SUMMARY
Helminths, allergens, and certain protists induce type 2 immune responses, but the underlying mechanisms of immune activation remain poorly understood. In the small intestine, chemosensing by epithelial tuft cells results in activation of group 2 innate lymphoid cells (ILC2s), which subsequently drive increased tuft cell frequency. This feed-forward circuit is essential for intestinal remodeling and helminth clearance. ILC2 activation requires tuft cell-derived interleukin-25 (IL-25), but whether additional signals regulate the circuit is unclear. Here we show that tuft cells secrete cysteinyl leukotrienes (cysLTs) to rapidly activate type 2 immunity following chemosensing of helminth infection. CysLTs cooperate with IL-25 to activate ILC2s, and tuft cell-specific ablation of leukotriene synthesis attenuates type 2 immunity and delays helminth clearance. Conversely, cysLTs are dispensable for the tuft cell response induced by intestinal protists. Our findings identify an additional tuft cell effector function and suggest context-specific regulation of tuft-ILC2 circuits within the small intestine.
INTRODUCTION
Parasitic helminth infection remains a significant global health issue, but the earliest sensing and signaling events that initiate “type 2” anti-helminth immunity are still poorly defined. Uncovering these pathways could inform vaccine design and identify targets for drugs that manipulate type 2 immune responses. Recently, three groups identified tuft cells as key initiators of type 2 immunity in the small intestine (SI) (Gerbe et al., 2016; Howitt et al., 2016; von Moltke et al., 2016). Tuft cells are rare epithelial cells found at mucosal barriers throughout the body. SI tuft cells employ a chemosensing pathway to recognize helminths such as Nippostrongylus brasiliensis, Heligmosomoides polygyrus, and Trichinella spiralis, as well as certain species of Tritrichomonad protists. Upon ligand sensing, tuft cells signal to and activate group 2 innate lymphoid cells (ILC2s) in the underlying lamina propria to initiate inflammation. Tuft cells are the sole source of intestinal interleukin 25 (IL-25), which they express constitutively (von Moltke et al., 2016). IL-25 activates ILC2s to produce the hallmark type 2 cytokines IL-13, IL-5 and IL-9 that propagate the type 2 response. IL-13, in particular, is important for tissue remodeling and is required for helminth expulsion (Urban et al., 1998). Among its many cellular targets, IL-13 signals directly on undifferentiated epithelial cells to bias their differentiation towards the tuft and goblet cell fates, resulting in a feed-forward loop of ILC2 activation and tuft and goblet cell hyperplasia (hereafter referred to as the tuft-ILC2 circuit). Given the 3-4 day turnover of intestinal epithelium (Barker, 2013), activation of the tuft-ILC2 circuit rapidly remodels the SI to promote helminth expulsion.
While progress has been made in elucidating the specific signals that activate SI tuft cells (Lei et al., 2018; Nadjsombati et al., 2018; Schneider et al., 2018), less is known about how such signals are converted into an effector response. The chemosensing pathways expressed in tuft cells converge on the calcium-gated membrane cation channel TRPM5, and type 2 immune responses are profoundly delayed or absent in Trpm5-deficient mice colonized with helminths or Tritrichomonads (Howitt et al., 2016; Nadjsombati et al., 2018). By analogy to taste receptor cells (Chaudhari and Roper, 2010), sodium influx through TRPM5 is presumed to depolarize tuft cell membranes, but the effector functions mobilized downstream of TRPM5 to activate the tuft-ILC2 circuit remain unknown. TRPM5 may regulate the release of IL-25 (Luo et al., 2019), the only intestinal tuft cell effector molecule identified to date; however based on our understanding of ILC2 activation (discussed below) we predict that physiologic levels of IL-25 are not sufficient to fully activate ILC2 responses.
ILC2s are tissue resident cells widely distributed throughout the body, and they express a specific set of surface receptors that attunes them to signals produced in the local environment (Ricardo-Gonzalez et al., 2018). ILC2s of the SI are particularly responsive to IL-25 signaling due to their high expression of the IL-25 receptor (Schneider et al., 2018). ILC2s resemble T helper 2 (Th2) CD4+ T cells in their dependence on the lineage-defining transcription factor GATA3 and expression of hallmark cytokines IL-13 and IL-5. Indeed, the chromatin landscapes of ILC2s and effector Th2 cells are nearly identical, suggesting that the same transcription factors are likely to regulate cytokine production in both cell types (Shih et al., 2016; Van Dyken et al., 2016). During T cell activation, T cell receptor (TCR) signaling simultaneously induces the AP-1, NF-κB, and NFAT transcription factors that cooperatively drive effector gene expression. ILC2s are in part defined by their lack of a TCR, and we and others demonstrated that lung ILC2s must therefore integrate multiple signaling pathways to replicate TCR signaling and achieve optimal activation (Lund et al., 2017; von Moltke et al., 2017). In the lung, cysteinyl leukotrienes (cysLTs) represent an important signal due to their ability to induce nuclear translocation of NFAT, which synergizes with IL-33-induced AP-1 and NF-κB to yield maximal cytokine production. Whether cysLTs are required for immunity to intestinal helminths, and which cells might produce them in this context, has not been investigated.
Leukotrienes are lipid signaling molecules generated by the conversion of arachidonic acid to LTA4 via the enzyme 5-lipoxygenase (5-LO) (Haeggström and Funk, 2011) (Figure 1A). LTA4 is rapidly converted into LTB4 by the enzyme LTA4 hydrolase, or into LTC4 by LTC4 synthase. Following secretion, LTC4 is further converted into LTD4 and LTE4, together comprising the cysLTs. Whereas LTB4 is primarily a chemoattractant, cysLTs are inflammatory mediators, with LTC4 and LTD4 having well-defined roles in promoting allergic pathology in the airways (Peters-Golden and Henderson, 2007). Leukotrienes are not preformed, but rather are synthesized and secreted on demand within minutes. In many cases, leukotriene synthesis is initiated by intracellular calcium flux that activates cytosolic phospholipase A2 (Uozumi et al., 1997). Once outside the cell, LTC4 and LTD4 are rapidly converted to the more stable LTE4 (Keppler et al., 1989).
Figure 1. Cysteinyl leukotrienes are a non-redundant signal for intestinal ILC2 activation.
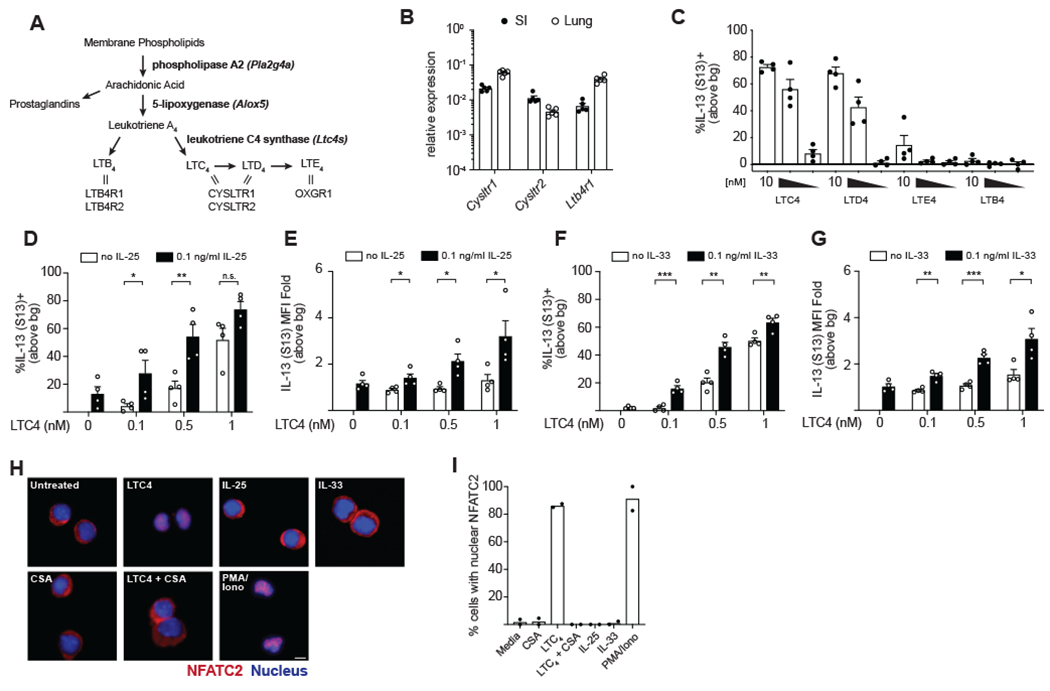
(A) Schematic of leukotriene synthesis. Enzymes in bold. Double lines represent ligand-receptor interactions. (B) Gene expression in ILC2s sorted from the SI lamina propria and lung. (C) Frequency of IL-13 (S13)+ SI ILC2s following 6 hour in vitro culture with the indicated leukotriene. Ten-fold dilutions from 10-0.1 nM are shown. (D,F) Frequency and (E,G) mean fluorescence intensity (MFI) of IL-13 (S13)+ SI ILC2s following 6 hour in vitro culture with the indicated combinations of LTC4 and IL-25 (D,E) or IL-33 (F,G). (H) SI ILC2s were treated for 30 min with 1 μM cyclosporin A (CSA) where indicated, followed by 90 min treatment with 100nM LTC4, 100ng/ml IL-25, 100ng/ml IL-33, or 30ng/ml PMA and 500ng/ml ionomycin. Cells were stained with anti-NFATC2 (red) and DAPI (blue). Scale bar: 20μm. (I) Quantification of cells with nuclear NFATC2. At least 30 cells were counted for each condition. In (B)-(G) each symbol represents an individual mouse pooled from two or more experiments. In (I) symbols are technical replicates representative of three independent experiments, bg, background. *p < 0.05, **p < 0.01, ***p < 0.001 by multiple t tests (D-G). n.s., not significant. Graphs depict mean + SEM. See also Figure S1.
Leukotriene synthesis is canonically thought to be restricted to hematopoietic cells, but tuft cells also express genes required for the synthesis of leukotrienes, including Pla2g4a, Alox5, and Ltc4s (Bezençon et al., 2008; Haber et al., 2017). Indeed, expression of these genes is one defining feature of a core tuft cell signature conserved across multiple tissues (Nadjsombati et al., 2018). We therefore hypothesized that tuft cells might generate leukotrienes to amplify type 2 inflammation in the SI.
RESULTS
Cysteinyl leukotrienes are a non-redundant signal for intestinal ILC2 activation
Leukotrienes drive ILC2 activation in the lung during allergy and helminth infection (Doherty et al., 2013; von Moltke et al., 2017), but less is known about their role in the SI. Given the tissue-specific imprinting of ILC2s (Ricardo-Gonzalez et al., 2018), we wanted to test if leukotrienes also regulate SI ILC2s. SI ILC2s express both the LTC4 and LTD4 receptors CYSLTR1 and CYSLTR2, similar to lung ILC2s (Figure 1B; gating strategies in Figures S1A–S1B). LTB4 binds to two receptors, the high-affinity LTB4R1 and lower-affinity LTB4R2. SI ILC2s also express Ltb4r1 (Figure 1B), whereas Ltb4r2 and the LTE4 receptor Oxgr1 are low or absent (data not shown). To confirm these findings functionally, we performed an in vitro activation assay using SI ILC2s sorted from the Il13Smart13 (S13) cytokine reporter mouse (Liang et al., 2011). In these mice, surface expression of non-signaling human CD4 allows for quantification of IL-13 protein expression by flow cytometry. LTC4 and LTD4 robustly activated SI ILC2s in a dose-dependent manner, in accordance with high expression of their receptors, whereas LTB4 and LTE4 had minimal effect in this assay (Figure 1C; gating in Fig S1C). LTC4-induced activation was abolished in the presence of the CYSLTR1-specific inhibitor montelukast (Figure S1D). In contrast, the CYSLTR2 inhibitor HAMI3379 only suppressed ILC2 activation by LTC4 at micromolar doses, likely reflecting inhibition of CYSLTR1 at high concentrations (Wunder et al., 2010). As in the lung, CYSLTR1-mediated sensing of LTC4 and/or LTD4 therefore appears to be the dominant mechanism of leukotriene-driven SI ILC2 activation.
CysLTs are known to cooperate with IL-33 to induce maximal lung ILC2 activation (Lund et al., 2017; von Moltke et al., 2017). Whereas IL-33 is the predominant innate cytokine found in the lung and lung ILC2s express the IL-33 receptor (Barlow et al., 2013), SI ILC2s are highly responsive to the cytokine IL-25, produced exclusively by tuft cells (von Moltke et al., 2016). We therefore hypothesized that cysLT signaling might also cooperate with IL-25 to drive ILC2 activation. To test this, we repeated in vitro stimulation of SI ILC2s, this time using sub-optimal doses of LTC4, IL-25, or both (Figures 1D–E and S1E). At these low concentrations, LTC4 or IL-25 alone minimally induced ILC2 activation. When LTC4 and IL-25 were used in combination, however, an additive effect was evident in both the frequency of responding cells and the amount of IL-13 expressed per cell. A similar effect was also observed with the combination of LTC4 and IL-33 (Figures 1F–G).
During lung ILC2 activation, cysLTs are non-redundant due to their ability to induce nuclear translocation of NFAT, which cooperates with IL-33-induced NF-κB and AP-1 (von Moltke et al., 2017). Specifically, signaling through CYSLTR1 induces a calcium flux that activates calcineurin, resulting in NFAT dephosphorylation and translocation to the nucleus (Lynch et al., 1999; von Moltke et al., 2017). IL-25 has also been shown to induce NF-κB and AP-1, but a role for NFAT induction has not been reported (Maezawa et al., 2006; Wong et al., 2005). Stimulation of SI ILC2s with LTC4 resulted in NFAT nuclear translocation, which was inhibited by the calcineurin inhibitor cyclosporin A (Figures 1H–I). Conversely, stimulation with IL-25 or IL-33 alone had no effect on NFAT localization. Thus, cysLTs are a non-redundant signal for SI ILC2 activation and cooperate with IL-25 and/or IL-33 to maximize effector cytokine production.
ILC2 homeostasis in the proximal small intestine is leukotriene-independent and minimally requires IL-25 and IL-33
We recently identified tuft cells as the sole source of IL-25 in the SI and found they express this cytokine constitutively (von Moltke et al., 2016). In that study we also found that ~20% of SI ILC2s from uninfected mice expressed IL-13, which was absent in IL-25-deficient mice. We therefore concluded that IL-25 is constitutively secreted and signals to SI ILC2s homeostatically. Subsequently, tuft cells were shown to be activated by Tritrichomonad protists that are common across vivariums (Howitt et al., 2016; Nadjsombati et al., 2018; Schneider et al., 2018); we determined in retrospect that the mice used in our original study were likely colonized by these protists as well. This finding prompted us to reevaluate the role of homeostatic IL-25 signaling using naïve, Tritrichomonad-free mice.
