ABSTRACT
We have recently shown that arginine methylation by protein arginine N-methyltransferase 1 (PRMT1) controls the response to cisplatin in ovarian cancer cells. In addition to increased methylation of chromatin proteins that favors senescence-associated secretory phenotype (SASP) activation, our study unraveled global hypo-methylation of RNA-binding proteins, which – we speculate – may promote their phase separation and stress granules formation.
KEYWORDS: Arginine methylation, mass spectrometry-based proteomics, PRMT1, SASP, cisplatin, stress granules, LLPS, phosphorylation
Main text
Arginine (R)-methylation is a protein post-translational modification implicated in the regulation of several cellular processes, including transcription, RNA splicing and metabolism, DNA damage response and growth factor-mediated signaling.1 R-methylation is catalyzed by the members of the protein arginine methyltransferase (PRMT) family, of which protein arginine N-methyltransferase 1 (PRMT1) is the predominant enzyme.
The recent optimization of biochemical workflows for the affinity enrichment of R-methyl peptides from cultured cells, coupled to high-resolution Mass Spectrometry (MS) analysis for their identification and quantification, has led to the expansion of the experimental R-methyl proteome annotation and to a better understanding of its plasticity in response to external stimuli.
Recently, our team has employed an MS-based proteomic approach to investigate the role of PRMT1-mediated R-methylation in response to the genotoxic stress induced by cisplatin (CDDP).2 We detected methylation changes in 92 R-sites on 56 proteins, with an overall methylation increase of histone H4 and chromatin-associated proteins, which was mirrored by the hypo-methylation of numerous soluble RNA-binding proteins (RBPs).
The increased methylation of chromatin-associated proteins following CDDP treatment was shown to be dependent on the activity of the PRKDC gene product, best known as DNA-dependent protein kinase catalytic subunit (DNA-PKcs), which promotes PRMT1 accumulation on chromatin. Here, PRMT1 induces the senescence-associated secretory phenotype (SASP) transcriptional program, a mechanism mediated by the activation of the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) pathway that, in dividing cells, serves to counteract the apoptosis triggered by sustained genotoxic stress. SASP leads to the expression and secretion of pro-inflammatory cytokines, which reinforce the DNA damage-induced cell cycle arrest and promote senescence.3
While our study mainly focused on the increase of PRMT1 activity on chromatin, in response to genotoxic stress, the observed global reduction of methylation of soluble RBPs is particularly intriguing and stimulates interesting speculations for future investigations.
In particular, we noticed that 82% of the down-regulated R-methyl sites localize within the PRMT1 recognition motif arginine-glycine-glycine/arginine-glycine (RGG/RG), which is highly enriched in low complexity (LC) regions of RBPs and is involved in liquid-liquid phase separation (LLPS) transition of this specific class of proteins.4 LLPS of RBPs is, in fact, mediated by the cationic interactions of arginines with aromatic (π) amino acids that lead to RBPs condensation and the consequent formation of membrane-less organelles. In this scenario, R-methylation of the RGG/RG motifs reduces LLPS by weakening cation–π interactions.5 In line with this, modulation of R-methylation in specific RBPs was shown to regulate stress granules (SGs), which are cytoplasmic membrane-less organelles assembled upon condensation of stalled messenger RNAs, RBPs and translation initiation factors when translation initiation is inhibited, by either drugs or stress.6 For instance, sodium arsenite induces the decrease of R-methylation of Ras GTPase-activating protein-binding protein 1 (G3BP1) and Ubiquitin-associated protein 2-like (UBAP2L), promoting SG formation, which is instead inhibited by PRMT1 overexpression.7,8
Intriguingly, when we intersected our RBPs hypo-methylated by CDDP treatment with the list of proteins annotated in the RNA Granule Database (http://rnagranuledb.lunenfeld.ca/), we found that 55% of them are functionally or structurally linked to SG formation.
Correlating all these facts with our observation that CDDP-induced DNA-PKcs-dependent phosphorylation of PRMT1 redirects PRMT1 activity toward histone H4 and chromatin-associated proteins at the cost of RBP methylation,2 we envisage a model whereby cells exposed to sustained genotoxic stress activate a signaling cascade leading to the switch of PRMT1 substrate preference toward chromatin-proteins and against soluble RBPs. These events have opposite consequences on gene expression, at different levels: SASP promotes transcriptional activation in the nucleus, and SG assembly maintains the translation inhibition of stalled messenger RNAs in the cytosol (Figure 1).
Figure 1.
