ABSTRACT
Armadillo (ARM) repeat proteins constitute a large protein family with diverse and fundamental functions in all organisms, and armadillo repeat domains share high structural similarity. However, exactly how these structurally similar proteins can mediate diverse functions remains a long-standing question. Vac8 (vacuole related 8) is a multifunctional protein that plays pivotal roles in various autophagic pathways, including piecemeal microautophagy of the nucleus (PMN) and cytoplasm-to-vacuole targeting (Cvt) pathways in the budding yeast Saccharomyces cerevisiae. Vac8 comprises an H1 helix at the N terminus, followed by 12 armadillo repeats. Herein, we report the crystal structure of Vac8 bound to Atg13, a key component of autophagic machinery. The 70-Å extended loop of Atg13 binds to the ARM domain of Vac8 in an antiparallel manner. Structural, biochemical, and in vivo experiments demonstrated that the H1 helix of Vac8 intramolecularly associates with the first ARM and regulates its self-association, which is crucial for Cvt and PMN pathways. The structure of H1 helix-deleted Vac8 complexed with Atg13 reveals that Vac8[Δ19–33]-Atg13 forms a heterotetramer and adopts an extended superhelical structure exclusively employed in the Cvt pathway. Most importantly, comparison of Vac8-Nvj1 and Vac8-Atg13 provides a molecular understanding of how a single ARM domain protein adopts different quaternary structures depending on its associated proteins to differentially regulate 2 closely related but distinct cellular pathways.
Abbreviations
Ape1: aminopeptidase I; ARM: armadillo repeat; Atg: autophagy-related; AUC: analytical ultracentrifugation; Cvt: cytoplasm-to-vacuole targeting; DIC: differential interference contrast; GFP: green fluorescent protein; GST: glutathione-S-transferase; ITC: isothermal titration calorimetry; NVJ: nucleus-vacuole junction; PDB: protein data bank; PMN: piecemeal microautophagy of the nucleus; prApe1: precursor Ape1; RMSD: root-mean-square deviation; SAXS: small-angle X-ray scattering; SD-N: nitrogen starvation medium; SEC: size-exclusion chromatography; tAtg13: Atg13 construct comprising residues 567–695; tNvj1: Nvj1 construct comprising residues 229–321; tVac8: Vac8 construct comprising residues 10–515; Vac8: vacuole related 8
KEYWORDS: Atg13, autophagy, crystal structure, cytoplasm-to-vacuole targeting (Cvt), Nvj1, piecemeal microautophagy of the nucleus (PMN), Vac8
Introduction
Yeast vacuoles, the equivalent of mammalian lysosomes, are membrane-bound organelles that serve a variety of pivotal functions including storage and digestion of waste products, regulation of cellular pH, and autophagy [1–7]. Vacuoles undergo constant fusion and fission that determines their inheritance during mitosis [8–10], and genetic screens of defects in vacuole inheritance have identified several genes specifically involved in the process [9,11–13], including Vac8 (vacuole related 8), a protein with armadillo repeats (ARMs) [14,15]. Vac8 binds to Myo2 (myosin 2) via the scaffold protein Vac17, and this ternary protein complex mediates vacuole migration toward the growing bud during cell division [16]. Vac8 is composed of ARMs, and myristoylation and palmitoylation lipid modification sites at the N-terminal region that anchor it into vacuolar membranes [14,15,17]. Consequently, the ARM domain is exclusively localized in the cytoplasm, where it is involved in the formation of membrane contact sites by acting as a scaffold to interact with various proteins from other organelles [14,15].
Vac8 also plays an important role in piecemeal microautophagy of the nucleus (PMN) [18–20], during which a portion of the nucleus is transported to and degraded in the vacuole during nutrition starvation. PMN occurs at the nucleus-vacuole junction (NVJ), a well-characterized membrane contact site [18–20]. Vac8 anchored in vacuolar membranes directly associates with the outer nuclear membrane protein Nvj1, and physical interaction between Vac8 and Nvj1 mediates the formation of nucleus-vacuole membrane contact sites [21], thereby promoting PMN [18–20]. In addition, a recent study revealed that Vac8 forms a complex with Lam6/Ltc1 (lipid transfer protein anchored at membrane contact site 6) at ER-vacuole contact sites and mediates sterol transport to generate sterol-enriched vacuolar membrane domains [22].
Furthermore, Vac8 is involved in the cytoplasm-to-vacuole targeting (Cvt) pathway, which transports cytoplasmic proteins to the vacuolar lumen in a manner similar to selective autophagy [15]. Upon nutrient starvation, Vac8 interacts with the key autophagy protein Atg13 (autophagy-related 13), and the resulting complex delivers cytoplasmic hydrolases such as Ape1 (aminopeptidase I) to the vacuole, where they become activated and functional [23]. Ape1 is synthesized as a precursor (prApe1) in the cytosol and transferred into the vacuole lumen through Cvt vesicles [15,23]. While much has been learned about the Vac8-Atg13 complex and its involvement in the Cvt pathway, the molecular mechanism by which Atg13 is specifically recognized by Vac8 remains elusive. In addition, our previous study suggested that Nvj1 shares a binding interface for Vac8 with Atg13; therefore, Nvj1 or Atg13 competes for binding to Vac8 [24]. However, it is still unknown how Vac8 can discriminate between Nvj1 or Atg13, and thereby differentially regulate PMN or Cvt pathways that occur simultaneously in yeast.
In the present study, we present 2 crystal structures of Atg13 bound to Vac8. These structures, along with biochemical and in vivo experiments, reveal that Atg13 binding may induce the self-association of Vac8 by regulating the H1 helix, and the resulting Vac8-Atg13 heterotetramer adopts a superhelical structure. Furthermore, we demonstrate that the quaternary structural organization of Vac8 caused by binding of Atg13 or Nvj1 selectively supports Cvt or PMN pathways, respectively.
Results
Structure of tVac8 in complex with tAtg13
The Saccharomyces cerevisiae Atg13 protein is composed of 738 residues (Figure 1A). The sequences from the N terminus to residue 521 are well conserved among mammalian ATG13 orthologs, and required for interaction with essential components of the autophagy machinery, including Atg1, Atg17, and Atg9 [25–27]. By contrast, the remaining residues from 522 to the end of the C terminus are present only in the yeast Atg13 protein. A previous yeast two-hybrid analysis and co-immunoprecipitation experiments revealed that the C-terminal region of Atg13 comprising residues 567–692 is required for association with Vac8 [23]. Based on these data, we made a truncated Atg13 construct (residues 567–695; designated as tAtg13) for structural analysis of the tVac8-tAtg13 complex. tVac8 (residues 10–515) was used in our previous study (Figure 1A) [24]. Analytical ultracentrifugation (AUC) experiment revealed that the tVac8-tAtg13 complex formed a heterodimer in solution (Figure 1B). Crystals of tVac8-tAtg13 were grown in a reservoir buffer containing polyethylene glycol (PEG) 400 as the main precipitant, and the structure was solved by the molecular replacement method using the Vac8 coordinates (PDB code: 5XJG) as a search model and refined to 2.9 Å resolution (Table S2).
Figure 1.
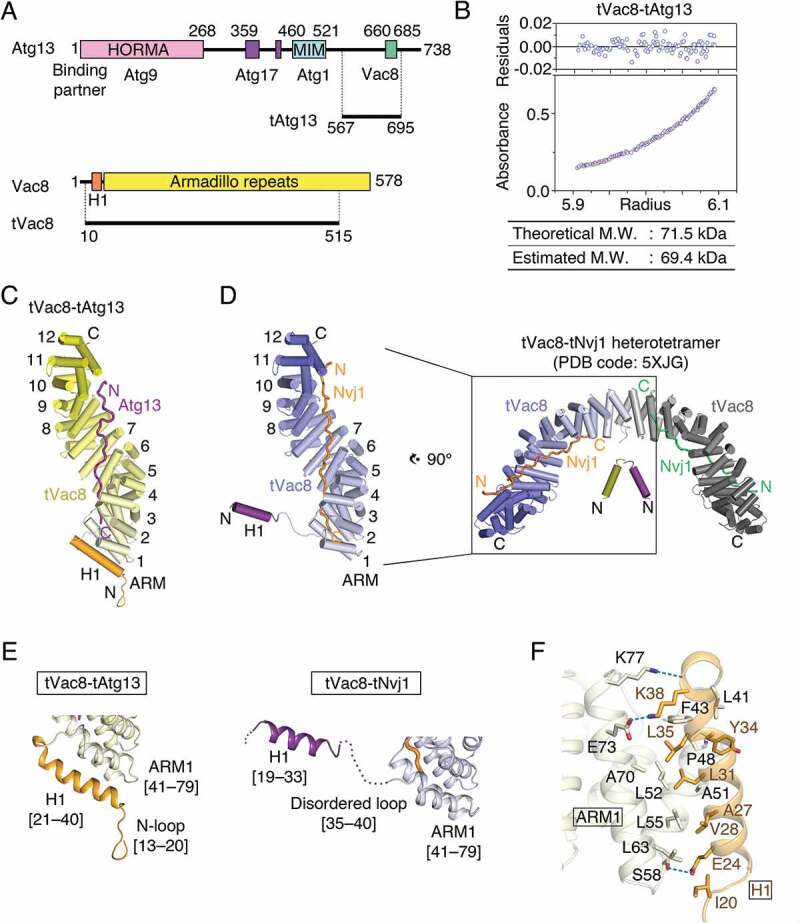
Structure of the tVac8-tAtg13 complex. (A) Domain structures of Saccharomyces cerevisiae Atg13 and Vac8. Previously identified conserved domains are marked by colored boxes, and the binding regions of associated proteins are indicated below. Residues 567–695 of Atg13 (designated as tAtg13) and residues 10–515 of Vac8 (designated as tVac8) were used for crystallization and biochemical experiments in this work. HORMA, Hop1, Rev7 and MAD2 domain; MIM, microtubule interacting and transport (MIT)-interacting motif. (B) Analytical ultracentrifugation (AUC) analysis of the molecular weight (M.W.) of the tVac8-tAtg13 complex in solution. The bottom panel indicates a representative plot of absorbance profiles of 2.5 μM tVac8-tAtg13 following a centrifugation run at 13,604 × g for 16 h at 20°C, and the top panel presents the residuals between the experimental data and the fitted line. (C) Overall structure of tAtg13 (purple) bound to the armadillo (ARM) repeat core of Vac8 (yellow). The structure was determined by molecular replacement using tVac8 as a search model (PDB code: 5XJG) and refined to 2.9 Å resolution (Table S2). The H1 regulatory helix of Vac8 is highlighted in orange (see the main text for an explanation of the role of the H1 helix). (D) Comparison of the overall structures of tVac8-tAtg13 in (C) and tVac8-tNvj1, and the structure of the tVac8 (blue)-tNvj1 (orange) complex displayed in the same orientation as in (C). A previous study revealed that the tVac8-tNvj1 complex forms a heterotetramer as shown on the right [24], while the tVac8-tAtg13 complex in (C) forms a heterodimer. The arrangement of the H1 helix in the tVac8-tNvj1 complex is emphasized as a purple cylinder. (E) Cartoon representation comparing the conformation of the H1 helices of tVac8-tAtg13 and tVac8-tNvj1 complexes. In the tVac8-tAtg13 structure (left), the H1 helix (orange) is composed of residues 21–40, and is directly associated with 2 helices of the first ARM (ARM1). By contrast, the H1 helix in the tVac8-tNvj1 complex (right, purple) consists of residues 19–33, which point away from ARM1 via a disordered loop comprising residues 35–40 (dotted line). (F) Cartoon representation showing the interactions between the H1 helix (orange) and ARM1 (pale yellow) of tVac8 in the structure of the tVac8-tAtg13 complex. Residues involved in the interaction are shown in ball-and-stick representation. Oxygen and nitrogen atoms are colored red and blue, respectively, and blue dotted lines indicate hydrogen bonds.