We compared IL-13 reporter expression in cells isolated from the proximal SI of naïve wildtype, Il25−/−, St2−/− and Alox5−/− mice. St2 constitutes part of the IL-33 receptor and is required for IL-33 signaling. Alox5 encodes 5-lipoxygenase, the enzyme that catalyzes the first step in all leukotriene synthesis (Figure 1A). We also included Trpm5−/− mice to assess the role of the TRPM5-dependent tuft cell chemosensing pathway during homeostasis. Defining ILC2s in the SI lamina propria as CD45+;Lineage(Lin)−; KLRG1+ cells (gating in Figure S1A), we found that IL-13 expression was generally low, especially when compared to previous measurements made in Tritrichomonad-colonized mice (Figure S2A) (von Moltke et al., 2016). IL-13 expression was comparable across genotypes, though some variability between mice was evident. We did note, however, that KLRG1 staining intensity appeared reduced in Il25−/− and Trpm5−/− mice. We therefore reexamined IL-13 expression in ILC2s identified as CD45+;Lin−;IL-17RB+. Again we found minimal IL-13 expression across all groups, but confirmed that KLRG1 MFI was indeed decreased in Il25−/− and Trpm5−/− mice (Figures 2A–C; gating in Fig S2B), suggesting some tuft cell-dependent regulation of ILC2s in the absence of any currently known tuft cell agonists.
Figure 2. ILC2 homeostasis in the proximal small intestine is leukotriene-independent and minimally requires IL-25 and IL-33.
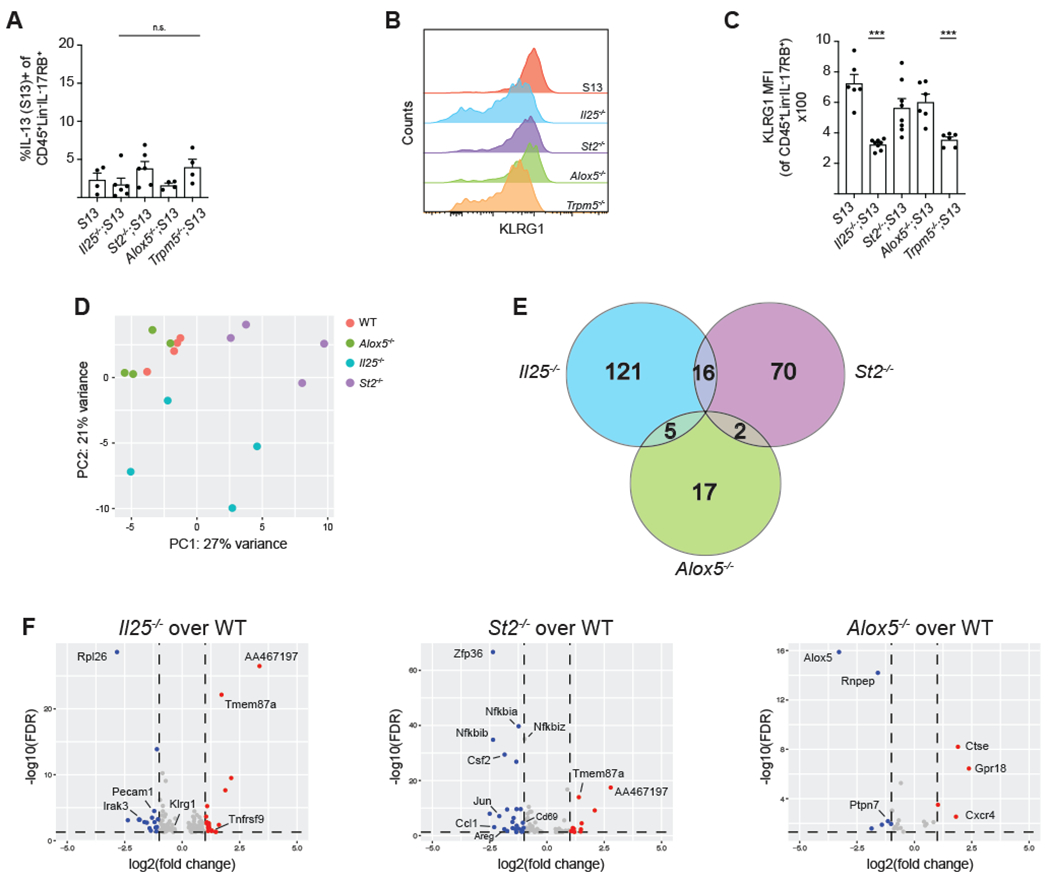
(A) Quantification of IL-13 (S13) expression by ILC2s (CD45+;Lineage−;IL-17RB+) in the proximal (first 5cm) SI of naïve mice. (B) Mean fluorescence intensity (MFI) of KLRG1 expression by ILC2s. (C) Quantification of MFI in (B). (D-F) mRNA sequencing of ILC2s of the indicated genotype sorted from the proximal SI of naïve mice. (D) PCA of top 500 differentially expressed genes (DEG). (E) Venn diagram depicting DEGs for each genotype as compared to wildtype (FDR <.05, row mean >10). (F) Volcano plots depicting DEGs as compared to wildtype (FDR <.05, row mean >10). Red = log2 fold change > 1, blue = log2 fold change < 1. In (A-D) each symbol represents an individual mouse pooled from two or more experiments. Samples in D were analyzed in one sequencing run. *p < 0.05, **p < 0.01, ***p < 0.001 by one way ANOVA (A, C) with comparison to S13. n.s., not significant. Graphs depict mean + SEM. See also Figure S2 and Tables S1–4.
To get a more complete picture of ILC2 activation states in naïve mice, we sorted CD45+;Lin−;IL-17RB+ cells from the proximal SI lamina propria of wildtype, Alox5−/−, Il25−/−, and St2−/− Tritrichomonad-free mice and performed bulk RNA sequencing (Table S1). We again found no evidence of a homeostatic role for leukotrienes: wildtype and Alox5−/− mice clustered together by PCA and only 24 genes, including Alox5−/−, were identified as differentially expressed (FDR <.05, row mean > 10) (Figures 2D–E). We did see more differences in the Il25- (142 genes) and St2- (88 genes) deficient ILC2s, but here too the homeostatic role of these cytokines was limited (Tables S2–4). In particular, the fold-change in gene expression was generally small, although we cannot rule out more significant post-transcriptional changes (Figure 2F). Klrg1 mRNA, for example, was downregulated only 0.8 fold in Il25−/− ILC2s, but clearly decreased at the protein level (Fig 2C). Also notable was the lack of effector cytokines on the list, with the exception of Areg, which was downregulated only in St2−/− ILC2s. Among the genes that were differentially expressed, we were surprised to see very little overlap between Il25- and St2-deficient ILC2s, despite related signaling pathways downstream of ST2 and the IL-25 receptor. Several NF-κB and AP-1 related genes were uniquely downregulated in the absence of St2, suggesting homeostatic and/or developmental defects that would be interesting to explore further. Overall, however, these results demonstrate that at homeostasis, leukotrienes do not modulate ILC2 gene expression in the proximal SI whereas IL-25 contributes minimally, but detectably.
Cysteinyl leukotrienes drive rapid ILC2 activation following helminth infection
ILC2s are tissue-resident cells that rapidly respond to local cues to initiate type 2 inflammation. To probe the earliest signaling events driving SI ILC2 activation we used helminth infection. H. polygyrus naturally infects mice through the oral route and transits directly to the proximal SI to establish infection, allowing us to deliver activating signals to the SI in a precisely timed manner. Sixteen hours after oral gavage with H. polygyrus L3 larvae, ILC2s in the proximal SI exhibited upregulated IL-13 expression (Figures 3A–B). This response was abolished in TRPM5-deficient and IL-25-deficient mice, placing tuft cell sensing of H. polygyrus upstream of ILC2 activation, as previously described (Howitt et al., 2016; von Moltke et al., 2016). Infection did not alter tuft cell Il25 expression at this time (Figure 3C; gating in Figure S3A). While IL-33 elicited in response to parasite damage has previously been shown to drive type 2 immunity in the SI (Molofsky et al., 2015), IL-33 signaling was not required for this initial tuft cell-dependent anti-helminth response (Figure 3B).
Figure 3. Cysteinyl leukotrienes drive rapid ILC2 activation following helminth infection.
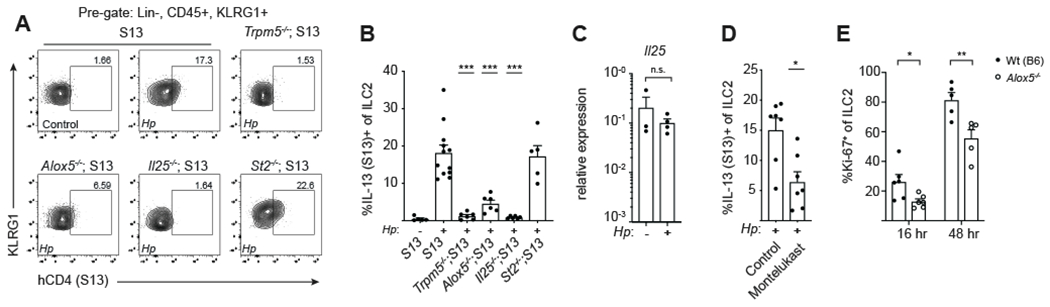
(A) Flow cytometry for IL-13 (S13) expression by ILC2s in the proximal (first 10cm) SI 16 hours after infection with H. polygyrus (H.p.). (B) Quantification of IL-13 (S13) expression in (A). (C) Il25 mRNA expression in tuft cells sorted from the proximal SI of naïve Wt(B6) mice and mice infected with H.p. for 16 hours. (D-E) Analysis of ILC2s from the proximal SI. (D) IL-13 (S13)+ ILC2s in mice treated with montelukast (10mg/kg) 60 min prior to 16 hours infection with H.p. (E) ILC2 Ki-67 expression at the indicated time points following infection with H.p. In (B)-(E) each symbol represents an individual mouse pooled from two or more experiments. *p < 0.05, **p < 0.01, ***p < 0.001 by one way ANOVA (B) with comparison to infected S13, by Mann-Whitney (C-D), or by multiple t tests (E). n.s., not significant. Graphs depict mean + SEM. See also Figure S3.
We found that Alox5−/− mice also had impaired ILC2 activation at this early time point, indicating that leukotrienes are required for ILC2 activation in vivo (Figure 3B). Consistent with in vitro findings, administration of the CYSLTR1 antagonist montelukast resulted in a similar defect in ILC2 activation, demonstrating that LTC4 and/or LTD4 serve a non-redundant function (Figure 3D). While there was no difference in the number of lamina propria ILC2s between wildtype and Alox5−/− mice at steady state (Figure S3B), we observed reduced proliferation of ILC2s early during H. polygyrus infection in Alox5−/− mice (Figure 3E). We were unable to isolate viable cells from infected tissue at later time points, so we examined mesenteric lymph nodes as a proxy. After four days of H. polygyrus infection, ILC2 number and activation were marginally increased compared to uninfected mice, whereas the response during N. brasiliensis infection was much more pronounced (Figures S3C–F). In both cases, ILC2 number and activation were decreased in Alox5−/− mice. Collectively, these results demonstrate that cysLTs are rapidly induced in the SI following helminth sensing by tuft cells and are required for optimal ILC2 activation.
Tuft-ILC2 circuit activation and worm clearance are delayed in the absence of cysteinyl leukotrienes
ILC2 activation following helminth infection is regulated by tuft cells through the feed-forward tuft-ILC2 circuit (von Moltke et al., 2016). As ILC2 activation was impaired in the initial response to H. polygyrus infection in Alox5−/− mice, we predicted that induction of the tuft-ILC2 circuit itself would also be disrupted. At homeostasis there was no difference in the number of tuft cells between wildtype and Alox5−/− mice, consistent with similar basal ILC2 activity (Figures 4A–B). Four days after H. polygyrus infection, however, wildtype mice developed tuft cell hyperplasia in the proximal SI, a response that was absent in Alox5−/− mice. Impaired tuft cell hyperplasia was also evident later during infection (Figure 4C). This response required signaling through CYSLTR1, further implicating LTC4 and LTD4 as the relevant mediators; however, we cannot rule out non-redundant contributions of other 5-LO-dependent leukotrienes (e.g. LTB4 from tuft cells or other lipids generated by transcellular biosynthesis) (Figure 4D). IL-25, but not IL-33, signaling was required for the early tuft cell response (Figure 4E).
Figure 4. Tuft-ILC2 circuit activation and helminth clearance are delayed in the absence of cysteinyl leukotrienes.