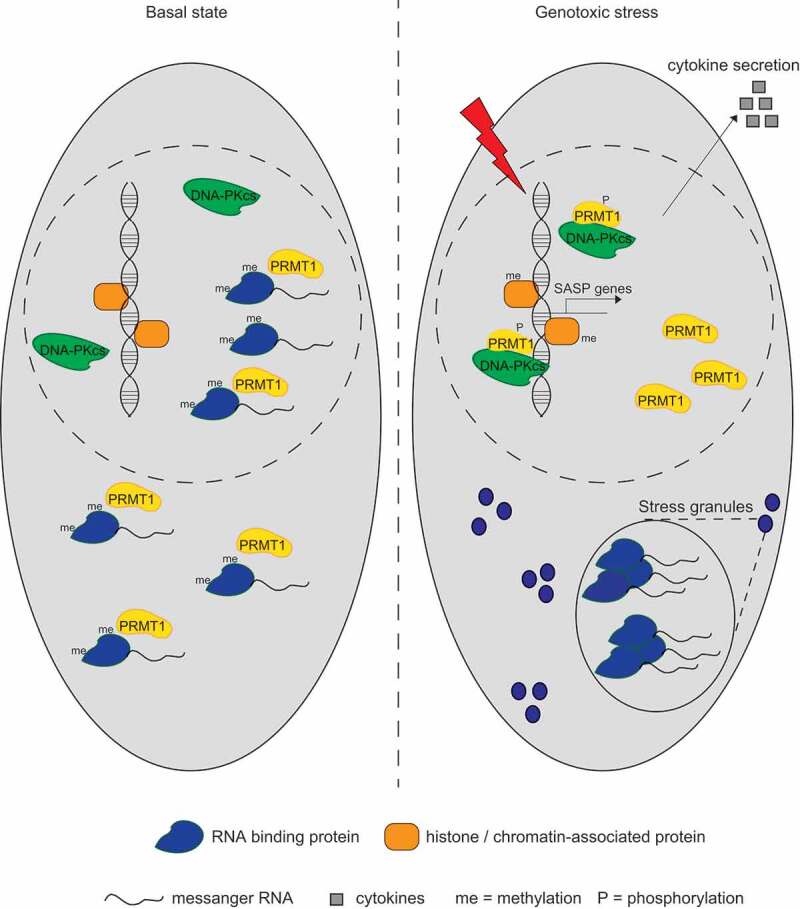
Dual role of PRMT1-dependent arginine methylation in cellular responses to genotoxic stress.
Last, as an additional layer of regulation, we noticed that several RBPs that are hypo-methylated in our settings and contain repetitive RGG/RG motifs (for example, heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) and RNA-binding protein EWS (EWSR1)) are phosphorylated during DNA Damage Response (DDR).9,10 This suggests that, upon CDDP treatment, DNA-PKcs may reduce the interaction of PRMT1 with its RBP substrates through their phosphorylation, in line with the evidence that phosphorylation can act as a molecular switch to rapidly modify protein functions and/or interactions during DDR.
Recent studies have demonstrated that RBPs undergoing LLPS are enriched not only in arginines, but also in serines (S), threonines (T), and tyrosines (Y), whose modification by methylation and phosphorylation, respectively, can regulate protein phase transitions.5 Hence, it will be very interesting, in the future, to investigate the crosstalk of ST/Y-phosphorylation and R-methylation of RBPs in the regulation of SG dynamics and – more in general – of LLPS during stress response.
Funding Statement
This work was supported by the Associazione Italiana per la Ricerca sul Cancro [IG-2018-21834].
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Guccione E, Richard S.. The regulation, functions and clinical relevance of arginine methylation. Nat Rev Mol Cell Biol. 2019;20(10):1–3. doi: 10.1038/s41580-019-0155-x. [DOI] [PubMed] [Google Scholar]
- 2.Musiani D, Giambruno R, Massignani E, Ippolito MR, Maniaci M, Jammula S, Manganaro D, Cuomo A, Nicosia L, Pasini D, et al. PRMT1 is recruited via DNA-PK to chromatin where it sustains the senescence-associated secretory phenotype in response to cisplatin. Cell Rep. 2020;30(4):1208–1222 e9. doi: 10.1016/j.celrep.2019.12.061. [DOI] [PubMed] [Google Scholar]
- 3.Faget DV, Ren Q, Stewart SA. Unmasking senescence: context-dependent effects of SASP in cancer. Nat Rev Cancer. 2019;19(8):439–453. doi: 10.1038/s41568-019-0156-2. [DOI] [PubMed] [Google Scholar]
- 4.Chong PA, Vernon RM, Forman-Kay JD. RGG/RG Motif regions in RNA binding and phase separation. J Mol Biol. 2018;430(23):4650–4665. doi: 10.1016/j.jmb.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 5.Hofweber M, Dormann D. Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J Biol Chem. 2019;294(18):7137–7150. doi: 10.1074/jbc.TM118.001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panas MD, Ivanov P, Anderson P. Mechanistic insights into mammalian stress granule dynamics. J Cell Biol. 2016;215(3):313–323. doi: 10.1083/jcb.201609081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang C, Chen Y, Dai H, Zhang H, Xie M, Zhang H, Chen F, Kang X, Bai X, Chen Z. UBAP2L arginine methylation by PRMT1 modulates stress granule assembly. Cell Death Differ. 2020;27(1):227–241. doi: 10.1038/s41418-019-0350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai WC, Gayatri S, Reineke LC, Sbardella G, Bedford MT, Lloyd RE. Arginine demethylation of G3BP1 promotes stress granule assembly. J Biol Chem. 2016;291(43):22671–22685. doi: 10.1074/jbc.M116.739573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klevernic IV, Morton S, Davis RJ, Cohen P. Phosphorylation of Ewing’s sarcoma protein (EWS) and EWS-Fli1 in response to DNA damage. Biochem J. 2009;418(3):625–634. doi: 10.1042/BJ20082097. [DOI] [PubMed] [Google Scholar]
- 10.Guil S, Long JC, Caceres JF. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol Cell Biol. 2006;26(15):5744–5758. doi: 10.1128/MCB.00224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


