Wang N, Majmudar CY, Pomerantz WC, Gagnon JK, Sadowsky JD, Meagher JL, Johnson TK, Stuckey JA, Brooks CL, Wells JA et al. : Ordering a dynamic protein via a small-molecule stabilizer. J Am Chem Soc 2013, 135:3363–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lodge JM, Justin Rettenmaier T, Wells JA, Pomerantz WC, Mapp AK: FP tethering: a screening technique to rapidly identify compounds that disrupt protein–protein interactions. MedChemComm 2014, 5:370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang N, Lodge JM, Fierke CA, Mapp AK: Dissecting allosteric effects of activator–coactivator complexes using a covalent small molecule ligand. Proc Natl Acad Sci 2014, 111:12061–12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramaswamy K, Forbes L, Minuesa G, Gindin T, Brown F, Kharas MG, Krivtsov AV, Armstrong SA, Still E, de Stanchina E et al. : Peptidomimetic blockade of MYB in acute myeloid leukemia. Nat Commun 2018, 9:110.• A peptidomimetic of the Myb transcriptional activation domain was shown bind to the CBP KIX domain with affinity comparable to the native transactivation domain and to have excellentin vivo activity including downregulation of Myb-dependent genes. MYBMIM is the first reversible inhibitor of the Myb binding site that does not have other known targets.
- 53.Yadav A, Thakur JK, Yadav G: KIXBASE: A comprehensive web resource for identification and exploration of KIX domains. Sci Rep 2017, 7:14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Guzman RN, Liu HY, Martinez-Yamout M, Dyson HJ, Wright PE: Solution structure of the TAZ2 (CH3) domain of the transcriptional adaptor protein CBP11. Edited by Summers MF. J Mol Biol 2000, 303:243–253. [DOI] [PubMed] [Google Scholar]
- 55.Wei J, Yang Y, Lu M, Lei Y, Xu L, Jiang Z, Xu X, Guo X, Zhang X, Sun H et al. : Recent advances in the discovery of HIF-1α-p300/ cbp inhibitors as anti-cancer agents. Mini Rev Med Chem 2018, 18:296–309. [DOI] [PubMed] [Google Scholar]
- 56.Kung AL, Zabludoff SD, France DS, Freedman SJ, Tanner EA, Vieira A, Cornell-Kennon S, Lee J, Wang B, Wang J et al. : Small molecule blockade of transcriptional coactivation of the hypoxia-inducible factor pathway. Cancer Cell 2004, 6:33–43. [DOI] [PubMed] [Google Scholar]
- 57.Cook KM, Hilton ST, Mecinović J, Motherwell WB, Figg WD, Schofield CJ: Epidithiodiketopiperazines block the interaction between hypoxia-inducible factor-1a (HIF-1α) and p300 by a zinc ejection mechanism. J Biol Chem 2009, 284:26831–26838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cherblanc FL, Chapman KL, Reid J, Borg AJ, Sundriyal S, Alcazar-Fuoli L, Bignell E, Demetriades M, Schofield CJ, DiMaggio PA et al. : On the histone lysine methyltransferase activity of fungal metabolite chaetocin. J Med Chem 2013, 56:8616–8625. [DOI] [PubMed] [Google Scholar]
- 59.Tibodeau JD, Benson LM, Isham CR, Owen WG, Bible KC: The anticancer agent chaetocin is a competitive substrate and inhibitor of thioredoxin reductase. Antioxid Redox Signal 2009, 11:1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi Q, Yin S, Kaluz S, Ni N, Devi NS, Mun J, Wang D, Damera K, Chen W, Burroughs S et al. : Binding model for the interaction of anticancer arylsulfonamides with the p300 transcription cofactor. ACS Med Chem Lett 2012, 3:620–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yin S, Kaluz S, Devi NS, Jabbar AA, de Noronha RG, Mun J, Zhang Z, Boreddy PR, Wang W, Wang Z et al. : Arylsulfonamide KCN1 inhibits in vivo glioma growth and interferes with HIF signaling by disrupting HIF-1α interaction with co-factors p300/CBP. Clin Cancer Rese: Off J Am Assoc Cancer Res 2012, 18:6623–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lao BB, Grishagin I, Mesallati H, Brewer TF, Olenyuk BZ, Arora PS: In vivo modulation of hypoxia-inducible signaling by topographical helix mimetics. Proc Natl Acad Sci 2014, 111:7531–7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kushal S, Lao BB, Henchey LK, Dubey R, Mesallati H, Traaseth NJ, Olenyuk BZ, Arora PS: Protein domain mimetics as in vivo modulators of hypoxia-inducible factor signaling. Proc Natl Acad Sci 2013, 110:15602–15607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ferguson JH, De Los Santos Z, Devi SN, Kaluz S, Van Meir EG, Zingales SK, Wang B: Design and synthesis of benzopyran-based inhibitors of the hypoxia-inducible factor-1 pathway with improved water solubility. J Enzyme Inhib Med Chem 2017, 32:992–1001.• The poor aqueous solubility of the TAZ1/HIF-1α inhibitor KCN1 can limit its use. In this paper, the authors developed a KCN1 analogue that is soluble in water at concentrations above 100 μM and has comparable potency to KCN1.