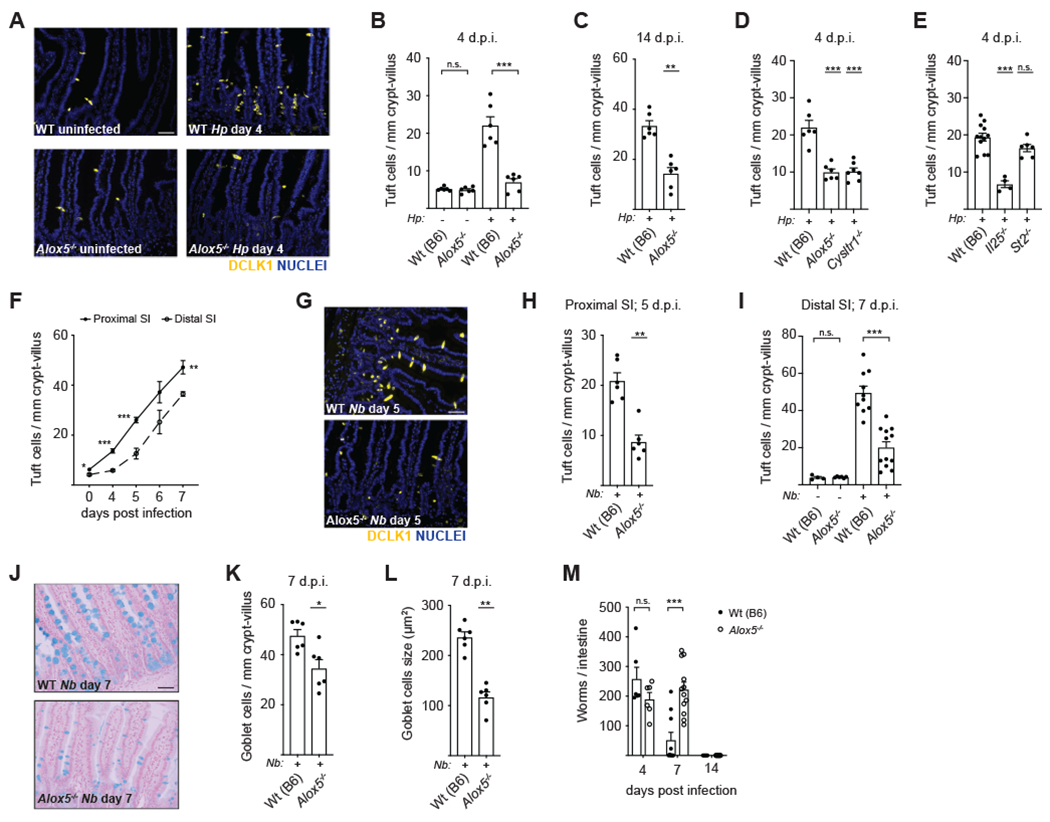
(A) Tuft cell frequency in the proximal (first 10cm) SI of mice infected with H. polygyrus (H.p.) for 4 days. DCLK1 (yellow) marks tuft cells. DAPI (blue) marks nuclei. Scale bar: 50μm. (B) Quantification of tuft cells in (A). (C) Tuft cell frequency in the proximal SI after 14 days of H.p. infection. (D-E) Tuft cell frequency in the proximal SI of the indicated genotypes after 4 days of H.p. infection. (F) Tuft cell frequency in the proximal and distal (last 10cm) SI of wildtype mice at the indicated time points post-N. brasiliensis (N.b.) infection. (G) Representative images of proximal SI on day 5 post-N. b. infection. Scale bar: 50μm. (H) Quantification of tuft cells in (G). (I) Tuft cell frequency in the distal SI after 7 days of N.b. infection. (J) Goblet cells identified in the jejunum (10-20cm from stomach) by alcian blue staining 7 days after infection with N.b. Representative images are shown. Scale bar: 50μm. (K-L) Quantification of goblet cell (K) number and (L) size in (J). (M) Worm burden across the entire SI at the indicated time points post-N.b. infection. In (B)-(E), (H)-(I), and (K)-(M) each symbol represents an individual mouse pooled from two or more experiments. In (F) each symbol represents the average of five mice pooled from two experiments, d.p.i., days post infection. *p < 0.05, **p < 0.01, ***p < 0.001 by Mann-Whitney (C, H, K-L), by multiple t tests (B, F, I, M), or by one way ANOVA (D-E) with comparison to Wt(B6). n.s., not significant. Graphs depict mean + SEM. See also Figure S4.
H. polygyrus establishes chronic infection in mice, making it a poor model for assessing the role of type 2 responses in worm clearance. To gauge whether leukotriene signaling contributes to restriction of helminths, we turned to N. brasiliensis, a rat-adapted helminth that causes a self-limiting infection in mice. In this model N. brasiliensis L3 larvae are injected subcutaneously, briefly transit through the lung, and reach the proximal SI around day 2-3 post-infection where they mature into adult worms. Accordingly, we first detected tuft cell hyperplasia in the proximal SI, with the distal SI displaying delayed kinetics in comparison (Figure 4F). At day 5 post-infection tuft cell numbers were noticeably increased in the proximal SI of N. brasiliensis-infected wildtype mice. As with H. polygyrus, Alox5−/− mice failed to induce tuft cell hyperplasia both at this early time point in the proximal SI (Figures 4G–H) and in the distal SI 7 days after infection (Fig 4I). Results were replicated in littermate controls, indicating that microbiome differences do not mediate observed differences (Figure S4A).
Intestinal organoids are three-dimensional cultures that contain all intestinal epithelial cell types and are commonly used to study epithelial biology in vitro (Sato et al., 2009). Using this system, we found that Alox5−/− intestinal stem cells have no intrinsic defect in their ability to differentiate into tuft cells following IL-13 stimulation (Figure S4B) and that cysLTs do not induce tuft cell expansion directly (Figure S4C). Further, the infection-induced increase in intestinal epithelial turnover, as assessed by in vivo BrdU pulse-chase, was marginally increased in Alox5−/− mice, confirming that the defect in tuft cell hyperplasia is not caused by a general delay in epithelial differentiation (Figure S4D). Arachidonic acid metabolism generates leukotrienes but can also produce prostaglandins, which have previously been implicated in anti-helminth immunity (Tait Wojno et al., 2015). To rule out a contribution of arachidonic acid being shunted into the prostaglandin pathway in the absence of 5-LO (Figure 1A), we treated leukotriene-deficient mice with ibuprofen to inhibit the cyclooxygenase enzymes that mediate prostaglandin synthesis; however, this had no effect on tuft cell numbers (Figure S4E).
Intestinal remodeling and increased goblet cell mucus secretion constitute part of the “weep and sweep” response that develops to promote worm expulsion. After 7 days of N. brasiliensis infection, Alox5−/− mice had fewer goblet cells than wildtype mice and these were smaller in size, indicating reduced mucus production (Figures 4J–L). The failure of Alox5−/− mice to efficiently induce the tuft-ILC2 circuit also resulted in sustained worm burdens at day 7, a time when wildtype mice begin to clear infection (Figure 4M). Despite this defect, both wildtype and Alox5−/− mice cleared infection by day 14, indicating that additional leukotriene-independent mechanisms of control exist. Nonetheless, these data collectively suggest a model in which cysLTs are rapidly induced in the SI following tuft cell sensing of helminths, and that this signal, along with IL-25, drives ILC2 activation to kickstart the tuft-ILC2 feed-forward circuit.
Tuft cells synthesize cysteinyl leukotrienes
ILC2 activation following helminth infection requires IL-25, produced exclusively by tuft cells in the SI (von Moltke et al., 2016). Accordingly, IL-25 deficiency impairs worm clearance following helminth infection (Fallon et al., 2006). When compared head-to-head, we found the clearance defect is more severe in tuft cell-deficient Pou2f3−/− mice than Il25−/− mice (Figure 5A), suggesting the existence of additional tuft cell effector functions. As published tuft cell transcriptomes include enzymes required for the synthesis of cysLTs (Bezençon et al., 2008; Haber et al., 2017; Nadjsombati et al., 2018), we hypothesized that tuft cells themselves might secrete leukotrienes.
Figure 5. Tuft cells synthesize cysteinyl leukotrienes.
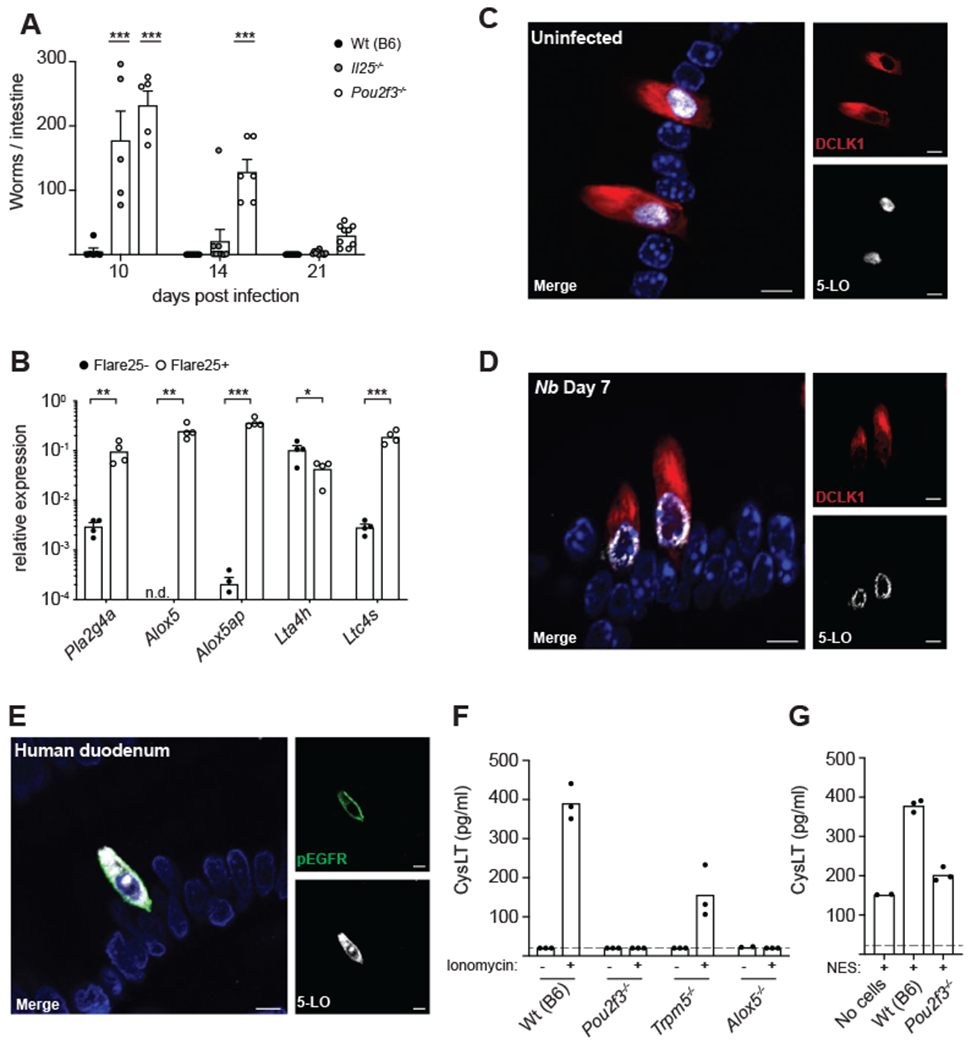
(A) Mice were infected with N. brasiliensis (N.b.) and SI worm burden determined at the indicated time points. (B) Gene expression in tuft cells (Flare25+) and non-tuft epithelial cells (Flare25+) sorted from the entire SI. (C) Colocalization of 5-LO (white) and DCLK1 (red) in the proximal (first 10cm) SI of a naïve mouse. DAPI (blue) marks nuclei. Representative images are shown. Scale bars: 5μm. (D) Example of 5-LO localization in tuft cells from the proximal SI of a mouse infected with N.b. for 7 days. Scale bars: 5μm. (E) Colocalization of 5-LO (white) and pEGFR (green) in human duodenum. Images are representative of three different patient samples. Scale bars: 5μm. (F-G) Cysteinyl leukotriene (cysLT) production in supernatants of intestinal monolayer cultures derived from mice of the indicated genotype following stimulation with 1μg/ml ionomycin (F) or 100μl NES (G) for 30 minutes. Dashed line represents limit of detection. In (A)-(B) each symbol represents an individual mouse from two or more pooled experiments. In (F)-(G) each symbol is a technical replicate and representative of three or more experiments, n.d., not detected. *p < 0.05, **p < 0.01, ***p < 0.001 by two way ANOVA (A) with comparison to Wt(B6) or by multiple t tests (B). n.s., not significant. Graphs depict mean + SEM. See also Figure S5.
To confirm previous transcriptomic data, we sorted tuft cells and non-tuft epithelium from the SI using the Flare25 reporter mouse and performed qPCR (Figure 5B; gating in Figure S5A). Tuft cells highly and specifically expressed Alox5 and Alox5ap, encoding 5-lipoxygenase (5-LO) and 5-lipoxygenase activating protein (FLAP), respectively, core components of the leukotriene synthesis pathway. Tuft cells additionally expressed Pla2g4a, a calcium-dependent phospholipase that liberates arachidonic acid from cell membranes; Ltc4s, required for the synthesis of all cysLTs; and Lta4h, the enzyme that generates LTB4. We confirmed expression of 5-LO at the protein level by microscopy (Figure 5C). 5-LO signal colocalized with the tuft cell marker DCLK1 and was not detected in non-tuft epithelial cells. Virtually all tuft cells had detectable 5-LO, regardless of infection status (Figures S5B). Consistent with previous studies of 5-LO (Haeggström and Funk, 2011), we found the protein localized to both the nucleus and the cytosol in naïve mice. We also observed instances of 5-LO translocating to the nuclear membrane in tuft cells of infected mice (Figure 5D), indicative of active leukotriene synthesis (Rouzer and Kargman, 1988; Wong et al., 1988). Additional 5-LO-expressing cells, likely of myeloid origin, were found in the lamina propria, but staining intensity was consistently highest in tuft cells. Human duodenal tuft cells, identified by pEGFR staining (McKinley et al., 2017), also consistently expressed 5-LO protein (Figures 5E and S5C).
To assess the capacity of tuft cells to generate cysLTs, we made twodimensional monolayers from primary epithelial cells (Liu et al., 2018; Thorne et al., 2018). Because the ligand that mediates helminth sensing is not yet known, we used ionomycin to simulate the intracellular Ca2+ flux that occurs downstream of chemosensing in tuft cells and activates TRPM5 (Deckmann et al., 2018; Hofmann et al., 2003; Prawitt et al., 2003). This was sufficient to induce cysLT production and secretion into the supernatant (Figure 5F). The response was entirely dependent on tuft cells, as cysLT production was absent in monolayers from Pou2f3-deficient mice that lack tuft cells but maintain all other epithelial lineages. Our ability to detect cysLTs was also partially TRPM5-dependent, demonstrating regulation of tuft-cell-derived leukotrienes by the chemosensing pathway. Wildtype and TRPM5-deficient cultures contained equivalent tuft cell numbers (data not shown). We further found that stimulation with N. brasiliensis excretory/secretory product (NES) induced cysLT secretion in a tuft cell-dependent manner (Figure 5G). Together these results confirm that tuft cells secrete cysLTs downstream of chemosensing.
Helminth infection results in cellular damage and release of endogenous ligands that can drive innate immune responses. For example, ATP was recently shown to be released from intestinal tissue isolated from H. polygyrus-infected mice (Shimokawa et al., 2017) and to activate airway tuft cells (Ualiyeva et al., 2020). We therefore considered whether danger-associated molecular pattern (DAMP) signaling might induce cysLT production. Using previously published tuft cell transcriptomic data (Nadjsombati et al., 2018) we identified ATP (P2rx1 and P2rx4) and adenosine (Adora1) receptors expressed by intestinal tuft cells. Unlike airway tuft cells, intestinal tuft cells do not express P2ry2. Stimulation of epithelial monolayers with either ATP or adenosine, however, failed to induce cysLT secretion (Figure S5D). We additionally tried to measure IL-25 protein secretion by ELISA following stimulation of monolayers with ionomycin or helminth products, but could not detect a signal in any condition. Thus, the specific helminth ligand(s) that elicit cysLT and perhaps IL-25 secretion by tuft cells remains to be determined.
Tuft cells are the physiologic source of leukotrienes for induction of anti-helminth immunity in the small intestine
Our data demonstrate that TRPM5-dependent sensing of helminths and cysLT signaling are both required for ILC2 activation and intestinal remodeling, and that tuft cells synthesize cysLTs. These findings suggest a model in which tuft cells respond to infection by synthesizing and secreting cysLTs that signal to ILC2s in the underlying lamina propria to initiate the feed-forward tuft-ILC2 circuit. Canonically, however, the hematopoietic compartment is thought to be the dominant source of leukotrienes, with mast cells, eosinophils, basophils, and macrophages being among the chief producers (Kanaoka and Boyce, 2004).