Figure 1C shows the overall structure of tVac8 complexed with tAtg13 in ribbon representation. In our previous study, we revealed that tVac8 is composed of an H1 helix (residues 19–33) at the N terminus, followed by 12 central armadillo repeats (ARMs; Figure 1D) [24]. The loop (residues 35–40) bridging the H1 helix and the first ARM (ARM1) is disordered in the structure (Figure 1D). The overall conformation of the 12 concerted ARMs of tVac8 observed in the crystal structure of the tVac8-tAtg13 complex was essentially identical to that observed in the tVac8-tNvj1 complex [24], with a root-mean-square deviation (RMSD) value of 0.88 Å for all Cα atoms. The 70-Å extended loop of Atg13 binds across the inner groove formed by the central ARMs of Vac8 (Figure 1C). Similar to the tVac8-tNvj1 complex, Atg13 binds to Vac8 in an antiparallel fashion (Figure 1C,D). Electron density for Atg13 was clearly visible only in the Vac8 contact region (residues 660–685), and all other residues were disordered in the structure, consistent with the structure of tNvj1 bound to tVac8 (Figure S1A) [24]. It is noteworthy that, while the tVac8-tNvj1 complex forms a heterotetramer comprising 2 tVac8-tNvj1 molecules that are organized via contacts with ARM1 of Vac8 (Figure 1D, right), such self-association of tVac8 was not observed in the structure of the tVac8-tAtg13 complex. Instead, as we proposed in a putative working model in our previous study [24], the H1 helix directly binds to ARM1 and eventually masks the dimeric interface (Figure 1E, left). The residues comprising the H1 helix in the tVac8-tAtg13 complex are different from those in the tVac8-tNvj1 complex. In particular, this helix is extended by 6 residues that are disordered in the structure of the tVac8-tNvj1 complex (Figure 1E). The side chain of Glu24 and the main chain carbonyl oxygen of Lys38 from the H1 helix engage in direct H-bonds with Ser58 and Lys77 of ARM1, respectively. In addition, the side chain of Lys38 of the H1 helix forms a salt bridge with the side chain of Glu73 of ARM1. Furthermore, Val28, Leu31, and Leu35 of H1 helix form a hydrophobic core with Ala51, Leu52, and Leu55 of ARM1 that are known to be key residues for self-association of Vac8 (Figure 1F).
Binding interface of tVac8 and tAtg13
Next, we further explored the interaction between tVac8 and tAtg13. Based on the structural conservation between tAtg13 and tNvj1, we divided the tVac8-binding region of tAtg13 into 2 parts (interface I and II) (Figure 2A). The N-terminal residues 660–671 (interface I) appear to interact loosely with ARMs 7–10 of tVac8, whereas the C-terminal residues 672–685 (interface II) are specifically recognized and tightly bound by ARMs 2–6 via both H-bonds and van der Waals interactions, resulting in a buried surface area of 956.28 Å2, which is nearly twice that of interface I (Figure 2B). In interface II, the structure of tAtg13 strongly resembles that of tNvj1 despite their negligible sequence homology (6% sequence identity between Atg13 and Nvj1; Figure 2A,B). In particular, Ile662 and Pro663 of tAtg13 in interface I form hydrophobic contacts with the side chains of Ala398, Ala401, Ile402, and Leu405 of tVac8, and His669 of tAtg13 forms intermolecular H-bonds with Arg317 and Glu395 of tVac8 (Figure 2C). In interface II, Phe678 of tAtg13 engages in extensive hydrophobic interactions with the side chains of Tyr226, Tyr227, and Leu192 of tVac8. Ala680, Phe684, and His685 at the C terminus of tAtg13 also form a tight hydrophobic network with Leu192, Ala107, and Val103 of tVac8 (Figure 2C). In addition to these extensive hydrophobic contacts, the side chains of Gln679 and Gln681 from tAtg13 form intermolecular H-bonds with the side chains of Lys160 and Asn152 from tVac8, respectively. The main chain atoms of Gln679, Phe684, and His685 of tAtg13 are recognized by the side chains of Asn193, Asn186, and Asn145 of tVac8 through intermolecular H-bonds (Figure 2C).
Figure 2.
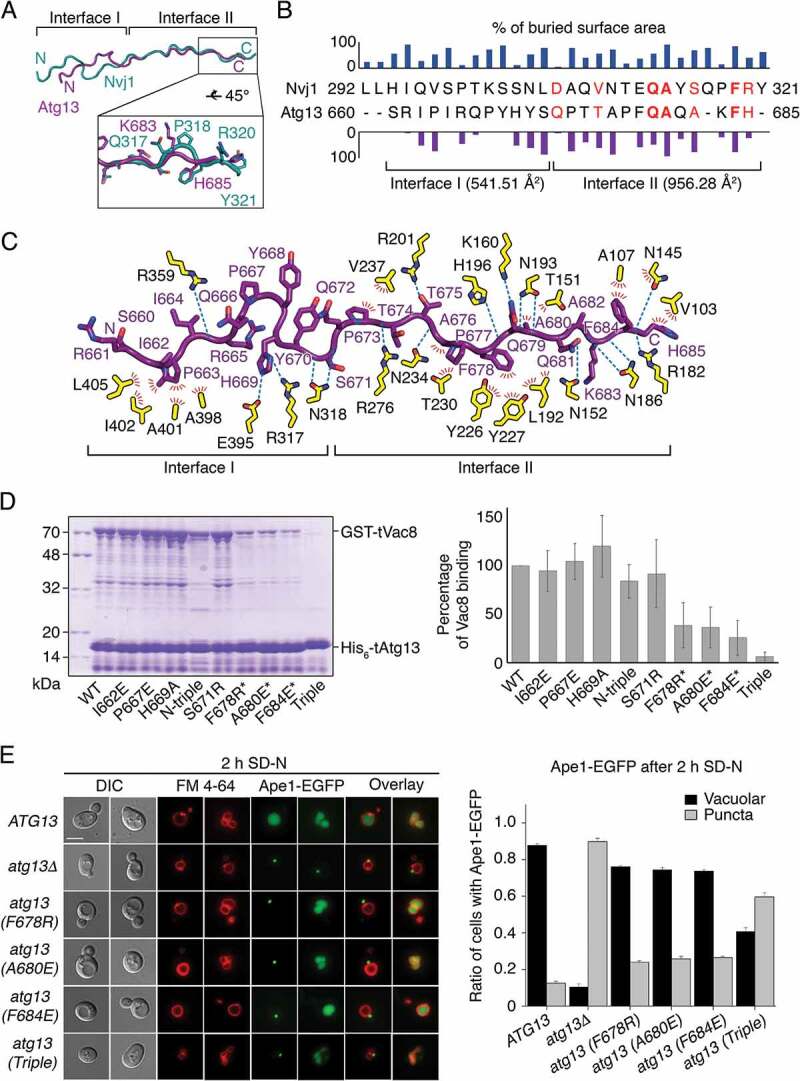
Binding interface between tAtg13 and tVac8. (A) Ribbon representation showing the superposition of tNvj1 (blue green) and tAtg13 (purple), which are bound to tVac8. Based on structural conservation, the structures of tAtg13 and tNvj1 can be divided into 2 subregions (designated as interface I and II). The bottom box shows a close-up view of the C terminus, highlighting the structural comparison. (B) Sequence alignment of tNvj1 and tAtg13 based on their structures shown in (A). Bar graphs indicate the percentage of buried surface area of each residue of tNvj1 (top) and tAtg13 (bottom), which were generated by the interaction with tVac8. Each buried surface area of interface I and II of tAtg13 to tVac8 is shown in parentheses. Identical and similar sequences are highlighted in bold red and normal red, respectively. (C) Ribbon representation showing protein-protein contacts identified in the crystal structure. Contacts between tVac8 (yellow) and tAtg13 (purple) are indicated by dotted lines (H-bonds) and red lines (hydrophobic interactions). Oxygen atoms are colored red, and nitrogen atoms are blue. (D) His6-tag affinity-isolation analysis of the interaction of tVac8 and tAtg13. N-terminal glutathione-S-transferase (GST)-fused tVac8 was co-expressed with wild-type (WT) and mutants of hexa histidine (His6)-tagged tAtg13, and immobilized on Ni2+-NTA resin. Proteins were separated by 12% SDS-PAGE and visualized by Coomassie Brilliant Blue staining. The N-triple mutant was generated by mutation of 3 residues in interface I (I662E P667E H669A). For the triple mutant, residues indicated by asterisks were simultaneously mutated. Quantification data are shown on the right (n = 3). (E) Interface II is important for the Atg13-Vac8-mediated Cvt pathway of Ape1. Vacuoles of yeast strains expressing wild-type or mutant Atg13 were labeled with FM 4–64 in YPD media at 30°C for 2 h. After free dye was removed, cells were resuspended in a nitrogen starvation medium (SD-N) and further cultured at 30°C for 2 h. GFP fluorescence from Ape1 and FM 4–64 fluorescence in vacuoles were then analyzed by fluorescence microscopy. Representative images from each cell type are shown (left). The graph shows quantification of cytoplasmic or vacuolar Ape1-EGFP (right). More than 100 cells with Ape1-EGFP were analyzed per strain. Scale bar: 5 μm.