- 65.De Guzman RN, Wojciak JM, Martinez-Yamout MA, Dyson HJ, Wright PE: CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding. Biochemistry 2005, 44:490–497. [DOI] [PubMed] [Google Scholar]
- 66.Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, Moon RT, Teo J-L, Oh SW, Kim HY et al. : A small molecule inhibitor of β-catenin/cyclic AMP response element-binding protein transcription. Proc Natl Acad Sci U S A 2004, 101:12682–12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ko AH, Chiorean EG, Kwak EL, Lenz H-J, Nadler PI, Wood DL, Fujimori M, Inada T, Kouji H, McWilliams RR: Final results of a phase Ib dose-escalation study of PRI-724, a CBP/beta-catenin modulator, plus gemcitabine (GEM) in patients with advanced pancreatic adenocarcinoma (APC) as second-line therapy after FOLFIRINOX or FOLFOX. J Clin Oncol 2016, 34 e15721–e15721. [Google Scholar]
- 68.El-Khoueiry AB, Ning Y, Yang D, Cole S, Kahn M, Zoghbi M, Berg J, Fujimori M, Inada T, Kouji H et al. : A phase I first-in-human study of PRI-724 in patients (pts) with advanced solid tumors. J Clin Oncol 2013, 31 2501–2501. [Google Scholar]
- 69.Higuchi Y, Nguyen C, Yasuda S-Y, McMillan M, Hasegawa K, Kahn M: Specific direct small molecule p300/β-catenin antagonists maintain stem cell potency. Curr Mol Pharmacol 2016, 9:272–279.•• The authors have used a series of counter screens to identify inhibitors that are selective for the p300 NRID. YH249 and YH250 are the first small molecule probes that show selectivity for p300 over CBP.
- 70.Ma H, Nguyen C, Lee K-S, Kahn M: Differential roles for the coactivators CBP and p300 on TCF/β-catenin-mediated survivin gene expression. Oncogene 2005, 24:3619. [DOI] [PubMed] [Google Scholar]
- 71.Teo J-L, Ma H, Nguyen C, Lam C, Kahn M: Specific inhibition of CBP/β-catenin interaction rescues defects in neuronal differentiation caused by a presenilin-1 mutation. Proc Natl Acad Sci U S A 2005, 102:12171–12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hasegawa K, Yasuda S-y, Teo J-L, Nguyen C, McMillan M, Hsieh C-L, Suemori H, Nakatsuji N, Yamamoto M, Miyabayashi T et al. : Wnt signaling orchestration with a small molecule DYRK inhibitor provides long-term xeno-free human pluripotent cell expansion. Stem Cells Transl Med 2012, 1:18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Banerjee ER, Laflamme MA, Papayannopoulou T, Kahn M, Murry CE, Henderson WR Jr: Human embryonic stem cells differentiated to lung lineage-specific cells ameliorate pulmonary fibrosis in a xenograft transplant mouse model. PLoS ONE 2012, 7:e33165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schürmann M, Janning P, Ziegler S, Waldmann H: Small-molecule target engagement in cells. Cell Chem Biol 2016, 23:435–441 [DOI] [PubMed] [Google Scholar]