To identify the relevant source of leukotrienes in vivo, we generated reciprocal bone marrow chimeras using wildtype and Alox5−/− mice and infected these with N. brasiliensis for 7 days. Wildtype mice reconstituted with Alox5-deficient bone marrow retained their ability to mount a type 2 response following infection, as measured by tuft cell expansion in the SI, whereas leukotriene-deficient hosts reconstituted with wildtype bone marrow failed to mount a response (Figure 6A). This defect was also observed in wildtype hosts reconstituted with bone marrow from Cysltr1−/−Cysltr2−/− mice (Figure 6B). These experiments identify a radio-resistant cell population as the relevant intestinal source of leukotrienes during helminth infection and implicate LTC4 and/or LTD4 as the primary mediators acting on bone marrow-derived cells.
Figure 6. Tuft cells are the physiologic source of leukotrienes for induction of type 2 immunity in the small intestine.
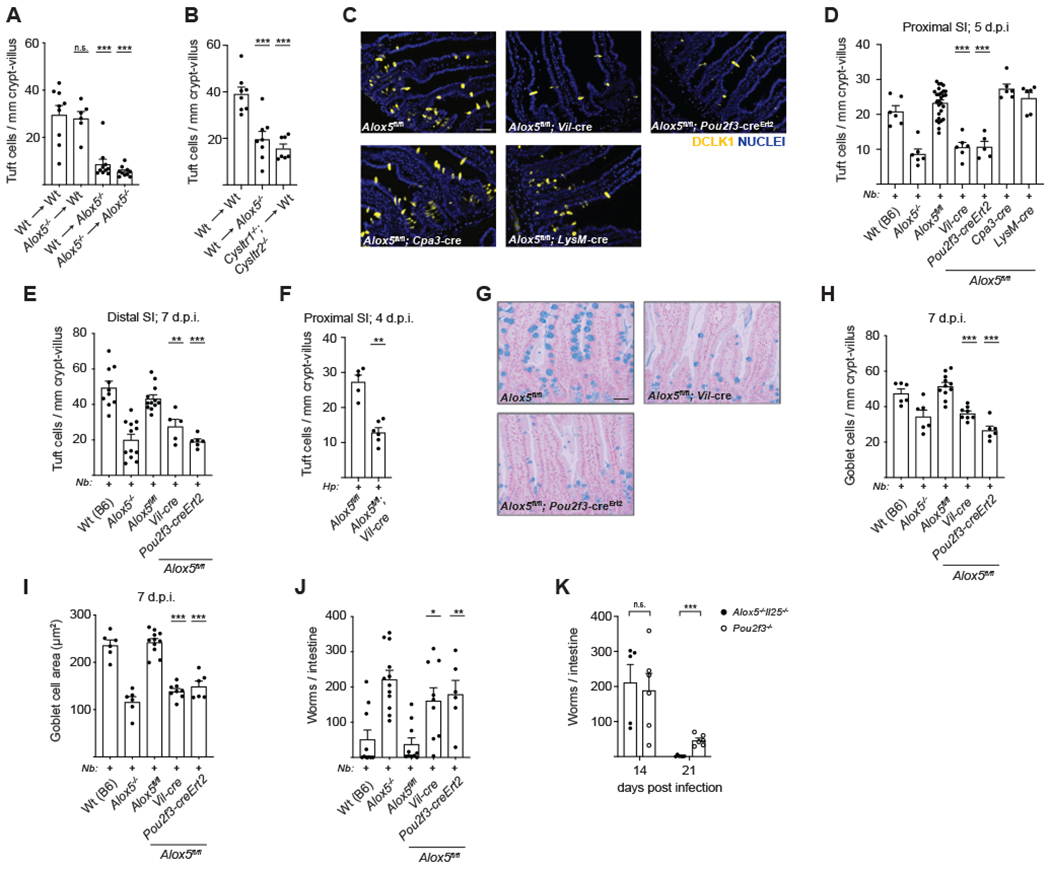
(A-B) Quantification of tuft cells in the distal (last 10cm) SI of chimeric mice of the indicated genotypes 7 days after N. brasiliensis (N.b.) infection. (C) Representative images of the proximal (first 10cm) SI 5 days post-N.b. infection. DCLK1 (yellow) marks tuft cells. DAPI (blue) marks nuclei. Scale bar: 50μm. (D) Quantification of tuft cells in (C). (E) Tuft cell frequency in the distal SI after 7 days of N.b. infection. (F) Tuft cell frequency in the proximal SI after 4 days of H. polygyrus (H.p.) infection. (G) Mice were infected with N.b. for 7 days and goblet cells identified in the jejunum (10-20cm from stomach) by alcian blue staining. Representative images are shown. Scale bar: 50μm. (H-I) Quantification of goblet cell (H) number and (I) size in (G). (J-K) Worm burden in the SI of N.b. infected mice at (J) day 7 or at (K) the indicated time points post-infection. In (A)-(B), (D)-(F), and (H)-(K) each symbol represents an individual mouse from two or more pooled experiments. In (D-E) and (H-J) Wt(B6) and Alox5−/− are the same as in Figure 4, shown for comparison, d.p.i., days post infection. *p < 0.05, **p < 0.01, ***p < 0.001 by one way ANOVA (A-B, D-E, H-J) with comparison to WT->WT or Alox5fl/fl, by Mann-Whitney (F), or by multiple t tests (K). n.s., not significant. Graphs depict mean + SEM. See also Figure S6.
To test the contribution of tuft cell-derived leukotrienes to the anti-helminth response, we generated Alox5-floxed mice to allow for cell-specific abrogation of leukotriene production (Figure S6A). We crossed these mice to the Villin1-cre strain to delete 5-LO expression in the intestinal epithelium, which we confirmed by immunofluorescence (Figure S6B). Epithelium-specific 5-LO deletion impaired the development of tuft cell hyperplasia in the proximal SI 5 days after infection with N. brasiliensis, indicating a defect in IL-13 signaling in these mice (Figures 6C–D). Similar results were observed in the distal SI after 7 days of N. brasiliensis infection (Figure 6E) and in the proximal SI 4 days following infection with H. polygyrus (Figure 6F). Within the intestinal epithelium, 5-LO expression is restricted to tuft cells. Nevertheless, to target tuft cells more specifically we generated and validated Pou2f3-creErt2-eGFP mice (Figures S6C–E). Tamoxifen-treated Alox5fl/fl;Pou2f3-creErt2 mice also showed impaired tuft cell expansion 5 days after N. brasiliensis infection. Both the Vil-cre+ and Pou2f3-creErt2+ strains phenocopied globally-deficient Alox5−/− mice, despite expression of 5-LO in many non-epithelial cells (Figures 6C–E). Consistent with these results and the bone marrow chimera experiments, deletion of 5-LO in mucosal mast cells using Cpa3-cre or macrophages and granulocytes using LysM-cre had no effect on the development of tuft cell hyperplasia.
While leukotrienes are required for airway type 2 responses, including ILC2 activation, deletion of tuft cells or 5-LO expression within tuft cells had no impact on the lung response following N. brasiliensis infection (Figures S6F–H), further underscoring the tissue-specific regulation of ILC2 activation and downstream inflammation. This finding also suggests that defects in SI type 2 responses following N. brasiliensis infection are tissue-intrinsic and not due to changes in the immune response during the lung phase of infection.
Returning to the SI, we further noted impaired goblet cell hyperplasia and hypertrophy in Alox5fl/fl; Vil-cre+ and Pou2f3-creErt2+ mice, which again phenocopied the defect found in Alox5−/− mice (Figures 6G–I). Consequently, mice lacking synthesis of leukotrienes in tuft cells failed to resolve helminth infection by 7 days-post infection (Figure 6J). Collectively, we identify tuft cells as the physiologically relevant source of cysLTs induced in the SI immediately following helminth infection and implicate cysLTs as a key effector that, in combination with IL-25, triggers the tuft-ILC2 circuit. We note that tuft cell-deficient Pou2f3−/− mice clear N. brasiliensis infection even more slowly than Alox5−/−Il25−/− double-deficient mice, suggesting that additional tuft cell effector functions remain to be discovered (Figure 6K).
Cysteinyl leukotrienes are dispensable for protist-induced type 2 immunity
In addition to sensing helminth infection, tuft cells in the SI are activated by the microbial metabolite succinate, secreted by certain Tritrichomonad protists and unidentified bacterial species that colonize the distal ileum and cecum (Lei et al., 2018; Schneider et al., 2018). We previously demonstrated that succinate administered in drinking water is sufficient to induce a type 2 immune response via the tuft-ILC2 circuit in the distal SI (Nadjsombati et al., 2018). Here we found that ILC2s isolated from the distal SI of wildtype and Alox5−/− mice had equivalent IL-13 expression following shortterm succinate treatment (Figures 7A–B). Accordingly, we found no differences in distal tuft cell hyperplasia after seven days of succinate treatment in Alox5−/− or Alox5fl/fl;Vil-cre+ mice (Figures 7C–E).
Figure 7. Cysteinyl leukotrienes are dispensable for protist-induced type 2 immunity.
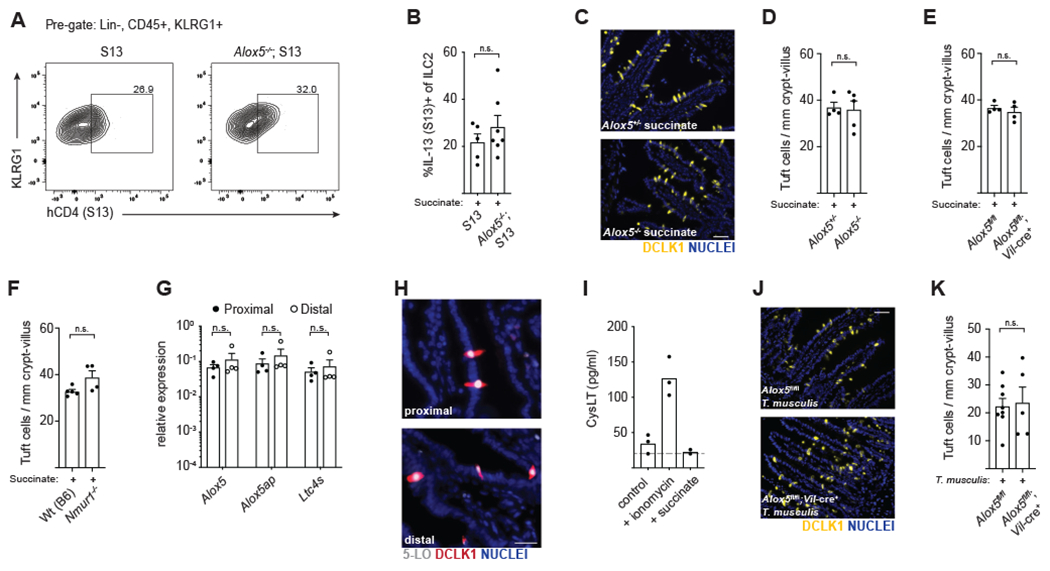
(A) Flow cytometry for IL-13 (S13) expression by ILC2s in the distal (last 10cm) SI after 36 hours of succinate treatment. (B) Quantification of IL-13 (S13) expression in (A). (C) Tuft cells in the distal SI after 7 days of succinate treatment. DCLK1 (yellow) marks tuft cells. DAPI (blue) marks nuclei. Scale bar: 50μm. (D) Quantification of tuft cells in (C). (E-F) Quantification of tuft cells in the distal SI of mice treated with succinate for 7 days. (G) Gene expression in tuft cells sorted from the proximal (first 10cm) or distal SI of naïve Wt(B6) mice. (H) 5-LO (white) and DCLK1 (red) in the proximal and distal SI of naïve mice. DAPI (blue) marks nuclei. Scale bar: 25μm. (I) Cysteinyl leukotriene (cysLT) production in supernatants of distal SI epithelial monolayer cultures following 30 minute stimulation with 500ng/ml ionomycin or 10mM succinate. Dashed line represents limit of detection. (J) Tuft cells in the distal SI 7 days post-colonization with T. musculis. Scale bar: 50μm. (K) Quantification of tuft cells in (J). In (B), (D-G) and (K) each symbol represents an individual mouse from two or more pooled experiments. In (I) symbols are technical replicates representative of two independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 by Mann-Whitney (B, D-F, K) or by multiple t tests (G). n.s., not significant. Graphs depict mean + SEM.
We considered whether the neuropeptide neuromedin U (NMU), recently shown to activate ILC2s in an NFAT-dependent manner (Cardoso et al., 2017; Klose et al., 2017; Wallrapp et al., 2017), might contribute to ILC2 activation in this context; however the response towards succinate was also independent of NMU receptor (NMUR1) signaling (Figure 7F).
Given that tuft cells in the distal SI have higher Sucnr1 expression and are more responsive to succinate than tuft cells in the proximal SI (Nadjsombati et al., 2018; Schneider et al., 2018), we hypothesized that the capacity to generate cysLTs might also vary between these sites. We found no difference in expression of genes related to leukotriene synthesis in tuft cells sorted from the proximal and distal SI, nor did we find changes in expression of 5-LO protein (Figures 7G–H). While epithelial monolayers derived from the distal SI were capable of generating cysLTs in vitro, succinate stimulation was insufficient to drive this response (Figure 7I). Colonization of Alox5fl/fl and Alox5fl/fl;Vil-cre+ mice with the succinate-producing protist Tritrichomonas musculis resulted in equivalent tuft cell hyperplasia seven days later (Figures 7J–K). Tuft cell-derived cysLTs therefore appear to be dispensable for the innate response directed against succinate-producing protists yet required for the response directed against helminths. These data suggest that tuft-ILC2 circuits in the SI are regulated in a context-dependent manner and that innate type 2 immune responses are qualitatively different for distinct classes of organisms, even within the same tissue.
DISCUSSION
Tuft cells have recently emerged as key initiators of type 2 immunity in the SI. Expression of a conserved chemosensing pathway enables tuft cells to sense luminal contents and activate the tuft-ILC2 circuit, leading to a protective immune response. Although production of IL-25 has been identified as a critical effector function, the greater delay in helminth clearance in tuft cell-deficient mice versus those lacking only IL-25 suggests additional mechanisms of immune regulation by tuft cells. In this study we demonstrate that tuft cells inducibly produce cysLTs following helminth infection and show that these leukotrienes cooperate with IL-25 to drive ILC2 activation. These findings are consistent with the recent report of cysLT production by airway tuft cells, although the receptor(s) and ligand(s) driving the response in these two tissues are distinct (Ualiyeva et al., 2020).