To characterize the interaction of Vac8 and Atg13 in solution, we generated a series of tAtg13 mutants conjugated with a hexa histidine (His6)-tag at the N terminus, and co-expressed each mutant with glutathione-S-transferase (GST)-tagged tVac8. As shown in Figure 2D, affinity-isolation experiments revealed that mutations in interface I did not affect the interaction, whereas single mutation of residues Phe678, Ala680, or Phe684 in interface II reduced association with tVac8. Consistently, triple mutation (F678R A680E F684E) of interface II completely abolished association with tVac8. We also measured binding constants (Kd) between Vac8 and tAtg13 mutants using isothermal titration calorimetry (ITC), and confirmed the results of affinity-isolation experiments (Figure S2). To investigate the importance of interface II in vivo, we examined the Cvt pathway of Ape1, which requires the Vac8-Atg13 interaction [23], by expressing one of the Atg13 mutants harboring a mutation in interface II in atg13Δ yeast cells (Figure 2E). While green fluorescent protein (GFP)-conjugated Ape1 was transported to the vacuolar lumen via the Cvt pathway in nearly 90% of wild-type yeast cells upon nitrogen starvation, only 10% of atg13Δ cells contained GFP-Ape1 in the vacuolar lumen. Cytoplasmic GFP puncta, indicative of defects in the Cvt pathway, were observed in most atg13Δ cells, confirming that Atg13 is essential for the Cvt pathway of Ape1 (Figure 2E). Furthermore, although each of the single mutations in interface II resulted in a minor defect in the Cvt pathway of Ape1, the defect was multiplied in cells expressing the Atg13 mutant with all 3 mutations (F678R A680E F684E), demonstrating that interface II is critical for the interaction between Atg13 and Vac8, and thereby their function in the Cvt pathway (Figure 2E). Consistently, vacuolar processing of prApe1, which requires the Cvt pathway, was impaired by these mutations in interface II (Figure S3A–C). By contrast, the mutant bearing all 3 mutations (I662E P667E H669A; referred to as N-triple) in interface I supported the Cvt pathway comparably to wild-type, consistent with the results of the in vitro affinity-isolation assay of the N-triple mutant (Figure 2D; Figure S3D).
In addition, a recent study elucidated 2 sets of motifs comprising positively charged residues (640KFK642 and 683KFHK686) in the Atg13 intrinsic disordered region (IDR, residues 571–700), which are responsible for interactions with phospholipids that are essential for efficient autophagy (Gatica et al., co-submitted manuscript). Among these positively charged residues, Lys683 and His685 are observable in the tVac8-tAtg13 structure, and their side chains are exposed to bulk solvent at the surface of the molecule (Figure 2C), further supporting their interactions with phospholipids. The presence of this second motif (683KFHK686) in interface II is also consistent with our observation that interface II is critical for the Cvt pathway (Figure 2E).
Structure of Vac8[Δ19–33] in complex with tAtg13
Based on combined structural and biochemical results, we proposed that a flexibly linked H1 helix is involved in regulating self-association of Vac8 through a folding-back mechanism in which the H1 helix binds ARM1 and masks the dimeric interface [24]. To advance our understanding of how the H1 helix regulates the self-association of Vac8, we generated an internal deletion construct lacking the H1 helix (residues 19–33; hereafter referred to as Vac8[Δ19–33]; Figure 3A). We measured the binding affinity using ITC and found that Vac8[Δ19–33] bound to tAtg13 with a Kd of 1.46 μM, indicating that deletion of the H1 helix only had a slight influence on the binding affinity (Figure 3B). Notably, AUC analysis confirmed that Vac8[Δ19–33]-tAtg13 indeed formed a heterotetramer in solution (Figure 3C). The Vac8[Δ19–33]-tAtg13 complex was crystallized in space group P212121 with 2 copies of the complex in the asymmetric unit, and the structure was solved by molecular replacement, and eventually refined at 3.2 Å resolution with Rwork/Rfree values of 0.22/0.25 (Figure S1B; Table S2 for data collection and refinement statistics).
Figure 3.
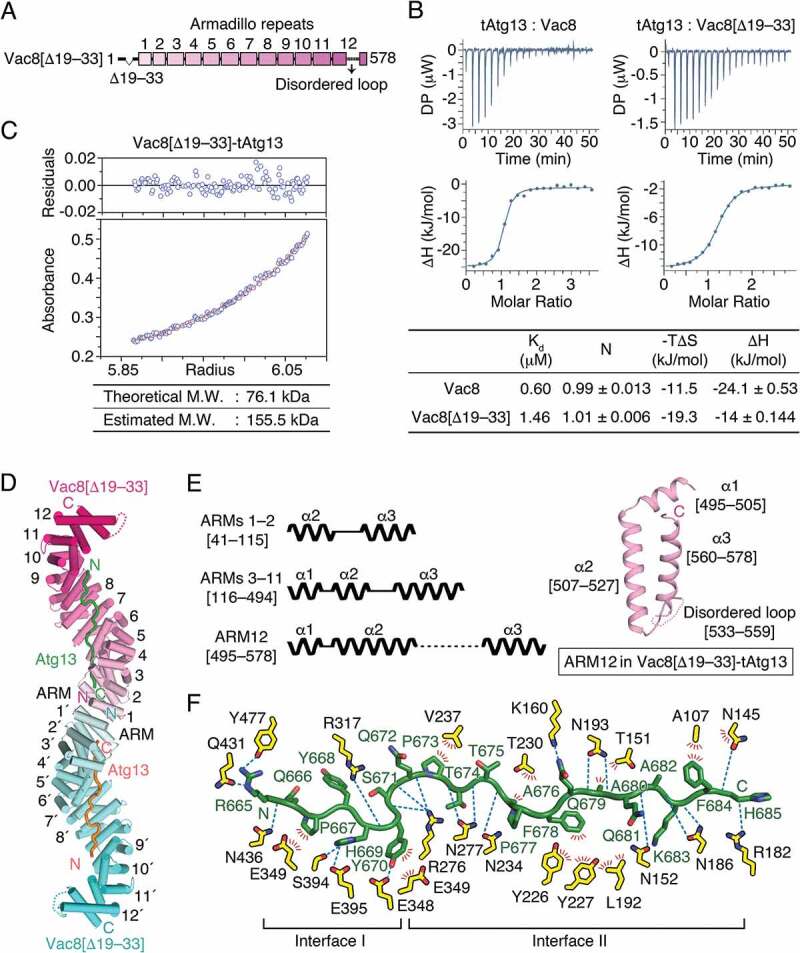
Structure of Vac8[Δ19–33] in complex with tAtg13. (A) Schematic diagram showing the organization of the ARMs of Vac8 revealed in this study. To investigate the role of the H1 helix in the self-association of Vac8, a truncated construct lacking the H1 helix (Vac8[Δ19–33]) was generated. (B) Isothermal titration calorimetry (ITC) analysis of tAtg13 binding to Vac8. The tAtg13 protein was titrated into full-length Vac8 (left) or Vac8[Δ19–33] (right) at 25°C. The upper panel shows raw data, and the lower panel shows data fitted to binding isotherms to calculate affinities. The table below thermograms summarizes the measured thermodynamic parameters. (C) AUC analysis of the molecular weight (M.W.) of the Vac8[Δ19–33]-tAtg13 complex performed as described in Figure 1B. (D) Overall structure of the Vac8[Δ19–33]-tAtg13 complex. Two copies of Vac8[Δ19–33] (cyan and pink) bound to tAtg13 (orange and green) are associated in the crystal asymmetric unit. Unlike tVac8-tAtg13 (Figure 1C), Vac8[Δ19–33]-tAtg13 forms a heterotetramer organized by self-association of ARM1 of Vac8[Δ19–33]. In the structure, Vac8[Δ19–33] has 12 intact ARMs, and tAtg13 binds to the inner groove generated by the ARMs of Vac8[Δ19–33] in an antiparallel manner. (E) Schematic diagram (left) illustrating the 3 types of ARMs present in the structure of Vac8[Δ19–33]. ARMs 1–2 consist of only 2 helices (α2 and α3; labeled to show the organization of helices in ARMs) that play important roles in Vac8 self-association. ARMs 3–11 have a typical ARM repeat comprising 2 short helices (α1 and α2) and a long helix (α3). ARM12 consists of a short helix (α1) and a relatively long helix (α2) followed by a long disordered region (residues 533–559) and helix α3. The right panel shows the structure of ARM12 in the Vac8[Δ19–33]-tAtg13 complex. (F) Close-up view of Atg13 (green) bound to Vac8[Δ19–33] (yellow) oriented as in Figure 2C.
A molecular model of Vac8[Δ19–33] bound to tAtg13 is presented in Figure 3D. As demonstrated by AUC experiment (Figure 3C), the Vac8[Δ19–33]-tAtg13 complex forms a heterotetramer, with 2 complexes self-associated through ARM1 of Vac8, adopting a superhelical shape with overall dimensions of 51 × 57 × 213 Å, that turns twice along the superhelical axis. Unlike tVac8-tAtg13 or tVac8-tNvj1 complexes that include a truncated Vac8 terminating at C-terminal residue 515, full-length Vac8 was used in the Vac8[Δ19–33]-tAtg13 complex, although the H1 helix was deleted. In the Vac8[Δ19–33]-tAtg13 structure, Vac8 N-terminal residues 1–18 remain disordered, indicating that this region is intrinsically flexible, even though corresponding sequences are highly conserved in other species. More importantly, the structure revealed that the C-terminal end is absolutely required to form an intact final armadillo repeat (ARM12). In our previous tVac8-tNvj1 study, we showed that C-terminal truncation of Vac8 results in an incomplete α2 helix of Vac8 ARM12 [24]. According to the structure revealed in the present study, the α2 helix is extended up to residue 527, and the α3 helix starts at residue 560 and finishes at residue 578, which is the C-terminal end (Figure 3E). The 27 residues from 533 to 559 that connect α2 and α3 helices of ARM12 are disordered in the structure. Based on this observation, we conclude that full-length sequences are absolutely required to form all 12 ARMs in Vac8. Given that proper organization of all ARMs might be critical for structural integrity, the whole sequence of Vac8 is likely important for performing biological functions. Indeed, the binding affinity of tVac8 for tNvj1 is reduced by 7-fold compared with the full-length Vac8 despite the fact that ARM12 does not directly contribute to the interaction [24].
The structure of tAtg13 bound to Vac8[Δ19–33] at interface I revealed a different conformation compared with the tVac8-tAtg13 complex (Figure 3F; Figure S4). In particular, the N-terminal 5 residues of tAtg13 are disordered in the structure of tAtg13 bound to Vac8[Δ19–33], and the interactions of Vac8[Δ19–33] and Atg13 in interface I differ from those in the tVac8-tAtg13 complex (Figure 2C, 3F; Figure S4A,B). Consistent with the tVac8-tAtg13 complex, contacts contributed by interface I do not appear to be strong, and only 525 Å2 of solvent-accessible surface area is buried. Detailed analysis in Figure S4C,D indicated that the conformation of tAtg13 bound to Vac8[Δ19–33] is more likely in solution because the N-terminal structure of tAtg13 bound to tVac8 appears to be affected by neighboring molecules due to crystal packing. The structure of tAtg13 at interface II in the 2 crystal structures superimposed precisely, suggesting that these interactions are orderly and functionally important (Figure S4B).
Dimerization of Vac8 in the Vac8-Atg13 complex
Self-association of Vac8 is required for formation of the nucleus-vacuole junction (NVJ) [24]. Herein, we also showed that Vac8 bound to Atg13 oligomerized through ARM1 of Vac8 (Figure 3D). Figure 4A highlights the 2-fold interface generated by pairwise arrangement of helices α2 and α3 from ARM1 of each subunit. Ser66 lies at the center of the 2-fold symmetry axis, and Glu73 engages in an H-bond with Asn62 of the other subunit. In addition, Ala51 and Leu52 form van der Waals contacts with Leu55, Leu63, and Ala70 of the other subunit (Figure 4A). To confirm that ARM1 is important for Vac8 dimerization in solution, we performed site-directed mutagenesis to generate the A51R and L55R mutants with altered contact sites, and conducted size-exclusion chromatography (SEC) with Vac8[40–515] lacking residues 1–39, also used in our previous study [24]. The Vac8[40–515]-tAtg13 complex migrated with a retention time consistent with the heterotetramer, while mutants eluted from the column as apparent heterodimers (Figure 4B).