CysLTs are inflammatory lipids to which SI ILC2s appear particularly responsive. Indeed, we observed cysLT-dependent ILC2 activation just 16 hours after helminth infection. Our finding that cysLTs and IL-25 cooperate to drive ILC2 activation is consistent with the emerging model that ILC2s must integrate multiple transcriptional pathways for activation, analogous to those engaged during T cell TCR signaling (Lund et al., 2017; McGinty and von Moltke, 2020; von Moltke et al., 2017). CysLTs are non-redundant due to their ability to induce NFAT nuclear translocation, which cooperates with IL-25-induced NF-κB and AP-1. We had hoped to test if exogenous cysLTs are sufficient to activate intestinal ILC2s in vivo, as they are in vitro and in the lung (von Moltke et al., 2017), but we did not detect ILC2 activation at low doses and higher doses were toxic due to pleiotropic effects. Therefore, we cannot make any definitive conclusions.
In addition to cysLTs, recent work also demonstrated that the neuropeptide NMU can activate ILC2s in an NFAT-dependent manner (Cardoso et al., 2017; Klose et al., 2017; Wallrapp et al., 2017). We consider it likely that cysLTs and NMU serve both redundant and non-overlapping roles in the SI, as is often the case in type 2 immunity (Neill et al., 2010; Vannella et al., 2016). Leukotriene deficiency delays helminth clearance, but infection is still eventually resolved, indicating that redundant mechanisms indeed exist. Additional studies are required to disentangle the contexts in which individual signals are most relevant. Given the central role tuft cells play in sensing helminth infection and initiating immune responses, however, we consider tuft cell-derived cysLTs as the likely earliest source for an NFAT-inducing signal driving ILC2 activation. Other sources of cysLTs may contribute later during the response.
In some respects, it is counterintuitive that a single cell type produces two ILC2 activating ligands. The selective mobilization of NF-κB and NFAT by IL-25 and cysLTs, respectively, can enable unique states of ILC2 activation. For example, cysLTs, but not IL-25 or IL-33, can drive IL-4 production by ILC2s (Doherty et al., 2013; Pelly et al., 2016). We suspect there are also important temporal aspects to the regulation of ILC2s by leukotrienes and IL-25. Specifically, we found little evidence of leukotriene signaling at homeostasis, while deletion of Il25 led to homeostatic changes in ILC2s, even in Tritrichomonad-free mice. The homeostatic status of tuft cells and ILC2s in wild mice and in humans remains uncertain, but to the extent that succinate sensing is involved, our findings suggest this would lead to IL-25 but not cysLT signaling. Once released, IL-25 has a much longer half-life than LTC4 and LTD4, further distinguishing the kinetic functions of these two signals. In sum, while IL-25 may provide both homeostatic and induced activation, cysLTs are well suited for rapid “on/off” regulation of the tuft-ILC2 circuit.
We have focused here on regulation of the tuft-ILC2 circuit, but many other leukotriene receptor-expressing cells populate the small intestine and may also respond to tuft cell-derived cysLTs (Bäck et al., 2011). Given the rapid extracellular degradation of LTC4 and LTD4, any responding cells would need to reside near or migrate in close proximity to tuft cells. Neurons, Th2 effector cells, intraepithelial lymphocytes, endothelial cells, and the smooth muscle surrounding lacteals are all candidates that warrant further investigation. IL-25 might also have additional targets aside from ILC2s. Most notably, tuft cells themselves express the IL-25 receptor and may therefore engage in autocrine signaling (Bezençon et al., 2008).
How IL-25 and cysLTs are secreted from tuft cells remains to be determined. TRPM5 is required for transduction of all tuft cell-activating signals identified to date, but its exact role remains elusive. In type 2 taste cells TRPM5 activation results in cellular depolarization and release of neuromediators that propagate the taste sensation (Chaudhari and Roper, 2010). TRPM5 may therefore regulate release of pre-formed IL-25 stored in tuft cells, as was suggested by one recent study (Luo et al., 2019) and by our finding that the MFI of KLRG1 on ILC2s was similarly decreased in naïve Il25−/− and Trpm5−/− mice. An alternative, but not mutually exclusive, model is that some IL-25 is secreted constitutively. Tuft cells constitutively express Il25 (Bezençon et al., 2008; von Moltke et al., 2016) and we found no evidence of transcriptional induction on a per cell basis following helminth infection. Given the canonical signal peptide encoded by Il25 mRNA, IL-25 protein should be delivered into the endoplasmic reticulum during translation, but how it is further trafficked remains uncertain. We had hoped to explore mechanisms of IL-25 secretion using our monolayer system or the scraped villi technique reported by Luo et al. but could not detect IL-25 under any conditions using commercially available IL-25 ELISAs. Clarification of the timing and mechanism of IL-25 secretion must therefore await development of improved in vitro tuft cell culture conditions and enhanced reagents for detection of IL-25 protein.
Similarly, it remains unclear how the TRPM5-dependent chemosensing pathway interacts with leukotriene production. Our in vitro data demonstrate a requirement for TRPM5 in the generation of cysLTs in response to calcium flux, but we were not able to distinguish between defects in synthesis versus secretion, or perhaps both. Further experiments will be required to dissect the precise interplay between chemosensing, TRPM5, and leukotriene generation.
Perhaps the most unexpected finding of this study was the differential requirement for cysLTs in the response to helminths versus succinate-producing protists. Tuft cell ligands may vary, but effector output has generally been considered stereotyped, at least within the SI. Indeed, all tuft cell-dependent immune responses identified in the SI to date also require IL-25. We find that tuft cell-derived cysLTs, on the other hand, are required to drive the innate anti-helminth response but are dispensable for the response directed against succinate-producing protozoans. The nature of innate type 2 responses in the SI therefore appears to be qualitatively different for distinct organisms. Succinate-producing protists like T. musculis colonize the distal SI and cecum and are common across vivariums. While such protists modulate immunological tone, they do not seem to impair host fitness nor do they themselves seem to be affected by a strong type 2 response (Chudnovskiy et al., 2016; Escalante et al., 2016; Howitt et al., 2016; Nadjsombati et al., 2018; Schneider et al., 2018). Generating inflammatory leukotrienes could therefore be considered a disproportionate response to a harmless commensal. Parasitic helminths, on the other hand, inflict significant damage, compete for nutrients, and generally decrease host fitness. A more robust inflammatory response might be appropriate in this context.
How this differential regulation is achieved at the cellular level remains to be determined since all tuft cells express the enzymes required for cysLT synthesis. Is succinate signaling alone insufficient to drive cysLT generation within tuft cells, as we observed in vitro? Or are cysLTs actually produced in response to succinate in vivo but are redundant for ILC2 activation in this context? Does tuft cell sensing of helminths rely on the recognition of one ligand that drives simultaneous production of IL-25 and cysLTs, or are there multiple signals that independently drive production of each effector? We favor the hypothesis that tuft cell effector output is in part stimulus-specific. Differential regulation of effectors would enable nuanced control over the immunological tone of the SI and consequently physiologic adaption appropriate to the type of organism sensed (e.g. parasite vs. commensal). Testing such hypotheses will likely require identification of additional tuft cell activating ligands and the receptors through which they signal, as well as additional effector molecules through which tuft cells orchestrate the innate type 2 response.
Lastly, how well murine tuft cell biology reflects that of humans remains to be determined. We found that human tuft cells in healthy duodenal tissue consistently expressed 5-LO protein, and others recently demonstrated that human intestinal tuft cells express the accessory protein FLAP (Schütz et al., 2019), together suggesting that these cells are primed to synthesize leukotrienes. In this regard it is notable that the commonly prescribed asthma and allergy medication montelukast, a CYSLTR1 inhibitor, is administered as an ingestible tablet. What effect, if any, this drug has on intestinal physiology or immunity is not clear but may prove relevant when considering type 2-associated pathologies in the intestine.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jakob von Moltke (jmoltke@uw.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Mice aged 6-12 weeks were used for all experiments. Mice were age-matched within each experiment, but pooled results include both male and female mice of varying ages. C57BL/6J mice were bred in house or purchased from Jackson Laboratories. B6. Alox5−/− (B6.129S2-Alox5tm1Fun/J), B6.Trpm5−/− (B6.129P2-Trpm5tm1Dgen/J), B6.Cysltrt1−/− (C57BL/6N-Cysltr1tm1Ykn/J), B6. Villin1-cre (B6.Cg-Tg(Vil1-cre)1000Gum/J), and B6.LysM-cre (B6.129P2-Lyz2tm1(cre)Ifo/J) mice were purchased from Jackson Laboratories. B6.Pou2f3−/− (Pou2f3 tm1.1(KOMP)Vlcg, Project ID #VG18280) were rederived from sperm stored in the Canadian Mouse Mutant Repository. B6.Il25Flare25/Flare25 and B6.Il13Smart13/Smart13 mice were generated as previously described (Liang et al., 2011; von Moltke et al., 2016). B6.Il25−/− mice were generously provided by A. McKenzie via R. Locksley. B6.Il1rl1−/− (St2−/−) mice were generously provided by S. Akira via R. Locksley. B6.Cpa3-cre (B6.Tg(Cpa3-cre)3Glli) mice were generously provided by A. Piliponsky. Nmur1tm1.1(KOMP)Vlcg mice were generously provided by V. Kuchroo. Alox5−/−, Il25−/−, St2−/−, and Trpm5−/− mice were crossed to the B6.Il13Smart13/Smart13 line. Il25−/− and St2−/− mice were further crossed to the B6.Arg1YARG reporter line (Reese et al., 2007). Cysltr1−/− (Maekawa et al., 2002) and Cysltr2−/− (Beller et al., 2004) mice were crossed to generate a double-deficient line and bone marrow was generously provided by N. Barrett. All mice were maintained in specific pathogen-free conditions at the University of Washington and were confirmed to be free of T. musculis and T. rainier by microscopy and qPCR, except those specifically colonized with T. musculis as noted.
METHOD DETAILS
Generation of Alox5-flox Mice
B6.Alox5fl/fl mice with loxP sites flanking the 6th exon of Alox5 were generated by homologous gene targeting in C57BL/6 embryonic stem cells. Exon 6, which contains critical iron-binding residues, was selected for deletion to replicate the targeting strategy used for B6.129S2-Alox5tm1Fun/J mice. A 2.0 kb 5’ homology arm containing the 5’ loxP site and spanning exon 6 of Alox5 was assembled using overlap extension PCR and cloned into pKO915-DT (Lexicon Genetics) using EcoRI and Xhol. Next, a 2.0 kb 3’ homology arm was amplified and inserted into the pK0915-DT vector containing the 5’ homology arm using Sacll and Smal. Lastly, a pre-assembled selection cassette containing frt-flanked neomycin and a 3’ loxP site was sub-cloned into the homology arm-containing pKO915-DT vector using Ascl. The selection cassette was screened for insertion in the proper orientation and the final construct was linearized with Notl and transfected by electroporation into C57BL/6 embryonic stem cells (PRXB6T). Cells were grown on irradiated feeders with the aminoglycoside G418 in the media, and neomycin-resistant clones were screened for 5’ and 3’ homologous recombination by PCR. Two positive clones were selected for injection (Gladstone Institutes, San Francisco) into albino C57BL/6 blastocysts to generate chimaeras. Male pups with highest ratios of black-to-white coat color from a single clone were selected to breed with homozygous Gt(Rosa26)FLP1/FLP1 females (Jackson Laboratories catalog # 009086) to excise the neomycin resistance cassette. Deletion of neomycin was confirmed by PCR. Alox5fl/fl genotyping primers were as follows: A5FL_F: TGTTTGGCTACCAGTTCCTGAATGG; A5FL_R: AGCAGATGACAGTTGGGTGACTATG (460 bp wildtype band; 577 bp knock-in band; no band after CRE-mediated excision).
Generation of Pou2f3-CreERT2-ires-EGFP Mice
A CreERT2-ires-EGFP followed by FRT-flanked PGK-Neo cassette was inserted by a homologous recombination in place of the initial ATG codon of Pou2f3 gene of C57BL/6J-derived ES cells. Chimeric mice were mated with an FLP strain to remove the PGK-Neo cassette. After excluding the FLP allele by mating with C57BL/6J, heterozygous Pou2f3-CreERT2-ires-EGFP mice were obtained.
Mouse Infection and Treatment
H. polygyrus and N. brasiliensis larvae were raised and maintained as previously described (Johnston et al., 2015; Liang et al., 2011). Mice were infected by oral gavage with 200 H. polygyrus L3 or subcutaneously with 500 N. brasiliensis L3 and euthanized at the indicated time points to collect tissues for staining or to count worm burden. In experiments using Pou2f3-CreErt2-eGFP mice, animals were administered 2.5mg tamoxifen by oral gavage at days 0, 2, and 5 post-infection. Worm burden was enumerated across the entire small intestine. For in vivo CYSLTR1-inhibition experiments, mice were injected intraperitoneally with 10mg/kg montelukast (Cayman Chemical) one hour prior to oral gavage with H. polygyrus. For ibuprofen experiments mice were given 1mg/ml ibuprofen ad libitum in drinking water starting one day prior to infection and continuing through the course of the experiment. For BrdU incorporation experiments mice were injected intraperitoneally with 1mg BrdU at the indicated time point and tissue harvested 24 hours later.
Succinate Treatment and Protist Colonization
For succinate experiments mice were given 150mM sodium succinate hexahydrate (Alfa Aesar) ad libitum in drinking water for the indicated amount of time. For protist colonization experiments, wildtype mice vertically colonized with Tritrichomonas musculis were used as a source of protists. Colonization of naïve mice was performed as previously described (Nadjsombati et al., 2018). Briefly, cecal contents were collected, washed in PBS, filtered through a 70μm strainer, and washed again. Total protist numbers were estimated using a hemocytometer. Naïve mice were colonized by oral gavage with 40-50 x106 protists. Intestinal tissue was collected seven days later for analysis.
Tissue Fixation and Staining
For tuft cell staining, intestinal tissues were flushed with PBS and fixed in 4% paraformaldehyde for 4 hours at 4° C. Tissues were washed with PBS and incubated in 30% (w/v) sucrose overnight at 4° C. Samples were then coiled into “Swiss rolls” and embedded in Optimal Cutting Temperature Compound (Tissue-Tek) and sectioned at 8 μm on a Microm HM550 cryostat (Thermo Scientific). Immunofluorescent staining was performed in Tris/NaCl blocking buffer (0.1 M Tris-HCL, 0.15 M NaCl, 5μg ml−1 TSA blocking reagents (Perkin Elmer), pH 7.5) as follows: 1 h 5% goat serum, 1 h primary antibody (αDCLKI, Abcam ab31704), 40 min goat anti-rabbit IgG F(ab’)2-AF594 secondary antibody (Invitrogen) and mounted with Vectashield plus DAPI (Vector Laboratories). Tuft cell frequency was calculated using ImageJ software to manually quantify DCLK1+ cells per millimeter of crypt-villus axis. Four 10x images of the swiss roll were analyzed for each replicate and at least 30 total villi were counted. Mouse 5-LO staining was performed as for DCLK1 above, with following modifications: 1 h 10% goat serum at room temperature (RT), overnight rabbit anti-5-LO (Sigma Aldrich, SAB1410449) at 4° C, 40 min goat anti-rabbit IgG F(ab’)2-AF594, 1 h 10% rabbit serum at RT, 2 h αDCLK1-AF488 (custom conjugation of ab31704) at RT. Human 5-LO staining was performed as above, replacing αDCLK1-AF488 with anti-pEGFR-AF488 (Abcam ab205827). Human duodenal tissue samples were collected during either Roux-en-Y or whipple procedures on three different deidentified patients and obtained through Northwest BioSpecimen. The University of Washington Institutional Review Board has determined that the deidentified tissue specimens used in this study do not meet the federal regulatory definition of research on human subjects.
For BrdU staining slides were rehydrated in PBS, incubated in 2 M HCI for 1 hr, and then neutralized in 0.1 M sodium borate for 10 min. Staining was performed as follows: 1 h Fc Block at RT, overnight rabbit-anti-BrdU-AF594 (Abcam ab220076) and Fc Block at 4° C, 1 min wash in PBS, 10 min fix in 0.2% paraformaldehyde at RT. BrdU incorporation was determined as the distance from the crypt to most distal BrdU+ cell per villus, counting at least 25 villi per sample.
For goblet cell staining, tissues were flushed with PBS, fixed in 10% buffered formalin at 4° C for 3 hours, coiled into “Swiss rolls” and returned to formalin. After 24 hours tissues were moved to 70% ethanol for storage. Tissue processing, paraffin embedding, sectioning, and staining were performed by the Pathology Research Services Laboratory at the University of Washington. Alcian blue staining was used to identify goblet cells. Goblet cell frequency was calculated as described above for tuft cells. Hypertrophy was quantified using ImageJ software to measure the area of at least 80 goblet cells for each biological replicate. Brightfield and fluorescent images were acquired on an Axio Observer A1 inverted microscope (Zeiss) and on an AR1 confocal microscope (Nikon).
Single-cell Tissue Preparation
For single cell epithelial preparations from small intestines, tissues were flushed with PBS, opened, and rinsed with PBS to remove intestinal contents. Intestinal tissue was cut into 2-5 cm pieces and incubated rocking at 37° C in 15ml HBSS (Ca+2/Mg+2-free) supplemented with 5mM dithiothreitol (DTT, Sigma-Aldrich), 5% fetal calf serum (FCS, VWR), and 10mM HEPES (Gibco). Tissues were vortexed vigorously and supernatant was discarded. Tissues were then incubated rocking at 37° C in 15ml HBSS (Ca+2/Mg+2-free) supplemented with 5mM EDTA (Invitrogen), 5% FCS, and 10mM HEPES. Tissues were vortexed thoroughly and released epithelial cells were passed through a 70 μm filter. Tissues were then incubated in fresh EDTA/HBSS solution for 15 minutes, vortexed, and filtered. Supernatants were pooled and washed once before staining for flow cytometry.
For lamina propria preparations small intestinal tissue was processed as above to remove the epithelial fraction. Tissues were then rinsed in 20ml HBSS (with Ca+2/Mg+2) supplemented with 5% FCS and 10mM HEPES, shaking at 37° C for 20 minutes. Supernatants were discarded and tissues were incubated in 5ml HBSS (with Ca+2/Mg+2) supplemented with 3% FCS, 10mM HEPES, 30 μg/ml DNase I (Sigma Aldrich), and 0.1 Wunsch/ml Liberase TM (Sigma Aldrich), shaking at 37° C for 30 minutes. Tissues were vortexed and cells were passed through a 70 μm filter and washed. The resulting pellet was resuspended in 40% Percoll (Sigma Aldrich), centrifuged for 5 minutes at 1500 rpm, and supernatant discarded. Pelleted cells were then washed and stained for flow cytometry. Note: to preserve IL-17RB staining, a modified protocol was used to isolate lamina propria cells for data in Figures 2 and S2. See “RNA Sequencing and Analysis” below for details.
For lung preparation tissue was dissociated by gentleMACS (Miltenyi Biotec) using program lung_01. Tissue was then incubated in HBSS (with Ca+2 and Mg+2) supplemented with 0.2mg/ml LiberaseTM (Roche) and 25 μg/ml Dnase I (Sigma) for 35 minutes shaking at 37° C, and subsequently run on gentleMACS using program lung_02. Cells were then filtered through a 70 μm strainer, washed, and stained for flow cytometry.
Flow cytometry and cell sorting
Single cell suspensions were prepared as described and stained in DPBS + 3% FBS with antibodies to surface markers for 20 min at 4° C, followed by DAPI (Roche) for dead cell exclusion. For intracellular cytokine staining cells were first washed in DPBS and stained with Zombie Violet fixable viability dye (BioLegend) for 20 min at 4° C and subsequently stained with antibodies to surface markers for 20 min at 4° C. Cells were then fixed and permeabilized using the FoxP3 Transcription Factor Staining Buffer kit (eBioscience) following manufacturer’s instructions. Cells were further stained with antibodies to intracellular proteins for 30 minutes at room temperature. Samples were FSC-A/SSC-A gated to exclude debris, SSC-FI/SSC-W gated to select single cells and gated to exclude dead cells. Samples were run on an LSR II (BD Biosciences) or Aurora (Cytek) and analyzed with FlowJo 10 (Tree Star). For cell sorting, single cell suspensions were prepared and stained as described and sorted on an Aria II (BD Biosciences).
Quantitative RT-PCR
For ILC2 qPCR, 10,000 small intestinal CD45+Lin−KLRG1+ cells and 10,000 lung CD45+Lin−Thy1.2+ST2+ cells from C57BL/6J mice were sorted into Buffer RLT (Qiagen) using an Aria II (BD Biosciences). Lineage dump included the markers CD3, CD4, CD5, CD8, CD11b, CD19, FcER1, and NK1.1. 10,000 tuft cells were sorted as CD45−EpCAM+ RFP+ from small intestinal epithelium of Il25Flare25/Flare25 mice or as CD45− EpCAM+CD24+SiglecF+from small intestinal epithelium of C57BL/6J mice. Previous RNA sequencing confirmed that these two gating strategies yield identical tuft cell transcriptional profiles. RNA was isolated using the Micro Plus RNeasy kit (Qiagen) and reverse transcribed using Superscript Vilo Master Mix (Life Technologies). cDNA was used as template for quantitative PCR with Power SYBR Green reagent on a StepOnePlus cycler (Applied Biosystems). Transcripts were normalized to Rps17 (40S ribosomal protein S17) expression.
RNA Sequencing and Analysis
In order to maintain IL-17RB staining that was lost with extended lamina propria tissue processing described above, single cell suspensions were prepared from the proximal small intestine of naïve mice using a modified protocol. All data in Figure 2 and Figure S2 were generated using this modified digestion protocol. Before tissue collection, mice were anesthetized with ketamine/xylazine. Next, the small intestine was nicked at the stomach and transected at the cecum and flushed with 20 mL 37°C HBSS (no Ca+2/Mg+2) + 10 mM HEPES. The mice were then perfused through the heart with 40 mL of 30 mM EDTA + 10 mM HEPES in HBSS (no Ca+2/Mg+2). Five minutes after initiating perfusion, the first 5 cm of the proximal SI were harvested, fileted open, transferred to 35 mL ice cold HBSS (no Ca+2/Mg+2) + 10 mM HEPES and shaken vigorously for 30 seconds to release epithelial cells. Intestinal pieces were stored in HBSS + 5% FCS on ice and then transferred into pre-warmed digest buffer composed of 5ml HBSS (with Ca+2/Mg+2) supplemented with 3% FCS, 10mM HEPES, 30 μg/ml DNase I (Sigma Aldrich), and 0.1 Wunsch/ml Liberase TM (Sigma Aldrich), shaking at 37° C for 30 minutes. Tissues were vortexed and cells were passed through a 70 μm filter and washed. The resulting pellet was resuspended in 40% Percoll (Sigma Aldrich), centrifuged for 5 minutes at 1500 rpm, and supernatant discarded. Pelleted cells were then washed and stained for flow cytometry and sorting.
500 ILC2s were sorted as CD45+Lin−IL-17RB+ directly into lysis buffer from the SMART-Seq v4 Ultra Low Input RNA Kit (Takara) and cDNA was generated following manufacturer’s instructions. Four biological replicates were collected for each genotype. Each biological replicate represents one mouse. Next-generation sequencing and analysis was performed by the Benaroya Research Institute Genomics Core. Sequencing libraries were generated using the Nextera XT library preparation kit with multiplexing primers, according to manufacturer’s protocol (Illumina), and library quality was assessed using the Tapestation (Agilent). High throughput sequencing was on HiSeq 2500 (Illumina), sequencing dual-indexed and single-end 58 base pair reads. All samples were in the same run with target depth of 5 million reads to reach adequate depth of coverage.
Sequencing was inspected by FASTQC (v0.11.3) and yielded a median read depth of 8.7 million reads per sample. The following analytic pipeline was managed on the Galaxy platform (Boekel et al., 2015). Reads were trimmed by 1 base at the 3’ end, and then trimmed from both ends until base calls had a minimum quality score of at least 30 (Galaxy FASTQ Trimmer tool v1.0.0). Sequence alignment was performed using STAR aligner (v2.4.2a) with the GRCm38 reference genome and gene annotations from Ensembl release 91. Gene counts were generated using HTSeq-count (v0.4.1). Quality metrics were compiled from PICARD (v1.134), FASTQC (v0.11.3), and HTSeq-count. Raw input from HTseq-count was normalized in DESeq2. Uniquely mapped Ensembl IDs (genes and non-coding RNAs) with a mean normalized read count <10 were excluded. The gene Arg1 was not included in the analysis as the Arg1Yarg reporter allele appeared to interfere with sequencing reads in the Il25−/− and St2−/− strains. The resulting datasets were deposited in the Gene Expression Omnibus (GEO: GSE144956).
ILC2 Stimulation Assay
Small intestinal lamina propria ILC2s were isolated from Il13Smart13 reporter mice and sorted as described. Sorted cells were plated at 4000-5000 cells per well in a 96 well plate and incubated overnight in 10 ng/ml IL-7 (R&D Systems) and basal media composed of high glucose DMEM supplemented with non-essential amino acids, 10% FBS, 100 U/mL penicillin, 100mg/mL streptomycin, 10mM HEPES, 1mM sodium pyruvate, 100μM 2-mercaptoethanol, and 2mM L-glutamine. The next morning media was replaced with fresh media and 10 ng/ml IL-7 and cells were stimulated with the indicated agonist. After a six-hour incubation at 37 °C, cells were stained with 1 μl/well of PE-conjugated anti-human CD4 for 20 minutes at 4 °C. Cells were washed, resuspended in DAPI, and analyzed on an LSRII (BD Biosciences).
NFAT Imaging
ILC2s were sorted from the small intestine lamina propria and cultured overnight in 10 ng/ml IL-7 (R&D Systems), as described above. The next morning cells were treated for 30 min with 1μM cyclosporin A (CSA) where indicated, followed by 90 min treatment with 100nM LTC4, 100ng/ml IL-25, 100ng/ml IL-33, or 30ng/ml PMA and 500ng/ml ionomycin. Cells were then affixed to slides by Cytospin and treated for 5 min with ice-cold methanol. Slides were stained as follows: 30 min block with 10% goat serum, 1 h rabbit-anti-NFATC2 (NFAT1: clone D43B1, Cell Signaling Technology), 40 min goat anti-rabbit IgG F(ab’)2-AF594 secondary antibody (Invitrogen). Cells were counterstained with DAPI and imaged on an Axio Observer A1 inverted microscope (Zeiss).
Bone Marrow Chimeras
Congenically-marked donor bone marrow cells were isolated from leg bones from mice of the indicated genotype. Bones were rinsed in 70% ethanol, rinsed three times in PBS, and then ground using a pestle. Extracted cells were washed in PBS and filtered through a 70 μm strainer. Cells were spun down and resuspended in ACK lysing buffer for five minutes to remove red blood cells. Cells were then washed in PBS and resuspended in RPMI. Recipient mice were irradiated (1000 rads) and retro-orbitally injected with 5x106 cells. Mice were placed on Baytril-medicated water for two weeks post-transfer and rested for at least six weeks to allow for hematopoietic reconstitution. Chimeric mice were infected with N. brasiliensis as described and intestinal tissue harvested on day 7 post-infection for enumeration of tuft cells. Chimerism was confirmed at time of harvest by staining splenocytes with congenic markers CD45.1 and CD45.2.
Organoid Culture
Small intestinal crypt-derived organoids were grown as described with modifications described below (Sato and Clevers, 2013). Briefly, proximal small intestine was isolated and villi manually scraped off with a glass coverslip. Tissue was then washed three times in cold PBS with vigorous shaking before 30 minute 4° C incubation in 2mM EDTA to release epithelial crypts, which were washed in PBS and filtered through a 70 μm strainer. Pelleted crypts were resuspended in Matrigel and plated at 400-500 crypts per well in a prewarmed plate, incubated at 37 °C for 5 minutes to allow for Matrigel solidification, and complete organoid media added. Organoid media was composed of DMEM/F12 supplemented with 2mM glutamine, 100 U/mL penicillin, 100mg/mL streptomycin, 10mM HEPES, 1X N2 supplement (Life Technologies), 1X B27 supplement (Life Technologies), 500mM N-acetylcysteine, 50μg/ml mEGF, and replacing recombinant R-spondin with supernatants from R-spondin expressing L-cells and replacing recombinant Noggin with supernatants from Noggin expressing cells. Crypts were harvested from the proximal (first 10cm) small intestine of naïve C57BL/6J mice and plated on day 0. On day 1 and day 4, media was replaced and organoids were treated with 20 ng/ml recombinant IL-13 or 10nM of the indicated leukotriene. On day 7 organoids were harvested and resuspended in Accutase (Corning). Organoids were sheared with a 28G insulin syringe, incubated for 1h at room temperature, washed, and then stained for flow cytometry as described above. Tuft cells were identified as CD45−EpCAM+SiglecF+CD24+.
Cysteinyl Leukotriene ELISA
Intestinal monolayer cultures were generated for ELISA experiments. Intact epithelial crypts were isolated from the small intestine following the protocol described for organoid culture. After crypts were isolated, however, they were resuspended in complete organoid media supplemented with 10μM Y27632 (Stemcell Technologies) and 10μM SB431542 (Stemcell Technologies) and plated in warm plates pre-coated with Matrigel. Plates were coated with Matrigel by adding 100 μl of 2% Matrigel in cold DMEM to each well in 48-well plates and incubating at least 30 minutes at 37 °C. Matrigel media was aspirated before plating crypts in organoid media. After plating, cells were incubated overnight at 37 °C to allow adherence and media containing unattached cells was replaced the next day, followed by an additional hour of incubation. Media was then aspirated, test stimuli added, and cells incubated for 30 minutes at 37 °C. Supernatants were collected and used to perform ELISA using the Cysteinyl Leukotriene Express ELISA kit (Cayman Chemical) following manufacturer’s instructions.