Figure 4.
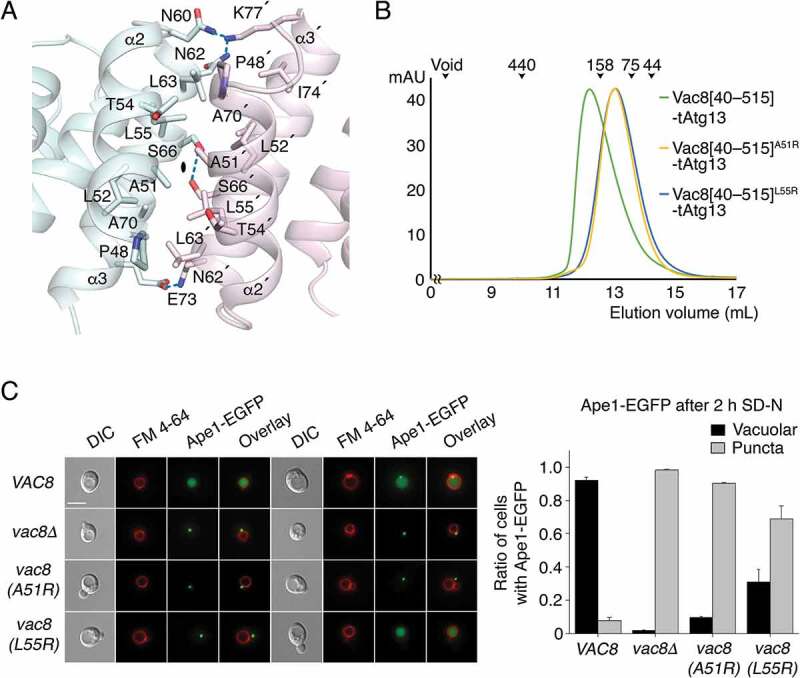
ARM1 of Vac8 mediates self-association of Vac8 in the Vac8-Atg13 complex. (A) Close-up view of the 2-fold dimeric interface of Vac8[Δ19–33]. Residues involved in the self-association of ARM1 are displayed in ball-and-stick representation. Intermolecular hydrogen bonds are indicated with blue dotted lines, and oxygen and nitrogen atoms are colored red and blue, respectively. (B) Size-exclusion chromatography (SEC) of wild-type and mutant (A51R or L55R) of Vac8[40–515] in complex with tAtg13. The indicated protein mixtures were applied to a Superdex 200 10/300 GL column equilibrated with buffer containing 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM DTT. The molecular weights of standard proteins are presented above the curves to compare the relative molecular mass (blue dextran 2000, void; ferritin, 440 kDa; aldolase, 158 kDa; conalbumin, 75 kDa; ovalbumin, 44 kDa). (C) Vac8 self-association in the Vac8-Atg13 complex is critical for the Cvt pathway of Ape1. Vacuoles of yeast strains expressing wild-type or mutant Vac8 were labeled with FM 4–64 in YPD media at 30°C for 2 h. Experiment was performed as described in Figure 2E. Representative images (left) are shown for each strain. The graph shows quantification of cytoplasmic or vacuolar Ape1-EGFP (right). Scale bar: 5 μm.
To further assess if Vac8 self-association is critical for the Cvt pathway, we replaced wild-type Vac8 with a mutant harboring A51R or L55R mutation, and examined whether the mutants support the Cvt pathway of Ape1 (Figure 4C). The results revealed that the Cvt pathway of Ape1 was impaired in yeast cells expressing the Vac8 mutants defective for self-association, clearly indicating that Vac8 dimerization in the Vac8-Atg13 complex is critical for the Cvt pathway. Consistently, conversion of the precursor form of Ape1 (prApe1) to the mature form of Ape1 in the vacuole was markedly impaired by A51R or L55R mutation (Figure S5). Triton X-114 phase partitioning confirmed that the Vac8 mutants did not affect myristoylation or palmitoylation, and thus the vacuolar targeting of Vac8 (Figure S6).
Structural comparison of Vac8-Atg13 and Vac8-Nvj1 complexes
As highlighted in Figure 5A, self-associated Vac8 exhibits striking differences in quaternary organization when binding to Atg13 or Nvj1. When bound to Nvj1, 2 Vac8 molecules are arranged via 2-fold symmetry around the axes of the ARM1 helices, resulting in an arch-shaped structure (Figure 5A,B, left). By contrast, when bound to Atg13, 2 Vac8 molecules adopt a superhelical structure in which the first ARMs are organized via 2-fold rotation symmetry around the axis perpendicular to the plane (Figure 5A,B, right).
Figure 5.
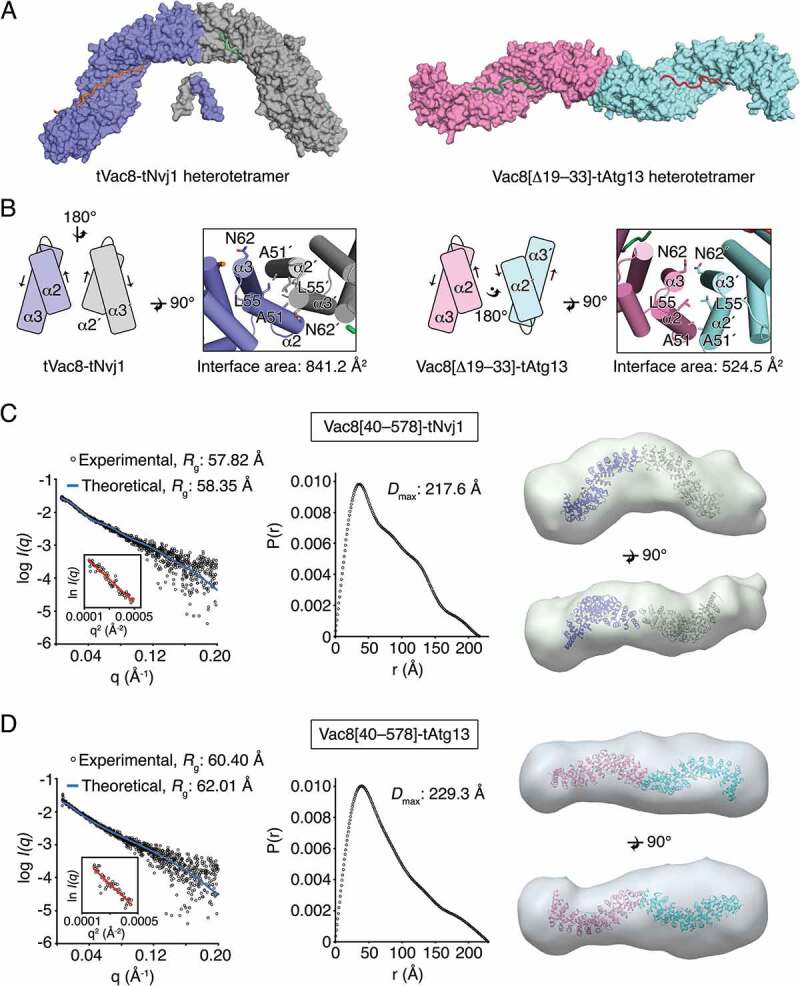
Binding of Atg13 or Nvj1 affects the quaternary structure of Vac8. (A) Surface representation of the tVac8-tNvj1 (left) and Vac8[Δ19–33]-Atg13 (right) heterotetrameric structures. In the crystal structures, 2 complexes of tVac8-tNvj1 adopt an arched shape. By contrast, 2 copies of Vac8[Δ19–33]-tAtg13 are arranged via 2-fold symmetry along the axis perpendicular to the plane, resulting in a superhelical shape. (B) The organization of ARM1 at the 2-fold interface in tVac8-tNvj1 (left) and Vac8[Δ19–33]-tAtg13 (right) heterotetramers. Arrows indicate the direction of the helical axis. (C and D) Small-angle X-ray scattering (SAXS) experimental data showing different quaternary structures of Vac8-Nvj1 (C) and Vac8-Atg13 (D). Experimental scattering data (hollow circles) of Vac8[40–578]-tNvj1 (C, left) or Vac8[40–578]-tAtg13 (D, left) agree well with theoretical scattering curves (blue line) calculated from structures of Vac8[Δ19–33]-tNvj1 or Vac8[Δ19–33]-tAtg13. A Guinier plot of Vac8[40–578]-tNvj1 (C) and Vac8[40–578]-tAtg13 (D) is shown on the left. The middle panels show the distance distributions and maximum dimensions (Dmax) of Vac8[40–578]-tNvj1 (C) and Vac8[40–578]-tAtg13 (D). Envelopes calculated from Vac8[40–578]-tNvj1 (C, right) and Vac8[40–578]-tAtg13 (D, right) SAXS data align well with structures of Vac8[Δ19–33]-tNvj1 and Vac8[Δ19–33]-tAtg13, respectively. The alignment was carried out using SUPCOMB [46], and model and envelope representations were generated by Chimera [48].
To confirm the distinct organization of Vac8 dimers in solution, we performed synchrotron small-angle X-ray scattering (SAXS) experiments using Vac8[40–578]-tNvj1 and Vac8[40–578]-tAtg13 complexes (Figure 5C,D; Table S3), in which Vac8 construct comprising residues 40–578 was used for minimalizing flexible regions and obtaining information from the complete ARM domain. The radius of gyration (Rg) calculated from one-dimensional experimental scattering plot of the Vac8[40–578]-tNvj1 complex (57.82 Å) was different from that of Vac8[40–578]-tAtg13 (60.40 Å; Figure 5C,D, left). However, experimental SAXS scattering data of Vac8[40–578]-tNvj1 or Vac8[40–578]-tAtg13 complexes fitted well to theoretical scattering curves calculated from the structure of Vac8[Δ19–33]-tNvj1, in which tVac8 coordinates of the tVac8-tNvj1 were replaced by that of Vac8[Δ19–33], or Vac8[Δ19–33]-tAtg13 crystal structures, respectively, in good agreement with theoretical Rg values (58.35 Å for Vac8[Δ19–33]-tNvj1 and 62.01 Å for Vac8[Δ19–33]-tAtg13; Figure 5C,D, left). The maximum dimension (Dmax) value for Vac8[40–578]-tNvj1 (217.6 Å) calculated from the P(r) distance distribution function was different from that of Vac8[40–578]-tAtg13 (229.3 Å; Figure 5C,D, middle). Most strikingly, the envelopes derived from Vac8[40–578]-tNvj1 and Vac8[40–578]-tAtg13 revealed distinctive quaternary structures in solution (Figure 5C,D, right). In particular, the SAXS envelope obtained from Vac8[40–578]-tNvj1 displayed an arched shape that superimposed well with the structure of the Vac8[Δ19–33]-tNvj1 heterotetramer (Figure 5C, right), whereas the envelope obtained from Vac8[40–578]-tAtg13 clearly adopted an extended rod shape that aligned well with the superhelical structure of the Vac8[Δ19–33]-tAtg13 complex (Figure 5D, right). In addition, the molecular weights of the Vac8[40–578]-tNvj1 and Vac8[40–578]-tAtg13 complexes measured by SAXS using Qp [28] were 132.16 kDa and 139.13 kDa, respectively, and these were also consistent with the heterotetramer complexes (2 × Vac8[40–578]-tNvj1 and 2 × Vac8[40–578]-tAtg13; Table S3). Taken together, the SAXS data strongly support the crystal structures, revealing that Vac8 adopts different quaternary structures in solution depending on its binding partners.
Quaternary structures of Vac8 differentially affect Cvt and PMN pathways
To confirm whether the characteristic organization of Vac8 is altered by binding of Atg13 or Nvj1 in solution, Vac8[40–515] mutations were generated. Ala51, involved in the association in both the arched and superhelical Vac8 structures, was mutated first, followed by Asn60 and Asn62 that contribute to the superhelical organization of Vac8 but project outward in the arched structure, and thus are not involved in the interaction (Figure 5B). Chemical cross-linking experiments followed by immunoblotting analysis with Nvj1 or Atg13 antibodies revealed that the A51R mutant failed to undergo self-association in the presence of tNvj1 or tAtg13, consistent with the SEC experiments (Figure 4B, 6A,B) [24]. Interestingly, the N62R single mutation or N60R N62R double mutation did not affect self-association of Vac8[40–515] bound to tNvj1, whereas the self-association of Vac8[40–515] was significantly reduced in the Vac8[40–515]N60R,N62R-tAtg13 complex (Figure 6A,B; Figure S7). Consistent with these results, the N62R single mutant of Vac8[40–515] complexed with tAtg13 eluted later from the SEC column, and the N60R N62R double mutation resulted in a more severe defect than the N62R single mutation while these mutants bound to tNvj1 eluted at heterotetrameric size (Figure 6C,D). Taken together, the results showed that mutation of residues Asn60 and Asn62 only affected the superhelical conformation generated by formation of the Vac8-Atg13 complexes, corroborating the idea that the quaternary organization of Vac8-Atg13 is clearly different from that of the Vac8-Nvj1 complex.
Figure 6.
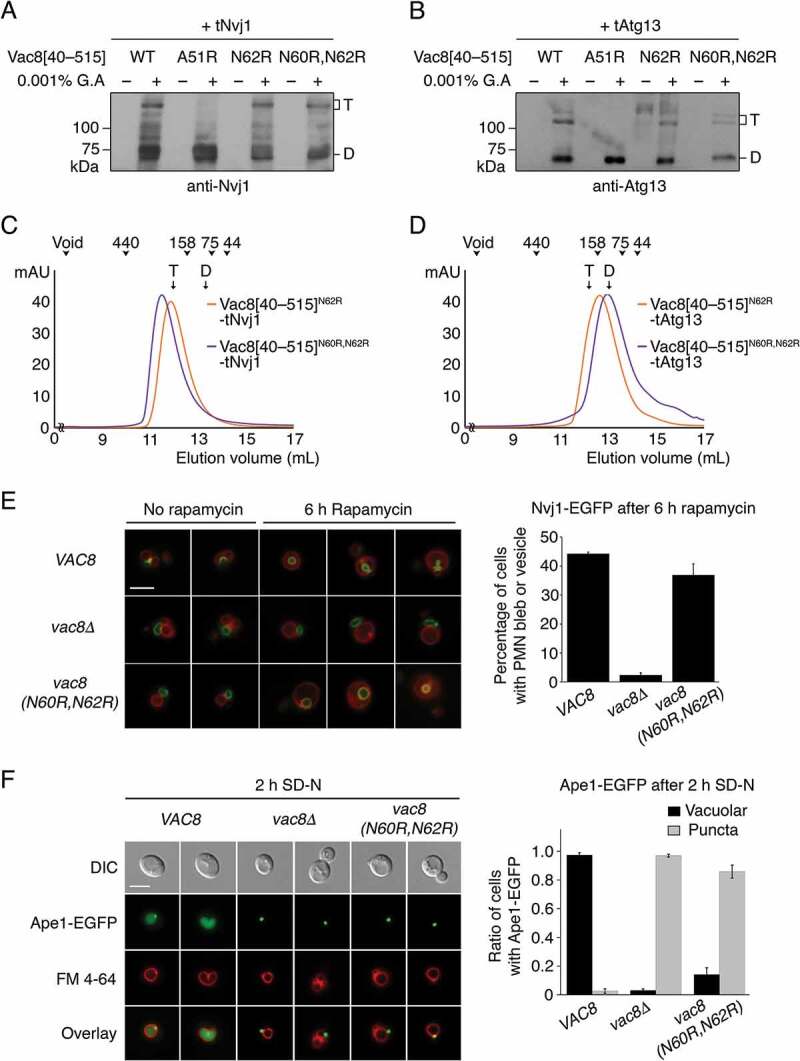
Different quaternary structures affect Cvt and PMN pathways. (A and B) Glutaraldehyde-mediated cross-linking of WT and mutant Vac8[40–515] complexed with tNvj1 (A) or tAtg13 (B). Proteins (0.5 mg/mL) were mixed with 0.001% glutaraldehyde (G.A) and incubated at 20°C for 30 min. Reaction products were separated by 8% SDS-PAGE, transferred to a PVDF membrane, and immunoblotted with anti-Nvj1 or anti-Atg13 antibody. T and D indicate Vac8[40–515]-tNvj1 (A) or Vac8[40–515]-tAtg13 (B) heterotetramer and heterodimer complexes, respectively. (C and D) SEC analysis of the N62R single mutant and N60R N62R double mutant of Vac8[40–515] complexed with tNvj1 (C) or tAtg13 (D). The experiment was performed as described in Figure 4B. T and D indicate the elution volumes of Vac8[40–515]-tNvj1 (C) or Vac8[40–515]-tAtg13 (D) heterotetramer and heterodimer complexes obtained from our previous study [24] and Figure 4B, respectively. (E) Piecemeal microautophagy of the nucleus (PMN) is largely intact in cells expressing the N60R N62R Vac8 double-mutant. Cells expressing Nvj1-EGFP were treated with 0.2 μM rapamycin for 6 h to induce PMN. GFP fluorescence from Nvj1 and red fluorescence (FM 4–64) from vacuoles were analyzed by fluorescence microscopy. Representative images (left) from each cell type with or without rapamycin (scale bar: 5 μm) and quantification (right) of cells with PMN bleb or PMN vesicles are shown. Yeast strains lacking the major vacuolar protease Pep4 were used to block the degradation of PMN blebs or vesicles in the vacuolar lumen. (F) The Vac8N60R,N62R mutant does not support the Cvt pathway. Vacuoles of wild-type yeast cells, vac8Δ cells, and vac8 (N60R,N62R) cells were labeled with FM 4–64 in YPD media at 30°C for 2 h. Experiment was performed as described in Figure 2E. Representative images (left) are shown for each strain. The graph shows quantification of cytoplasmic or vacuolar Ape1-EGFP (right). Scale bar: 5 μm.
To investigate the biological relevance of Atg13-specific Vac8 organization, we examined whether the Vac8N60R,N62R mutant supported the Cvt or PMN pathways (Figure 6E,F). As shown in Figure 6F, the Vac8N60R,N62R mutant failed to support the Cvt pathway, but interestingly, the PMN pathway was not affected (Figure 6E). These results strongly suggest that Vac8 adopts different quaternary structures depending on its binding partners, and in doing so mediates different cellular processes.
Discussion
Vac8 is composed of 12 ARMs across the whole sequence, and has a myristoylation and 3 palmitoylation sites in the N-terminal region for anchoring into vacuolar membranes, followed by an H1 helix, and ARM1 starts at residue 41 (Figure 1E) [14,15,17,24]. In a previous study, we suggested that the H1 helix plays a pivotal role in regulating the dynamic organization of Vac8 [24]. The present study confirmed this putative regulation mechanism by showing that the H1 helix directly interacts with ARM1 and hinders the dimeric interface (Figure 1C,E). In more detail, the N-terminal H1 helix is placed next to ARM1 that also serves as a binding interface for the formation of the 2-fold dimer of Vac8 (Figure 1E). Intramolecular association of the H1 helix and ARM1 regions blocks exposure of the 2-fold interface, thereby hindering self-association of Vac8, which is crucial for the PMN and Cvt pathways. We previously reported that disruption of Vac8 dimerization markedly affects both PMN and Cvt pathways [24] (also see Figure 4C). Structural comparison of tVac8-tNvj1, in which the H1 helix is dissociated from ARM1 (referred to as “active”; Figure 1D), and tVac8-tAtg13 in which the H1 helix is bound to ARM1 (referred to as “inactive”; Figure 1C), revealed that the residues comprising the H1 helix are altered between active and inactive conformations. In the “inactive” conformation, the C terminus is extended by 6 residues that are disordered in the “active” conformation (Figure 1E). The dynamic organization of the sequence and structure of the H1 helix could in principle regulate the conversion from the “inactive” to the “active” states. The regulatory importance of the H1 helix is further supported by high sequence conservation across Vac8 homologs; even the residues in the disordered region (residues 35–40) of the “active” state are highly conserved among other species [24].
Given that the self-association of Vac8 is important for both PMN and Cvt pathways, it is noteworthy that one role of the H1 helix might be to initiate these pathways. We therefore attempted to identify factors triggering conformational changes in the H1 helix by focusing on a possible covalent modification in the vicinity of this secondary structural element. Interestingly, global analysis of phosphorylation sites in the cyclin-dependent kinase Cdk1 in S. cerevisiae identifies residues Ser11 and Ser16 of Vac8 as strong candidate sites for Cdk1-mediated phosphorylation [29]. Ser11 is disordered in the structure of both tVac8-tAtg13 and Vac8[Δ19–33]-tAtg13 complexes, suggesting that this region is structurally flexible. Meanwhile, Ser16 is visible in the tVac8-tAtg13 complex, and the side chain is exposed to the surface and hence solvent-accessible (Figure S8). Moreover, because its side-chain hydroxyl group does not participate in any intermolecular interactions with other residues, this residue could potentially be phosphorylated by kinases. More importantly, Ser16 is located just beneath the N terminus of the H1 helix, indicating that modification of this residue could affect the conformation of the H1 helix (Figure S8).