N. brasiliensis Excretory-Secretory Product (NES) Preparation
To generate NES, Lewis rats were infected subcutaneously with 5000 N. brasiliensis L3. Mature (L5) worms were collected from the entire small intestine 7 days post infection. Worms were washed 10 times in Wash Solution I (PBS with 200 U mL−1 Pen-Strep), followed by a 40 minute incubation in Wash Solution 1 at room temperature. Worms were allowed to equilibrate in Wash Solution II (RPMI 1640 with 200 U mL−1 Pen-Strep) for 1 hr at 37°C, before being transferred to a tissue culture flask in NES culturing media (RPMI 1640, with 100 U mL−1 Pen-Strep, 2mM L-Glutamine and 1% glucose) and cultured at 37°C. Supernatant was collected at 24, 48, and 72 hr and filtered prior to use as NES.
QUANTIFICATION AND STATISTICAL ANALYSIS
All experiments were performed using randomly assigned mice without investigator blinding. All data points reflect biological replicates (i.e. mice), except in Figures 1I, S4B–C, 5F–G, S5D, and 7I where they represent technical replicates. No data were excluded. Statistical analysis was performed as noted in figure legends using Prism 7 (GraphPad) software. Graphs show mean + SEM.
DATA AND CODE AVAILABILITY
RNA-seq data are available at the NCBI Gene Expression Omnibus under accession number GEO: GSE144956.
Supplementary Material
Document S1. Figures S1-6.
Table S1. Normalized RNA-seq results, Related to Figure 2.
Table S2. Differentially expressed genes in ILC2s from Alox5−/− mice, Related to Figure 2.
Table S3. Differentially expressed genes in ILC2s from Il25−/− mice, Related to Figure 2.
Table S4. Differentially expressed genes in ILC2s from St2−/− mice, Related to Figure 2.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD326 (EpCAM) APC, clone G8.8 | BioLegend | Cat#118214, RRID: AB_1134102 |
| Human CD4 PE, clone RPA-T4 | BioLegend | Cat#300508, RRID: AB_314076 |
| CD45 BV605, clone 30-F11 | BioLegend | Cat#103155, RRID: AB_2650656 |
| CD45 BV650, clone 30-F11 | BioLegend | Cat#103151, RRID: AB_2565884 |
| KLRG1 PECy7, clone 2F1/KLRG1 | BioLegend | Cat#138416, RRID: AB_2561736 |
| CD3 Percp-cy5.5, clone 145-2C11 | BioLegend | Cat#100328, RRID: AB_893318 |
| CD4 Percp-cy5.5, clone RM4-5 | BioLegend | Cat#100540, RRID: AB_893326 |
| CD5 Percp-cy5.5, clone 53-7.3 | BioLegend | Cat#100624, RRID: AB_2563433 |
| CD8 Percp-cy5.5, clone 53-6.7 | BioLegend | Cat#100724, RRID: AB_389326 |
| CD11b Percp-cy5.5, clone M1-70 | BioLegend | Cat#101228, RRID: AB_893232 |
| CD11 b AlexaFluor 700, clone M1-70 | BioLegend | Cat#101222, RRID: AB_493705 |
| CD19 Percp-cy5.5, clone 1D3/CD19 | BioLegend | Cat#152406, RRID: AB_2629815 |
| FceRI Percp-cy5.5, clone MAR-1 | BioLegend | Cat#134320, RRID: AB_10641135 |
| NK1.1 Percp-cy5.5, clone PK136 | BioLegend | Cat#108728, RRID: AB_2132705 |
| Siglec F APC-Cy7, clone E50-2440 | BD Pharmingen | Cat#565527, RRID: AB_2732831 |
| CD24 Percp-cy5.5, clone M1/69 | BioLegend | Cat#101824 RRID: AB_1595491 |
| Ki67 AlexaFluor 488, clone 16A8 | BioLegend | Cat#652417 RRID: AB_2564236 |
| Rabbit anti-DCAMKL1 | Abcam | Cat#ab31704, RRID: AB_873537 |
| F(ab’)2 goat anti-rabbit IgG AF594 | Invitrogen | Cat#A11072, RRID: AB_2534116 |
| Rabbit anti-5-LO | Sigma Aldrich | Cat# SAB1410449 RRID: AB_2819149 |
| Thy1.2 (CD90.2) BV605, clone 53-2.1 | Biolegend | Cat#140318 RRID: AB_2650924 |
| ST2(Il1rl1) biotin, clone DJ8 | MD Bioscience | Cat#101001B RRID: AB_947551 |
| Rabbit anti-BrdU-AF594 | Abcam | Cat#ab220076 RRID: AB_2819150 |
| Rabbit anti-NFATC2 (NFAT1), clone D43B1 | Cell Signaling Technology | Cat#5861T RRID: AB_10834808 |
| Rabbit anti-pEGFR (Y1068), clone EP774Y | Abcam | Cat#ab205827 RRID: AB_732110 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant IL-13 | Peprotech | Cat#210-13 |
| Recombinant IL-25 | R & D Systems | Cat#1399-IL-025 |
| Recombinant IL-33 | R & D Systems | Cat#3626-ML-010 |
| Recombinant IL-7 | R & D Systems | Cat#h407-ML-005 |
| Strepavidin PE/Dazzle | Biolegend | Cat#405248 |
| Vectashield with DAPI | Vector Laboratories | Cat#H-1200 |
| DNase I | Sigma Aldrich | Cat#D4513 |
| LiberaseTM | Sigma Aldrich | Cat#5401127001 |
| TSA Blocking reagent | Perkin Elmer | Cat#FP1020 |
| DAPI | Roche | Cat#10236276001 |
| Fc Block | BD Biosciences | Cat#553141 |
| Accutase | Corning | Cat# 25058C1 |
| Recombinant murine EGF | Peprotech | Cat# 31509A |
| Y27632 | Stemcell Technologies | Cat# 72302 |
| SB431542 | Stemcell Technologies | Cat# 72232 |
| Leukotriene C4 | Cayman Chemical | Cat# 20210 |
| Leukotriene D4 | Cayman Chemical | Cat# 20310 |
| Leukotriene E4 | Cayman Chemical | Cat# 20410 |
| Leukotriene B4 | Cayman Chemical | Cat# 20110 |
| Montelukast | Cayman Chemical | Cat# 10008318 |
| BrdU | BioLegend | Cat# 423401 |
| Sodium succinate hexahydrate | Alfa Aesar | Cat#41983 |
| Critical Commercial Assays | ||
| FoxP3 Transcription Factor Staining Kit | ThermoFisher | Cat#00-5523-00 |
| RNeasy Plus Micro Kit | Qiagen | Cat#74034 |
| SuperScript VILO cDNA Synthesis Kit | Invitrogen | Cat#11766050 |
| SMARTseq V4 Ultra Low Input RNA Kit | Takara | Cat# 634894 |
| Cysteinyl Leukotriene Express ELISA Kit | Cayman Chemical | Cat#10009291 |
| Deposited Data | ||
| RNAseq data files | This paper | GEO: GSE144956 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | Jackson Laboratory | Cat# 000664 |
| Mouse: B6.129P2-Trpm5tm1Dgen/J | Jackson Laboratory | Cat# 005848 |
| Mouse: B6.129S2-Alox5tm1Fun/J | Jackson Laboratory | Cat# 004155 |
| Mouse: B6.Cysltr1tm1Ykn/J | Jackson Laboratory | Cat#030814 |
| Mouse: B6.Cg-Tg(Vil1-cre)1000Gum/J | Jackson Laboratory | Cat# 021504 |
| Mouse: B6.129P2-Lyz2tm1(cre)Ifo/J | Jackson Laboratory | Cat# 004781 |
| Mouse: B6.Il25Flare25/Flare25 | von Moltke et al., 2016 | N/A |
| Mouse: B6.Il13Smart13/Smart13 | Liang et al., 2012 | N/A |
| Mouse: B6.Arg1YARG/YARG | Reese et al. 2007 | N/A |
| Mouse: B6.Pou2f3−/− | Provided by M. Anderson: Project ID #VG18280 | N/A |
| Mouse: B6.Tg(Cpa3-cre)3Glli | Provided by A. Piliponsky | N/A |
| Mouse: B6.Il25−/− | Provided by A. McKenzie, via R. Locksley | N/A |
| Mouse: B6.Il1rl1−/− | Provided by S. Akira, via R. Locksley | N/A |
| Mouse: Nmur1tm1.1(KOMP)Vlcg | Provided by V. Kuchroo | N/A |
| Mouse: B6.Cysltr1−/− | Provided by N. Barrett | N/A |
| Mouse: B6.Cysltr2−/− | Provided by N. Barrett | N/A |
| Mouse: Alox5-flox | This paper | N/A |
| Mouse: Pou2f3-creErt2-eGFP | This paper | N/A |
| Oligonucleotides | ||
| Rps17qRT-PCR primers: 5’-CGCCATTATCCCCAGCAAG-3’, 5’-TGTCGGGATCCACCTCAATG-3’ | Von Moltke et al., 2017 | N/A |
| Cysltr1 qRT-PCR primers: 5’-TTGAGCCTCCACAGAACAATC-3’, 5’-TTCCTACGACTTGGCATGTTTT-3’ | Von Moltke et al., 2017 | N/A |
| Cysltr2 qRT-PCR primers: 5’-TGTCACCAGTGTCAGGAGTG-3’, 5’-ACTTTTGAGGACTCAGCTCCAA-3’ | https://pga.mgh.harvard.edu/primerbank/ | PrimerBank ID# 19526902a1 |
| Ltb4r1 qRT-PCR primers: 5’-CTGATCTGCGCTCCGAACTAT-3’, 5’-TGCGCCCGATGTCAGAGTA-3’ | https://pga.mgh.harvard.edu/primerbank/ | PrimerBank ID# 161760677C3 |
| Ltb4r2 qRT-PCR primers: 5’-ATGTCTGTCTGCTACCGTCC-3’, 5’-AGCTCCATACTACGAAGCCAT-3’ | Von Moltke et al., 2017 | N/A |
| Oxgr1 qRT-PCR primers: 5’-CAGCCAGTGATTCGGATTTCC-3’, 5’-GCTGGTCAGATACAGCAAGTC-3’ | Von Moltke et al., 2017 | N/A |
| Il25 qRT-PCR primers: 5’-ACAGGGACTTGAATCGGGTC-3’, 5’-TGGTAAAGTGGGACGGAGTTG-3’ | Von Moltke et al., 2016 | N/A |
| Pla2g4a qRT-PCR primers: 5’-TGGTGGGATTCTCTGGTGTGA-3’, 5’-GGAAAATCGGGGTGAGAGTACA-3’ | https://pga.mgh.harvard.edu/primerbank/ | PrimerBank ID# 133891677c3 |
| Alox5 qRT-PCR primers: 5’-ACTACATCTACCTCAGCCCATT-3’, 5’-GGTGACATCGTAGGAGTCCAC-3’ | https://pga.mgh.harvard.edu/primerbank/ | PrimerBank ID# 886333a1 |
| Alox5ap qRT-PCR primers: 5’-AGCATGAAAGCAAGGCGCATA-3’, 5’-GTACGCATCTACGCAGTTCTG-3’ | https://pga.mgh.harvard.edu/primerbank/ | PrimerBank ID# 33563242a1 |
| Ltc4s qRT-PCR primers: 5’-ATGAAGGACGAAGTGGCTCTT-3’, 5’-CCTGTAGGGAGAAGTAGGCTTG-3’ | https://pga.mgh.harvard.edu/primerbank/ | PrimerBank ID# 20380551a1 |
| Lta4h qRT-PCR primers: 5’-CTGACTTTGGACACAAAAGACCT-3’, 5-AGATTTCCATCGGTGACCCTT-3’ | https://pga.mgh.harvard.edu/primerbank/ | PrimerBank ID# 116734869C2 |
| Software and Algorithms | ||
| GraphPad Prism 7 | GraphPad Software | N/A |
| FlowJo 10.4.2 | Tree Star | https://www.flowjo.com/solutions/flowjo/downloads |
ACKNOWLEDGEMENTS
We thank D. Hailey and the Garvey Cell Imaging Lab in the Institute for Stem Cell & Regenerative Medicine for microscopy support, M. Black and the UW Cell Analysis Facility for flow cytometry support, the mouse husbandry staff in the UW SLU vivarium, Northwest BioSpecimen for human tissue samples, V. Gersuk, M. Rosasco, and the Benaroya Research Institute Genomics Core for help with RNA sequencing, V. Kuchroo at Brigham and Women’s Hospital for mice, and M. Fontana for helpful comments on the manuscript. JWM was supported by the University of Washington Immunology Training Grant (T32 AI106677). MSN was supported by the UW Immunology Department’s Titus Fellowship. JVM is a Searle Scholar. Work at the University of Washington was supported by NIH 1DP2 OD024087 and a Kenneth Rainin Foundation Synergy Award. The Pou2f3-CreERT2-ires-eGFP strain was generated at Monell Chemical Senses Center with funding support from NIH R01 DC015491 and NIH R01 DC017503. The Alox5-flox strain was generated at the University of California, San Francisco with generous funding support from R. Locksley(NIH R01 AI026918, UCSF Diabetes Research Center, and Sandler Asthma Basic Research Center at UCSF for support of H-EL, targeting, and mouse costs).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Bäck M, Dahlén S-E, Drazen JM, Evans JF, Serhan CN, Shimizu T, Yokomizo T, and Rovati GE (2011). International Union of Basic and Clinical Pharmacology. LXXXIV: Leukotriene Receptor Nomenclature, Distribution, and Pathophysiological Functions. Pharmacol. Rev 63, 539–584. [DOI] [PubMed] [Google Scholar]
- Barker N (2013). Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol 15, 19. [DOI] [PubMed] [Google Scholar]
- Barlow JL, Peel S, Fox J, Panova V, Hardman CS, Camelo A, Bucks C, Wu X, Kane CM, Neill DR, et al. (2013). IL-33 is more potent than IL-25 in provoking IL-13-producing nuocytes (type 2 innate lymphoid cells) and airway contraction. J. Allergy Clin. Immunol 132, 933–941. [DOI] [PubMed] [Google Scholar]
- Beller TC, Maekawa A, Friend DS, Austen KF, and Kanaoka Y (2004). Targeted gene disruption reveals the role of the cysteinyl leukotriene 2 receptor in increased vascular permeability and in bleomycin-induced pulmonary fibrosis in mice. J. Biol. Chem 279, 46129–46134. [DOI] [PubMed] [Google Scholar]
- Bezençon C, Fürholz A, Raymond F, Mansourian R, Métairon S, Le Coutre J, and Damak S (2008). Murine intestinal cells expressing Trpm5 are mostly brush cells and express markers of neuronal and inflammatory cells. J. Comp. Neurol 509, 514–525. [DOI] [PubMed] [Google Scholar]
- Boekel J, Chilton JM, Cooke IR, Horvatovich PL, Jagtap PD, Käll L, Lehtiö J, Lukasse P, Moerland PD, and Griffin TJ (2015). Multiomic data analysis using Galaxy. Nat. Biotechnol 33, 137–139. [DOI] [PubMed] [Google Scholar]