To explore whether Ser11 and/or Ser16 are phosphorylated in vivo, we attempted to identify phosphorylation sites of Vac8 by LC-MS/MS analysis (Figure 7A). The results revealed that Ser16 was indeed phosphorylated, but we failed to detect phosphorylation of Ser11. Although these results do not necessarily rule out the possibility that Ser11 is a phosphorylation site, we focused on investigating the importance of Ser16 in the regulation of Cvt and PMN pathways by mutating this residue to alanine or glutamate. As shown in Figure 7C,D, both PMN and Cvt pathways were severely impaired when Ser16 was replaced with alanine. Intriguingly, these pathways were similarly impaired when Ser16 was replaced with glutamate, a phosphomimetic amino acid (Figure 7E,F). Given that the alanine or glutamate mutation did not impair the vacuolar localization of Vac8 (Figure 7B), these results strongly suggest that Ser16 is essential for the PMN and Cvt pathways, and its phosphorylation must be elaborately controlled in a spatiotemporal manner for these pathways [30]. Our results imply that the fundamental functions of Vac8 may potentially be regulated by signal-mediated processes through phosphorylation, although it remains elusive whether phosphorylation of Ser16 per se might directly cause structural changes in the H1 helix, resulting in switching between its active and inactive conformations (Figure 7G). In the same vein, our findings also imply that the formation of membrane contact sites, such as NVJs, may be regulated by external signaling pathways. Activation or inhibition of these pathways may trigger formation or disruption of NVJs, and thereby regulate PMN by modulating the structures and functions of Vac8.
Figure 7.
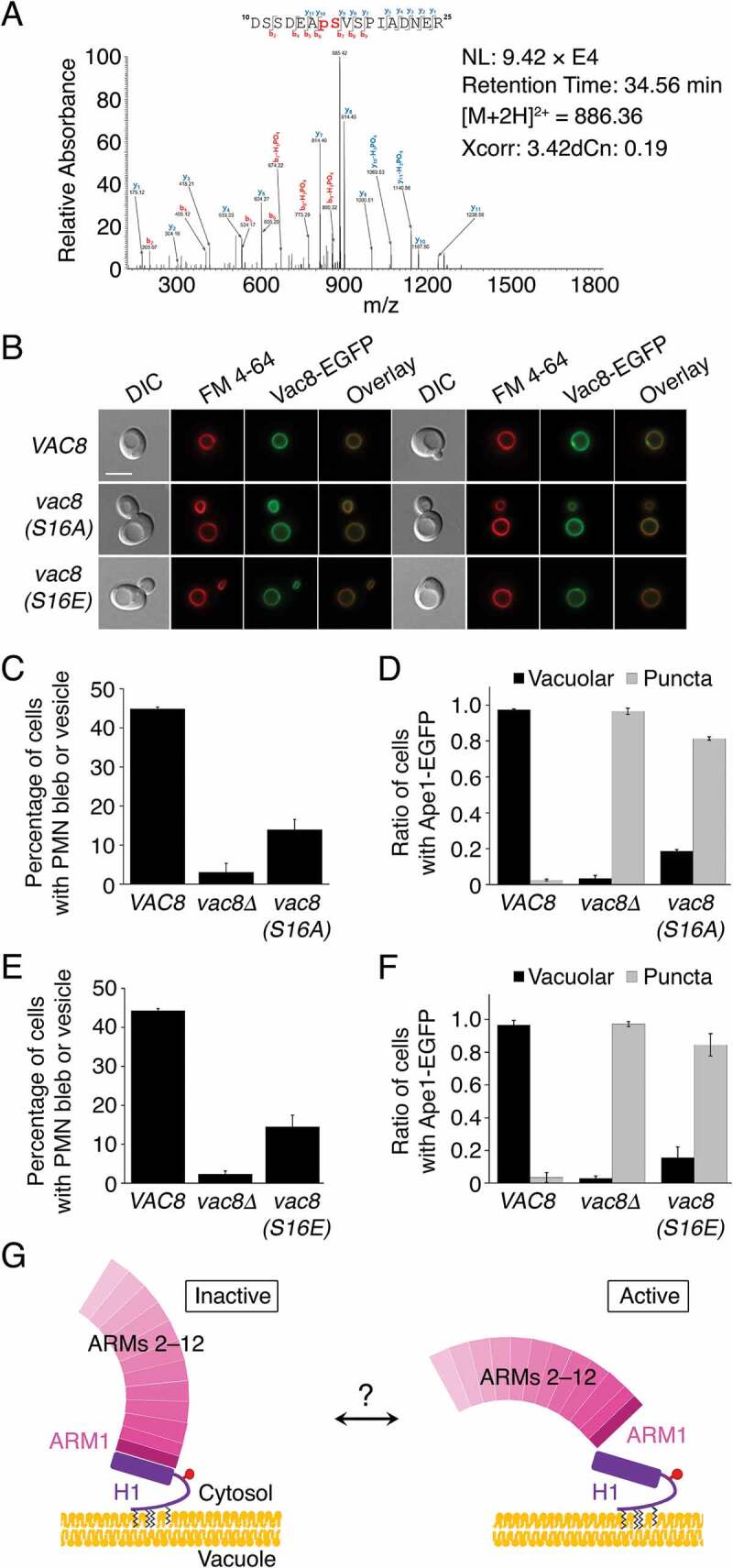
Phosphorylation of Ser16 in Vac8 may regulate the PMN and Cvt pathways. (A) Mass spectrometric analysis of phosphorylated peptides from trypsin digestion of Vac8 reveals phosphorylation of residue Ser16 of Vac8. (B) Replacement of Ser16 with alanine or glutamate does not influence the vacuolar localization of Vac8. GFP fluorescence from Vac8 and FM 4–64 fluorescence from vacuoles were analyzed by fluorescence microscopy. (C and D) Vac8S16A mutant does not support the PMN (C) or Cvt (D) pathways. (E and F) Replacement of Ser16 with glutamate, a phosphomimetic amino acid, largely abolishes the function of Vac8 in both the PMN (E) and Cvt (F) pathways. (G) Putative model of H1 helix-mediated regulation of Vac8. Vac8 is anchored in the vacuolar membrane via lipidation at the N terminus. The H1 helix (purple) directly binds to ARM1 and hinders the self-association of Vac8 by masking the dimeric interface (left; “inactive” conformation). The H1 helix can be released from ARM1, resulting in exposure of the dimeric interface, allowing its self-association (right; “active” conformation). The cartoon representation proposes a putative model in which conformational changes of the H1 helix might be induced by phosphorylation (red circle) of Ser16 or unknown factors (question mark).
An intramolecular folding-back mechanism acting on ARM-containing proteins has been suggested for the CTNNB1/β-catenin-CDH1/E-cadherin complex that plays essential roles in cell-cell adhesion [31]. The structure of CDH1/E-cadherin bound to CTNNB1/β-catenin, which possesses 12 ARMs [32], strongly resembles those of the Vac8-Atg13 and Vac8-Nvj1 complexes determined in the present study (Figure 1C) [24]. The additional secondary structural elements at the N- and C-termini of CTNNB1/β-catenin, rather than within the central ARM domain, are involved in interactions with other binding partners or self-association with the ARM domain itself, and thereby differentially regulate molecular functions in cell adhesion and WNT signaling [31,33]. Although it has never been reported that the H1 helix of Vac8 mediates interactions with other proteins, we herein propose a similar versatile role for the H1 helix in regulating the function of Vac8 by modulating inter- and/or intramolecular binding in a comparable manner. Interestingly, however, expression of Vac8 mutant lacking the H1 helix, Vac8[Δ19–33], in vac8Δ yeast cells fully restored the PMN pathway, but not the Cvt pathway, suggesting that the H1 helix plays a complex functional role, possibly acting as a switch elaborately regulated by phosphorylation (Figure S9).
Vac8 also plays an important role in forming NVJs, and therefore represents novel membrane contact sites. NVJs are formed by direct interaction between Vac8 and Nvj1, and we demonstrated that disruption of this interaction prevents the formation of NVJs, and consequently blocks the PMN pathway [24]. Structural comparison of tVac8-tAtg13 and tVac8-tNvj1 complexes revealed several common features; both tNvj1 and tAtg13 form extended loops in the Vac8-binding region that competitively bind in an antiparallel manner to the inner groove generated by the Vac8 ARM domain (Figure 1C,D) [24]. While obvious structural variations between the 2 extended loops were apparent at interface I of the N-terminal region, interface II in the C-terminal region of the 2 structures superimposed well, despite the low sequence identity shared between Nvj1 and Atg13 (Figure 2A,B). Interestingly, the contact region at interface II is very close to ARM1 that is critical for self-association of Vac8. The most remarkable structural variation between Vac8[Δ19–33]-tAtg13 and tVac8-tNvj1 was the clear difference in conformation of the quaternary structures of Vac8 bound to Nvj1 or Atg13. When bound to Nvj1, the Vac8 heterotetramer forms an arch-shaped structure, whereas it adopts a superhelical conformation in the Vac8-Atg13 complex (Figure 5). We experimentally confirmed that the organization of the quaternary structures of Vac8 selectively affected Cvt or PMN pathways in yeast (Figure 6E,F). We therefore investigated how the quaternary conformation of Vac8 is altered, and structure alignment of Nvj1 and Atg13 revealed an additional visible residue (Tyr321) in tNvj1, in interface II at the C terminus, that is clearly involved in dimerization of Vac8 (Figure 2A,B; Figure S10). Consistently, we previously proposed a significant role for Tyr321 in self-association of Vac8 and formation of NVJs [24]. The absence of this residue in Atg13 could prevent Vac8 from adopting an arch-shaped structure, favoring a different Vac8 conformation. It is still unknown whether there are other proteins that specifically recognize the different quaternary structures of Vac8, and future work should address how the quaternary structures may contribute to the cellular functions of Vac8 at the molecular level. Nevertheless, our work provides a seminal framework for understanding the molecular mechanism by which the armadillo repeat protein Vac8 can differentially regulate the 2 distinct Cvt and PMN pathways at the molecular level.
Materials and methods
Cloning, expression, and protein purification
Overexpression constructs for glutathione-S-transferase (GST)-fused tVac8 and His6-tagged tAtg13 proteins have been described previously [34]. For the Vac8[Δ19–33]-tAtg13 complex, DNA encoding Vac8[Δ19–33] was generated by PCR and cloned into the vector pGEX-6P-1 (GE Healthcare Life Sciences). For co-expression and purification of tVac8-tAtg13 and Vac8[Δ19–33]-tAtg13, plasmids were transformed into Escherichia coli BL21 (DE3) competent cells, and cells were grown in Luria-Bertani medium (MP Biomedicals, 113002032) to an absorbance at 600 nm (A600) of 0.6. Protein expression was then induced by the addition of 0.3 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG; GoldBio, I2481), and culturing was continued at 18°C overnight. Cultured cells were harvested by centrifugation at 3,200 × g for 18 min and resuspended in buffer A containing 25 mM sodium phosphate, pH 7.8, 400 mM sodium chloride (NaCl), 10 mM imidazole. Proteins were first purified on a HiTrap chelating HP column (GE Healthcare Life Sciences, 17040901) charged with Ni2+ in buffer A. The GST-tag and His6-tag were removed using PreScission (GE Healthcare Life Sciences, 27084301) and tobacco etch virus (New England BioLabs, P8112S) proteases, respectively, during dialysis overnight against buffer containing 25 mM Tris-HCl, pH 7.8, 150 mM NaCl, 4 mM β-mercaptoethanol (β-ME) at 4°C. Non-cleaved proteins and cleaved tags were removed by a second round of Ni2+-immobilized metal affinity chromatography (IMAC) and GST affinity chromatography. Proteins were further purified by size-exclusion chromatography (SEC) using a HiLoad 16/600 Superdex 200 pg column (GE Healthcare Life Sciences, 28989335) in buffer B consisting of 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM dithiothreitol (DTT). Proteins were concentrated to 10 mg/mL for later use. Mutant variants of Vac8 and tAtg13 used in this study were obtained by site-directed mutagenesis and confirmed by DNA sequencing.
Crystallization and structure determination
The tVac8-tAtg13 complex was crystallized as described previously [34] from a reservoir solution comprising 25% (v:v) polyethylene glycol (PEG) 400 (Sigma-Aldrich, 91893), 100 mM Tris-HCl, pH 8.5, 2% (v:v) ethylene glycol (Sigma-Aldrich, 324558), 2% (w:v) PEG 3350 (Sigma-Aldrich, 88276), 5 mM DTT. Crystals were transferred into reservoir buffer supplemented with 30% (v:v) PEG 400 for cryoprotection, and flash-frozen in liquid nitrogen. X-ray diffraction data were collected at a wavelength of 0.979 Å on beamline 5C of the Pohang Accelerator Laboratory [35]. Data were processed with the program HKL2000 [36]. The structure was solved by molecular replacement using Phaser in the Phenix suite [37] with published coordinates of Vac8 (PDB code: 5XJG) as a search model. Following rigid-body and positional refinement of the model using Phenix [38] and Coot [39], the sequence of tAtg13 (residues 660–685) could be positioned into the resulting electron density (Figure S1A). Subsequently, restrained refinement reduced the free R-factor to 24.6% (and the crystallographic R-factor to 20.6%) for data between 50 and 2.9 Å resolution (Table S2). Residues 10–12 and 514–515 of tVac8, and residues 567–659 and 686–695 of tAtg13, were not modeled due to disordered electron density.
The Vac8[Δ19–33]-tAtg13 complex was crystallized at 4°C by mixing 1 μL of protein solution with the same volume of reservoir solution comprising 15% (w:v) PEG 3350, 0.1 M N-(2-acetamido)iminodiacetic acid (ADA), pH 6.5, 0.2 M ammonium sulfate. Crystals (space group P212121, a = 69.475 Å, b = 85.268 Å, c = 272.820 Å) contain 2 copies of the Vac8[Δ19–33]-tAtg13 complex in the asymmetric unit, and diffract synchrotron X-rays to 3.2 Å resolution. Crystals were transferred to well solution plus 30% (v:v) ethylene glycol for cryoprotection and flash-frozen in liquid nitrogen. X-ray diffraction data collection and phase calculation were performed as described above. The final model of the Vac8[Δ19–33]-tAtg13 complex was refined to Rwork/Rfree values of 21.6/25.5% following iterative rounds of refinement and model building. The model contains residues 47–528 and 561–576 of Vac8[Δ19–33] for first copy, residues 48–532 and 560–578 of Vac8[Δ19–33] for second copy, and residues 665–685 of tAtg13 (Figure S1B). Diffraction data and refinement statistics are summarized in Table S2. All structural images in figures were generated by PyMOL (http://www.pymol.org/).
Analytical ultracentrifugation
The absolute molecular weights of tVac8-tAtg13 (Figure 1B) and Vac8[Δ19–33]-tAtg13 (Figure 3C) complexes were measured by sedimentation equilibrium analysis using an XL-A analytical ultracentrifuge (Beckman Coulter) as described previously [40]. Protein samples were prepared in buffer C comprising 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM β-ME.
Affinity-isolation assay
To assess the interaction between Vac8 and Atg13 (Figure 2D), supernatants of E. coli cells co-expressing GST-tVac8 and wild-type or mutant His6-tAtg13 were incubated with 15 μL of Ni-NTA agarose (Qiagen, 30210) for 1 h at 4°C. Beads were washed 3 times with buffer consisting of 25 mM sodium phosphate, pH 7.4, 400 mM NaCl, 50 mM imidazole, 0.1% (v:v) Triton X-100 (Sigma-Aldrich, X100). Proteins were eluted with 4× sodium dodecyl sulfate (SDS) loading buffer, separated by 12% SDS-polyacrylamide gel electrophoresis (PAGE), and stained with Coomassie Brilliant Blue. Protein bands of Vac8 were quantified using ImageJ software. Data are shown as the mean ± SD of 3 independent experiments.
Isothermal titration calorimetry
The apparent dissociation constant (Kd) for Vac8 (full-length and Vac8[Δ19–33]) binding to tAtg13 proteins (Figure 3B) and full-length Vac8 binding to mutants of tAtg13 (Figure S2) was measured by isothermal titration calorimetry (ITC) using a MicroCal iTC200 instrument (GE Healthcare Life Sciences). A 0.7 mM sample of tAtg13 was titrated into 0.05 mM Vac8 protein in buffer C at 25°C. Measurements were obtained over 20 injections of 2 μL with a reference power of 5 μcal/s and a stirring speed of 1000 rpm. The “one set of sites” model was used to generate titration curves using MicroCal PEAQ-ITC analysis software (Malvern panalytical).
Size-exclusion chromatography assay
To analyze the relative molecular weight of wild-type and mutant Vac8-Atg13 complexes (Vac8[40–515]-tAtg13 and Vac8[40–515]A51R or L55R-tAtg13 in Figure 4B) and Vac8[40–515]N62R or N60R,N62R complexed with tNvj1 or tAtg13 (Figure 6C,D), purified protein complexes were loaded onto a calibrated Superdex 200 10/300 GL column (GE Healthcare Life Sciences, 17517501) in buffer B, and eluted at a flow rate of 0.5 mL/min.
Small-angle X-ray scattering
Small-angle X-ray scattering (SAXS) data were collected at beamline 6D at Pohang Accelerator Laboratory (PAL). Scattering data were collected using 30 s exposures for each protein samples (4 mg/mL) in buffer B, followed by background-subtraction. Further data processing and analysis were performed using the ATSAS package [28]. The radius of gyration (Rg) determined by Guinier analysis was analyzed by PRIMUS [41], and the distance distribution function P(r) and maximum dimension of the particle (Dmax) were obtained by the indirect Fourier transform method using GNOM [42]. Ab initio modeling of Vac8[40–578]-tNvj1 and Vac8[40–578]-tAtg13 was performed using DAMMIF [43] with 10 independent runs and averaged by DAMAVER [44]. Theoretical scattering curves were generated by CRYSOL [45] using the structure of Vac8[Δ19–33]-tNvj1, in which tVac8 coordinates of the tVac8-tNvj1 were replaced by that of Vac8[Δ19–33], and Vac8[Δ19–33]-tAtg13 crystal structures. Envelopes generated by DAMAVER were superimposed with structures using the SUPCOMB program [46]. Experimental parameters from SAXS analysis are summarized in Table S3.
Cross-linking
For cross-linking analysis, 0.5 mg/mL wild-type or mutant Vac8 (Vac8[40–515] or Vac8[40–515]A51R, Vac8[40–515]N62R, or Vac8[40–515]N60R,N62R) in complex with tAtg13 or tNvj1 was prepared in buffer comprising 25 mM sodium phosphate, pH 7.5, 150 mM NaCl, 5 mM DTT. Each protein complex was incubated with 0.001% (w:v) glutaraldehyde (Sigma-Aldrich, G7651) for 30 min at 20°C. The reaction was stopped by the addition of 100 mM Tris-HCl, pH 7.6 and reaction products were analyzed by 8% SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane using a Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad). The identity of cross-linked species was analyzed by immunoblotting with anti-Vac8, anti-Atg13, and anti-Nvj1 antibodies.
Yeast strains and vacuole staining with FM 4-64
Yeast strains used in this study are listed in Table S1. Vacuoles of growing yeast cells were labeled with FM 4–64 as previously described [24] with minor modifications. Briefly, cells were grown at 30°C to an optical density at 600 nm (OD600) of 0.8 in YPD (10 g/L yeast extract, 20 g/L peptone, 20 g/L dextrose) medium. Cells were then incubated with aeration in YPD medium containing 20 μM FM 4–64 (Molecular Probes, T13320) for 30 min at 30°C, harvested by centrifugation (2,400 × g) at room temperature for 1 min, washed, resuspended in fresh medium, and cultured for a further 3 h at 30°C. After cells were harvested by centrifugation, they were mounted on a pad of 2% low-melting agarose formed in a slide and analyzed using a fluorescence microscope (Nikon Eclipse Ti-U) with a Nikon Plan Apo 100×, 1.45/NA oil immersion objective.
Cvt and PMN pathway induction
Cells were grown overnight at 30°C with shaking in YPD medium, cultures were adjusted to OD600 = 0.5, FM 4–64 (0.5 μM) was added to visualize vacuoles, and cells were further cultured at 30°C for 2 h. For Cvt pathway induction, cells grown in YPD were washed with sterile water 3 times, resuspended in nitrogen starvation media (SD-N) (1.7g/L yeast nitrogen base, 20g/L dextrose), and further cultured at 30°C for 2 h. For PMN induction, 0.2 μM rapamycin (Sigma-Aldrich, R8781) was added and cells were further incubated at 30°C for 6 h.
Precursor Ape1 processing assay
Yeast cells were grown overnight at 30°C with shaking in YPD medium. Cells were washed with sterile water 3 times, resuspended in SD-N media, and further cultured at 30°C for 2 h. Cells were harvested by centrifugation for 10 s at 11,400 × g at room temperature, and the pellet was resuspended in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 · 7H20, 1.8 mM KH2PO4). Cells were vortexed with glass beads for 3 min, mixed with 2× SDS sample buffer, and boiled for 5 min. After centrifugation, the resulting supernatant was separated by SDS-PAGE and analyzed by immunoblotting with anti-Ape1 antiserum (a generous gift from Dr. Daniel Klionsky, University of Michigan) or anti-actin antibody (a generous gift from Dr. Bill Wickner, Dartmouth College).