- Cardoso V, Chesné J, Ribeiro H, García-Cassani B, Carvalho T, Bouchery T, Shah K, Barbosa-Morais NL, Harris N, and Veiga-Fernandes H (2017). Neuronal regulation of type 2 innate lymphoid cells via neuromedin U. Nature 549, 277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari N, and Roper SD (2010). The cell biology of taste. J. Cell Biol 190, 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudnovskiy A, Mortha A, Kana V, Kennard A, Ramirez JD, Rahman A, Remark R, Mogno I, Ng R, Gnjatic S, et al. (2016). Host-Protozoan Interactions Protect from Mucosal Infections through Activation of the Inflammasome. Cell 167, 444–456.el4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckmann K, Rafiq A, Erdmann C, Illig C, Durschnabel M, Wess J, Weidner W, Bschleipfer T, and Kummer W (2018). Muscarinic receptors 2 and 5 regulate bitter response of urethral brush cells via negative feedback. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 32, 2903–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty TA, Khorram N, Lund S, Mehta AK, Croft M, and Broide DH (2013). Lung Type 2 innate lymphoid cells express CysLTIR that regulates Th2 cytokine production. J. Allergy Clin. Immunol 132, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante NK, Lemire P, Cruz Tleugabulova M, Prescott D, Mortha A, Streutker CJ, Girardin SE, Philpott DJ, and Mallevaey T (2016). The common mouse protozoa Tritrichomonas muris alters mucosal T cell homeostasis and colitis susceptibilityTritrichomonas muris alters T cell homeostasis. J. Exp. Med 213, 2841–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon PG, Ballantyne SJ, Mangan NE, Barlow JL, Dasvarma A, Hewett DR, Mcllgorm A, Jolin HE, and McKenzie ANJ (2006). Identification of an interleukin (IL)-25-dependent cell population that provides IL-4, IL-5, and IL-13 at the onset of helminth expulsion. J. Exp. Med 203, 1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V, Cesses P, Gamier L, Pouzolles M, Brulin B, et al. (2016). Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, et al. (2017). A single-cell survey of the small intestinal epithelium. Nature 551, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeggström JZ, and Funk CD (2011). Lipoxygenase and Leukotriene Pathways: Biochemistry, Biology, and Roles in Disease. Chem. Rev. Ill, 5866–5898. [DOI] [PubMed] [Google Scholar]
- Hofmann T, Chubanov V, Gudermann T, and Montell C (2003). TRPM5 is a voltage-modulated and Ca(2+)-activated monovalent selective cation channel. Curr. Biol. CB 13, 1153–1158. [DOI] [PubMed] [Google Scholar]
- Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV, Gallini CA, Redding K, Margolskee RF, Osborne LC, et al. (2016). Tuft cells, taste chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351, 1329–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CJC, Robertson E, Harcus Y, Grainger JR, Coakley G, Smyth DJ, McSorley HJ, and Maizels R (2015). Cultivation of Heligmosomoides Polygyrus: An Immunomodulatory Nematode Parasite and its Secreted Products. J. Vis. Exp. JoVE [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka Y, and Boyce JA (2004). Cysteinyl Leukotrienes and Their Receptors: Cellular Distribution and Function in Immune and Inflammatory Responses. J. Immunol 173, 1503–1510. [DOI] [PubMed] [Google Scholar]
- Keppler D, Huber M, Baumert T, and Guhlmann A (1989). Metabolic inactivation of leukotrienes. Adv. Enzyme Regul 28, 307–319. [DOI] [PubMed] [Google Scholar]
- Klose CSN, Mahlakoiv T, Moeller JB, Rankin LC, Flamar A-L, Kabata H, Monticelli LA, Moriyama S, Putzel GG, Rakhilin N, et al. (2017). The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature 549, 282–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei W, Ren W, Ohmoto M, Urban JF, Matsumoto I, Margolskee RF, and Jiang P (2018). Activation of intestinal tuft cell-expressed Sucnr1 triggers type 2 immunity in the mouse small intestine. Proc. Natl. Acad. Sci. U. S. A 115, 5552–5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H-E, Reinhardt RL, Bando JK, Sullivan BM, Ho l.-C., and Locksley RM (2011). Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat. Immunol 13, 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Qi Z, Li X, Du Y, and Chen Y-G (2018). Monolayer culture of intestinal epithelium sustains Lgr5 + intestinal stem cells. Cell Discov. 4, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund SJ, Portillo A, Cavagnero K, Baum RE, Naji LH, Badrani JH, Mehta A, Croft M, Broide DH, and Doherty TA (2017). Leukotriene C4 Potentiates IL-33-lnduced Group 2 Innate Lymphoid Cell Activation and Lung Inflammation. J. Immunol. Baltim. Md 1950 199, 1096–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X-C, Chen Z-H, Xue J-B, Zhao D-X, Lu C, Li Y-H, Li S-M, Du Y-W, Liu Q, Wang P, et al. (2019). Infection by the parasitic helminth Trichinella spiralis activates a Tas2r-mediated signaling pathway in intestinal tuft cells. Proc. Natl. Acad. Sci 116, 5564–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KR, O’Neill GP, Liu Q, Im D-S, Sawyer N, Metters KM, Coulombe N, Abramovitz M, Figueroa DJ, Zeng Z, et al. (1999). Characterization of the human cysteinyl leukotriene CysLT 1 receptor. Nature 399, 789. [DOI] [PubMed] [Google Scholar]
- Maekawa A, Austen KF, and Kanaoka Y (2002). Targeted gene disruption reveals the role of cysteinyl leukotriene 1 receptor in the enhanced vascular permeability of mice undergoing acute inflammatory responses. J. Biol. Chem 277, 20820–20824. [DOI] [PubMed] [Google Scholar]
- Maezawa Y, Nakajima H, Suzuki K, Tamachi T, Ikeda K, Inoue J, Saito Y, and Iwamoto I (2006). Involvement of TNF receptor-associated factor 6 in IL-25 receptor signaling. J. Immunol. Baltim. Md 1950 176, 1013–1018. [DOI] [PubMed] [Google Scholar]
- McGinty JW, and von Moltke J (2020). A three course menu for ILC and bystander T cell activation. Curr. Opin. Immunol 62, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley ET, Sui Y, Al-Kofahi Y, Millis BA, Tyska MJ, Roland JT, Santamaria-Pang A, Ohland CL, Jobin C, Franklin JL, et al. (2017). Optimized multiplex immunofluorescence single-cell analysis reveals tuft cell heterogeneity. JCI Insight 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AB, Savage AK, and Locksley RM (2015). Interleukin-33 in Tissue Homeostasis, Injury, and Inflammation. Immunity 42, 1005–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadjsombati MS, McGinty JW, Lyons-Cohen MR, Jaffe JB, DiPeso L, Schneider C, Miller CN, Pollack JL, Nagana Gowda GA, Fontana MF, et al. (2018). Detection of Succinate by Intestinal Tuft Cells Triggers a Type 2 Innate Immune Circuit. Immunity 49, 33–41.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TKA, Bucks C, Kane CM, Fallon PG, Pannell R, et al. (2010). Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464, 1367–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelly VS, Kannan Y, Coomes SM, Entwistle LJ, Rückerl D, Seddon B, MacDonald AS, McKenzie A, and Wilson MS (2016). IL-4-producing ILC2s are required for the differentiation of T H 2 cells following Heligmosomoides polygyrus infection. Mucosal Immunol. 9, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters-Golden M, and Henderson WR (2007). Leukotrienes. N. Engl. J. Med 357, 1841–1854. [DOI] [PubMed] [Google Scholar]
- Prawitt D, Monteilh-Zoller MK, Brixel L, Spangenberg C, Zabel B, Fleig A, and Penner R (2003). TRPM5 is a transient Ca2+-activated cation channel responding to rapid changes in [Ca2+]i. Proc. Natl. Acad. Sci 100, 15166–15171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese TA, Liang H-E, Tager AM, Luster AD, Rooijen NV, Voehringer D, and Locksley RM (2007). Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature 447, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo-Gonzalez RR, Van Dyken SJ, Schneider C, Lee J, Nussbaum JC, Liang H-E, Vaka D, Eckalbar WL, Molofsky AB, Erle DJ, et al. (2018). Tissue signals imprint ILC2 identity with anticipatory function. Nat. Immunol 19, 1093–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzer CA, and Kargman S (1988). Translocation of 5-lipoxygenase to the membrane in human leukocytes challenged with ionophore A23187. J. Biol. Chem 263, 10980–10988. [PubMed] [Google Scholar]
- Sato T, and Clevers H (2013). Primary mouse small intestinal epithelial cell cultures. Methods Mol. Biol. Clifton NJ 945, 319–328. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. [DOI] [PubMed] [Google Scholar]
- Schneider C, O’Leary CE, von Moltke J, Liang H-E, Ang QY, Turnbaugh PJ, Radhakrishnan S, Pellizzon M, Ma A, and Locksley RM (2018). A Metabolite-Triggered Tuft Cell-ILC2 Circuit Drives Small Intestinal Remodeling. Cell 174, 271–284.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz B, Ruppert A-L, Strobel O, Lazarus M, Urade Y, Büchler MW, and Weihe E (2019). Distribution pattern and molecular signature of cholinergic tuft cells in human gastrointestinal and pancreatic biliary tract. Sci. Rep 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih H-Y, Sciumè G, Mikami Y, Guo L, Sun H-W, Brooks SR, Urban JF, Davis FP, Kanno Y, and O’Shea JJ (2016). Developmental Acquisition of Regulomes Underlies Innate Lymphoid Cell Functionality. Cell 165, 1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa C, Kanaya T, Hachisuka M, Ishiwata K, Hisaeda H, Kurashima Y, Kiyono H, Yoshimoto T, Kaisho T, and Ohno H (2017). Mast Cells Are Crucial for Induction of Group 2 Innate Lymphoid Cells and Clearance of Helminth Infections. Immunity 46, 863–874.e4. [DOI] [PubMed] [Google Scholar]
- Tait Wojno ED, Monticelli LA, Tran SV, Alenghat T, Osborne LC, Thome JJ, Willis C, Budelsky A, Farber DL, and Artis D (2015). The prostaglandin D 2 receptor CRTH2 regulates accumulation of group 2 innate lymphoid cells in the inflamed lung. Mucosal Immunol. 8, 1313–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne CA, Chen IW, Sanman LE, Cobb MH, Wu LF, and Altschuler SJ (2018). Enteroid Monolayers Reveal an Autonomous WNT and BMP Circuit Controlling Intestinal Epithelial Growth and Organization. Dev. Cell 44, 624–633.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ualiyeva S, Hallen N, Kanaoka Y, Ledderose C, Matsumoto I, Junger WG, Barrett NA, and Bankova LG (2020). Airway brush cells generate cysteinyl leukotrienes through the ATP sensor P2Y2. Sci. Immunol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozumi N, Kume K, Nagase T, Nakatani N, Ishii S, Tashiro F, Komagata Y, Maki K, Ikuta K, Ouchi Y, et al. (1997). Role of cytosolic phospholipase A 2 in allergic response and parturition. Nature 390, 618. [DOI] [PubMed] [Google Scholar]
- Urban JF, Noben-Trauth N, Donaldson DD, Madden KB, Morris SC, Collins M, and Finkelman FD (1998). IL-13, IL-4Ralpha, and Stat6 are required for the expulsion of the gastrointestinal nematode parasite Nippostrongylus brasiliensis. Immunity 8, 255–264. [DOI] [PubMed] [Google Scholar]
- Van Dyken SJ, Nussbaum JC, Lee J, Molofsky AB, Liang H-E, Pollack JL, Gate RE, Haliburton GE, Ye CJ, Marson A, et al. (2016). A tissue checkpoint regulates type 2 immunity. Nat. Immunol 17, 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannella KM, Ramalingam TR, Borthwick LA, Barron L, Hart KM, Thompson RW, Kindrachuk KN, Cheever AW, White S, Budelsky AL, et al. (2016). Combinatorial targeting of TSLP, IL-25, and IL-33 in type 2 cytokine–driven inflammation and fibrosis. Sci. Transl. Med 8, 337ra65–337ra65. [DOI] [PubMed] [Google Scholar]
- von Moltke J, Ji M, Liang H-E, and Locksley RM (2016). Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J, O’Leary CE, Barrett NA, Kanaoka Y, Austen KF, and Locksley RM (2017). Leukotrienes provide an NFAT-dependent signal that synergizes with IL-33 to activate ILC2s. J. Exp. Med 214, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrapp A, Riesenfeld SJ, Burkett PR, Abdulnour R-EE, Nyman J, Dionne D, Hofree M, Cuoco MS, Rodman C, Farouq D, et al. (2017). The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature 549, 351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A, Hwang SM, Cook MN, Hogaboom GK, and Crooke ST (1988). Interactions of 5-lipoxygenase with membranes: studies on the association of soluble enzyme with membranes and alterations in enzyme activity. Biochemistry 27, 6763–6769. [DOI] [PubMed] [Google Scholar]
- Wong CK, Cheung PFY, Ip WK, and Lam CWK (2005). Interleukin-25-induced chemokines and interleukin-6 release from eosinophils is mediated by p38 mitogen-activated protein kinase, c-Jun N-terminal kinase, and nuclear factor-kappaB. Am. J. Respir. Cell Mol. Biol 33, 186–194. [DOI] [PubMed] [Google Scholar]
- Wunder F, Tinel H, Kast R, Geerts A, Becker E, Kolkhof P, Hütter J, Ergüden J, and Härter M (2010). Pharmacological characterization of the first potent and selective antagonist at the cysteinyl leukotriene 2 (CysLT2) receptor. Br. J. Pharmacol 160, 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Figures S1-6.
Table S1. Normalized RNA-seq results, Related to Figure 2.
Table S2. Differentially expressed genes in ILC2s from Alox5−/− mice, Related to Figure 2.
Table S3. Differentially expressed genes in ILC2s from Il25−/− mice, Related to Figure 2.
Table S4. Differentially expressed genes in ILC2s from St2−/− mice, Related to Figure 2.
Data Availability Statement
RNA-seq data are available at the NCBI Gene Expression Omnibus under accession number GEO: GSE144956.