Phosphopeptide enrichment and LC-MS/MS analysis of Vac8
Protein bands corresponding to Vac8 were excised from the SDS-PAGE gel and digested in-gel with trypsin for LC-MS/MS analysis. Enrichment of phosphopeptides was performed as previously described [47]. Briefly, trypsin-digested proteins were resuspended in equilibration buffer (50% acetonitrile, 0.1% trifluoroacetic acid) before injection onto the Fe-IMAC column (Propac IMAC-10 4 × 50 mm, Thermo Scientific, 063276). Samples were injected (0.1 mL/min over 10 min) onto the Fe-IMAC column, and the column was washed with equilibration buffer (0.3 mL/min over 16 min). Phosphopeptides were eluted with a linear gradient from 0% to 45% Fe-IMAC elution buffer (0.5% NH4OH; 0.2 mL/min over 60 min). The Fe-IMAC column was re-equilibrated with equilibration buffer (0.5 mL/min over 30 min). Flow-through and eluate fractions were collected according to the UV signal (214 nm), dried, and stored at −80°C. LC-MS/MS experiments were performed on an Ultimate 3000 nanoHPLC system (Thermo Scientific) coupled online to a Q Exactive mass spectrometer (Thermo Scientific). Peptides were injected onto a 20 mm × 75 μm Acclaim PepMap C18 nano-trap column (Thermo Scientific, 164535) at a flow rate of 300 nL/min in 98% solvent A (0.1% formic acid in water). After washing, peptides were separated on a 150 mm × 75 μm Acclaim PepMap C18 reverse phase analytical column (Thermo Scientific, ES800) using a linear gradient from 2% to 40% solvent B (acetonitrile with 0.1% formic acid) over a 65 min gradient with a flow rate of 300 nL/min. Data-dependent scans consisting of one full MS scan (450–2,000 m/z, AGC target 1e6) and 10 data-dependent MS/MS scans were used to generate MS/MS spectra of eluted peptides using the dynamic exclusion option.
Mass spectrometric data analysis
Mass spectrometry-derived data were searched against the canonical S. cerevisiae proteome database downloaded from UniProtKB (http://www.uniprot.org/) using Proteome Discoverer software (Thermo Scientific). Mass spectrometry data were searched using the SEQUEST algorithm with carbamidomethylation of cysteine as a fixed modification, and N-terminal acetylation, oxidation of methionine and phosphorylation at serine, threonine, and tyrosine as variable modifications. MS/MS spectra were searched with a precursor mass tolerance of 10 ppm and fragment mass tolerance of 0.02 Da. The search was limited to fully tryptic peptide candidates and a maximum of 2 missed cleavages were allowed. Data were searched against the target-decoy database and peptide spectral matches (PSM) were validated using percolator based on q-values at a 1% false discovery rate (FDR).
Immunoblotting
Samples were separated by SDS-PAGE and analyzed by immunoblotting with indicated antibodies. ECL exposed films were quantitated using ImageJ software. All data are representative of experiments performed at least 3 times. Actin levels were analyzed and confirmed comparable between samples by comparing band intensities of actin from increasing concentrations of yeast cytosol.
Statistical analysis
Statistical significance was determined from at least 3 independent experiments using Student’s t-test. All data represent the mean ± SEM.
Supplementary Material
Acknowledgments
We thank staff from beamlines 5C, 11C, and 6D at the Pohang Accelerator Laboratory (PAL) for assistance with X-ray diffraction data collection and SAXS experiments. We also thank Dr. Daniel Klionsky (University of Michigan, Ann Arbor) for kindly providing Atg13 and Ape1 antisera and Dr. Bill Wickner (Dartmouth College) for his generous gift of rabbit antisera against GFP and yeast actin.
Funding Statement
This research was supported by the Cell Logistics Research Center (grant number 2016R1A5A1007318) funded by the National Research Foundation of Korea. This research was also supported by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number HI18C1395), the U-K Brand Research Fund (1.180064.01) of UNIST, and the GIST Research Institute (GRI) at GIST.
Accession Numbers
The atomic coordinates and crystallographic structure factors have been deposited in the Protein Data Bank under the following accession codes: 6KBM and 6KBN for tVac8-tAtg13 and Vac8[Δ19–33]-tAtg13, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental Material
Supplementary material for this article can be accessed here.
References
- [1].Jones EW, Webb GC, Hiller MA.. Biogenesis and function of the yeast vacuole. Cold Spring Harbor Monogr Archive. 1997;21:363–470. [Google Scholar]
- [2].van Den Hazel HB, Kielland‐Brandt MC, Winther JR. Biosynthesis and function of yeast vacuolar proteases. Yeast. 1996;12(1):1–16. [DOI] [PubMed] [Google Scholar]
- [3].Thumm M, Wolf DH. From proteasome to lysosome: studies on yeast demonstrate the principles of protein degradation in the eukaryote cell. Adv Mol Cell Biol. 1998;27:43–70. Elsevier. [Google Scholar]
- [4].Forgac M. Structure and properties of the vacuolar (H+)-ATPases. J Biol Chem. 1999;274(19):12951–12954. [DOI] [PubMed] [Google Scholar]
- [5].Graham LA, Powell B, Stevens TH. Composition and assembly of the yeast vacuolar H (+)-ATPase complex. J Exp Biol. 2000;203(1):61–70. [DOI] [PubMed] [Google Scholar]
- [6].Baba M, Takeshige K, Baba N, et al. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol. 1994;124(6):903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Takeshige K, Baba M, Tsuboi S, et al. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119(2):301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Weisman LS, Bacallao R, Wickner W. Multiple methods of visualizing the yeast vacuole permit evaluation of its morphology and inheritance during the cell cycle. J Cell Biol. 1987;105(4):1539–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Weisman LS, Emr SD, Wickner WT. Mutants of Saccharomyces cerevisiae that block intervacuole vesicular traffic and vacuole division and segregation. Proc Nat Acad Sci. 1990;87(3):1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Weisman LS. Yeast vacuole inheritance and dynamics. Annu Rev Genet. 2003;37(1):435–460. [DOI] [PubMed] [Google Scholar]
- [11].Shaw JM, Wickner WT. vac2: a yeast mutant which distinguishes vacuole segregation from Golgi‐to‐vacuole protein targeting. EMBO J. 1991;10(7):1741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gomes de Mesquita DS, van Den Hazel HB, Bouwman J, et al. Characterization of new vacuolar segregation mutants, isolated by screening for loss of proteinase B self-activation. Eur J Cell Biol. 1996;71(3):237–247. [PubMed] [Google Scholar]
- [13].Wang Y-X, Zhao H, Harding TM, et al. Multiple classes of yeast mutants are defective in vacuole partitioning yet target vacuole proteins correctly. Mol Biol Cell. 1996;7(9):1375–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pan X, Goldfarb DS. YEB3/VAC8 encodes a myristylated armadillo protein of the Saccharomyces cerevisiae vacuolar membrane that functions in vacuole fusion and inheritance. J Cell Sci. 1998;111(15):2137–2147. [DOI] [PubMed] [Google Scholar]
- [15].Wang Y-X, Catlett NL, Weisman LS. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J Cell Biol. 1998;140(5):1063–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tang F, Kauffman EJ, Novak JL, et al. Regulated degradation of a class V myosin receptor directs movement of the yeast vacuole. Nature. 2003;422(6927):87–92. [DOI] [PubMed] [Google Scholar]
- [17].Subramanian K, Dietrich LEP, Hou H, et al. Palmitoylation determines the function of Vac8 at the yeast vacuole. J Cell Sci. 2006;119(12):2477–2485. [DOI] [PubMed] [Google Scholar]
- [18].Roberts P, Moshitch-Moshkovitz S, Kvam E, et al. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14(1):129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kvam E, Goldfarb D. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy. 2007;3(2):85–92. [DOI] [PubMed] [Google Scholar]
- [20].Dawaliby R, Mayer A. Microautophagy of the nucleus coincides with a vacuolar diffusion barrier at nuclear–vacuolar junctions. Mol Biol Cell. 2010;21(23):4173–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pan X, Roberts P, Chen Y, et al. Nucleus–vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol Biol Cell. 2000;11(7):2445–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Murley A, Sarsam RD, Toulmay A, et al. Ltc1 is an ER-localized sterol transporter and a component of ER–mitochondria and ER–vacuole contacts. J Cell Biol. 2015;209(4):539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Scott SV, Nice DC, Nau JJ, et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275(33):25840–25849. [DOI] [PubMed] [Google Scholar]
- [24].Jeong H, Park J, Kim H-I, et al. Mechanistic insight into the nucleus–vacuole junction based on the Vac8p–Nvj1p crystal structure. Proc Nat Acad Sci. 2017;114(23):E4539–E4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fujioka Y, Suzuki SW, Yamamoto H, et al. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat Struct Mol Biol. 2014;21(6):513–521. [DOI] [PubMed] [Google Scholar]
- [26].Yamamoto H, Fujioka Y, Suzuki SW, et al. The intrinsically disordered protein Atg13 mediates supramolecular assembly of autophagy initiation complexes. Dev Cell. 2016;38(1):86–99. [DOI] [PubMed] [Google Scholar]
- [27].Suzuki SW, Yamamoto H, Oikawa Y, et al. Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc Nat Acad Sci. 2015;112(11):3350–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Franke D, Petoukhov MV, Konarev PV, et al. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J Appl Crystallogr. 2017;50(4):1212–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Holt LJ, Tuch BB, Villén J, et al. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325(5948):1682–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dephoure N, Gould KL, Gygi SP, et al. Mapping and analysis of phosphorylation sites: a quick guide for cell biologists. Mol Biol Cell. 2013;24(5):535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gottardi CJ, Gumbiner BM. Distinct molecular forms of β-catenin are targeted to adhesive or transcriptional complexes. J Cell Biol. 2004;167(2):339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Huber AH, Weis WI. The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell. 2001;105(3):391–402. [DOI] [PubMed] [Google Scholar]
- [33].Xu W, Kimelman D. Mechanistic insights from structural studies of β-catenin and its binding partners. J Cell Sci. 2007;120(19):3337–3344. [DOI] [PubMed] [Google Scholar]
- [34].Park J, Song K, Oh S, et al. Purification, crystallization, and X-ray crystallographic analysis of Vac8p complexed with Atg13p from Saccharomyces cerevisiae. Biodesign. 2017;5(3):114–117. [Google Scholar]
- [35].Park S-Y, Ha S-C, Kim Y-G. The protein crystallography beamlines at the pohang light source II. Biodesign. 2017;5(1):30–34. [Google Scholar]
- [36].Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. Elsevier. [DOI] [PubMed] [Google Scholar]
- [37].McCoy AJ, Grosse-Kunstleve RW, Adams PD, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40(4):658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Adams PD, Afonine PV, Bunkóczi G. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66(2):213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Emsley P, Lohkamp B, Scott WG. et al. Features and development of Coot. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66(4):486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jeong H, Park J, Lee C. Crystal structure of Mdm12 reveals the architecture and dynamic organization of the ERMES complex. EMBO Rep. 2016;17(12):1857–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Konarev PV, Volkov VV, Sokolova AV, et al. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36(5):1277–1282. [Google Scholar]
- [42].Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25(4):495–503. [Google Scholar]
- [43].Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2009;42(2):342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36(3):860–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Svergun D, Barberato C, Koch MHJ. CRYSOL–a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28(6):768–773. [Google Scholar]
- [46].Kozin MB, Svergun DI. Automated matching of high-and low-resolution structural models. J Appl Crystallogr. 2001;34(1):33–41. [Google Scholar]
- [47].Ruprecht B, Koch H, Medard G, et al. Comprehensive and reproducible phosphopeptide enrichment using iron immobilized metal ion affinity chromatography (Fe-IMAC) columns. Mol Cell Proteomics. 2015;14(1):205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


