ABSTRACT
Hepatocellular carcinoma (HCC) has a poor prognosis, owing to its high potential for growth and metastasis. In this study, we aimed to investigate the roles of Phospholysine Phosphohistidine Inorganic Pyrophosphate Phosphatase (LHPP)in human HCCcell growth and metastasis. We analyzed the LHPP expression level in human HCC tissues paired normal tissues in the Oncomine database, and assessed the relationship between the LHPP expression levels with HCC patient’s overall survival and the prognostic value of LHPP in human HCC by Kaplan-Meier survival analysis. Real-time PCR and Western Blot were used to examine the expression levels of LHPP in normal liver cell line (LO2) and human HCC cell lines (SMCC-7721, HepG2, Huh7, MHCC-97 H, and LM3). Through lentivirus infection, we established human HCC stable cell lines (Huh7 and LM3) overexpressing LHPP. Then, we detected these cell viability, colony , and invasion. Subsequently, we performed the gene set enrichment analysis (GSEA) for the RNA-seq data of HCC patients from TCGA. Finally, we examined the expression level of several oncogenes, including CCNB1, PKM2, MMP7, and MMP9, in these cells via real-time PCR assay. Here, we found thatLHPPis significantly downregulated in the human HCC tissues paired normal tissues. Furthermore, the high expression level of LHPP is associated with better clinical outcomes in human HCC. Overexpression of LHPPinhibitscell growth and metastasis in human HCC cells, and LHPP expression levels negatively correlate with cell cycle and metastasis in HCC tissues. Moreover, the level of LHPP is negatively correlated with CCNB1, PKM2, MMP7, and MMP9 in human HCC cells and HCC tissues. These findings highlight a novel tumor suppressor in human HCC growth and metastasis, and provide a promising diagnostic and prognostic factor for humanHCC.
KEYWORDS: HCC, LHPP, cell growth, metastasis
Introduction
Hepatocellular carcinoma (HCC), also known as hepatoma, is the most common primary liver malignancy and also the second most common cause of cancer-related death worldwide. HCC accounts for 90% of primary liver cancers [1–3]. There are diverse etiologies involves in HCC development, which is a multistep complicated process. And several crucial factors contribute to the process, such as hepatitis B virus (HBV) infection, hepatitis C virus (HCV) infection, cirrhosis, diabetes mellitus, alcohol abuse, and aflatoxin [4–6]. So far, five conventional methods in still widely used in clinical: surgical resection, liver transplantation, radiofrequency ablation (RFA), percutaneous ethanol injection (PEI), and transarterial chemoembolization (TACE) [2]. Of note, due to the high incidence of recurrence, the long-term survival is still unsatisfactory and the5-year recurrence rate exceeds 70%. However, the molecular mechanisms for the occurrence and development of HCC are still largely unclear [7]. Therefore, it is imperative to identify novel genes or pathways that are involved during HCC progression.
Phosphohistidine phosphate inorganic pyrophosphatase (LHPP) is a member of the haloacid dehalogenase like hydrolase domain-containing (HDHD) gene family. It is also a new kind of inorganic pyrophosphatase (PPase), which is considered to be essential for life and closely related to cell survival, growth, and differentiation. Histidine phosphatase can dephosphorylate proteins containing histidine phosphate. The effect is opposite to that histidine-kinase. Their interaction plays an important role in tumor proliferation, invasion, metastasis, and poor prognosis [8–10]. Three histidine phosphatases were well known: , and LHPP, and the role of the former two in tumor progression have been widely reported, but not LHPP. Moreover, there is still lack of evidence addressing LHPP’sfunction as a phosphatase in cancer development [11–14].
LHPP gene locates at chromosome 10 q26.13, and consists of three leucine zipper domains. It ubiquitously expresses in brain, kidney, and liver [15]. Early studies mainly focus on its psychiatric phenotypes and found an SNP at the LHPP gene (rs35936514/rs34997829) correlated with MDD and AD-RSB interaction [16–18]. Recently, increasing evidence showed that LHPP is closely linked to cancer development. Overexpression of LHPPisclosely related to hyperthyroidism, while its low expression is related to tumor stage in Graves’ disease and AFTN [19,20]. A genome–wideassociation study identifies risk loci in acute lymphoblastic leukemia [21]. And LHPP (rs201982221) acts as the significantly associated loci in oral and pharyngeal cancers [22]. Recent studies also showed LHPPacting as a tumor suppressor in cervical cancer and HCC (mainly by using HCC mouse model) [23–25]. Interestingly, LHPPoverexpression can prevent liver function damage in L-dKO mice. L-dKO mice is an HCC mouse model by liver-specific deletion of PTEN and TSC1, and the L-dKO liver tumors mimicked poorly differentiated human HCC [23]. In their research, they just introduce the role of mouse LHPPin the HCC mouse model, but not mention humans. Yet, the role and mechanism ofLHPPin human HCC are still largely unknown.
In the present study, we aimed to investigate the roles of LHPP in human HCC cell growth and metastasis. We showed that LHPP was down-regulated in human HCC tissues. Further, we demonstrated the reduction of LHPP in human HCC cell lines. Functional studies identified that overexpression of LHPP attenuated the cell viability, colony formation, and invasion in human HCC cells. Moreover, we also found that the expression of several oncogenes, including CCNB1, PKM2, MMP7, and MMP9, is suppressed by LHPP forceful expression in HCC.
Results
High expression of LHPP is associated with better clinical outcomes in HCC
Firstly, we analyzed the expression of LHPPfrom the Oncomine database (https://www.oncomine.org/resource/main.html) and we found that LHPP expression was significantly downregulated in the HCC tissues paired normal tissues (Figure 1(a,b)). Furthermore, we analyzed the LHPP expression level in the tumor tissues derived from 359 postoperative HCC patients in the TCGA data (https://tcga-data.nci.nih.gov/publications/tcga). These tissues were grouped under LHPP low expression or LHPPhigh expression according to their mRNA levels. We next assessed the relationship between the LHPPexpression level and patient overall survival by Kaplan-Meier survival analysis. The overall survival was extremely down-regulation in the LHPP-Low expression group compared with the LHPP-High expression group (Figure 1(c)). Consistently, data analysis from the Kaplan–Meier Plotter database (http://kmplot.com/analysis/) also revealed lower LHPPexpression in HCC patients showed unfavorable overall survival, progression-free survival, and relapse-free survival times compared with the higher LHPP expression HCC patients (Figure 1(d–f)). Taken together, these data suggest that LHPPmay play a vital role in HCCprogression.
Figure 1.
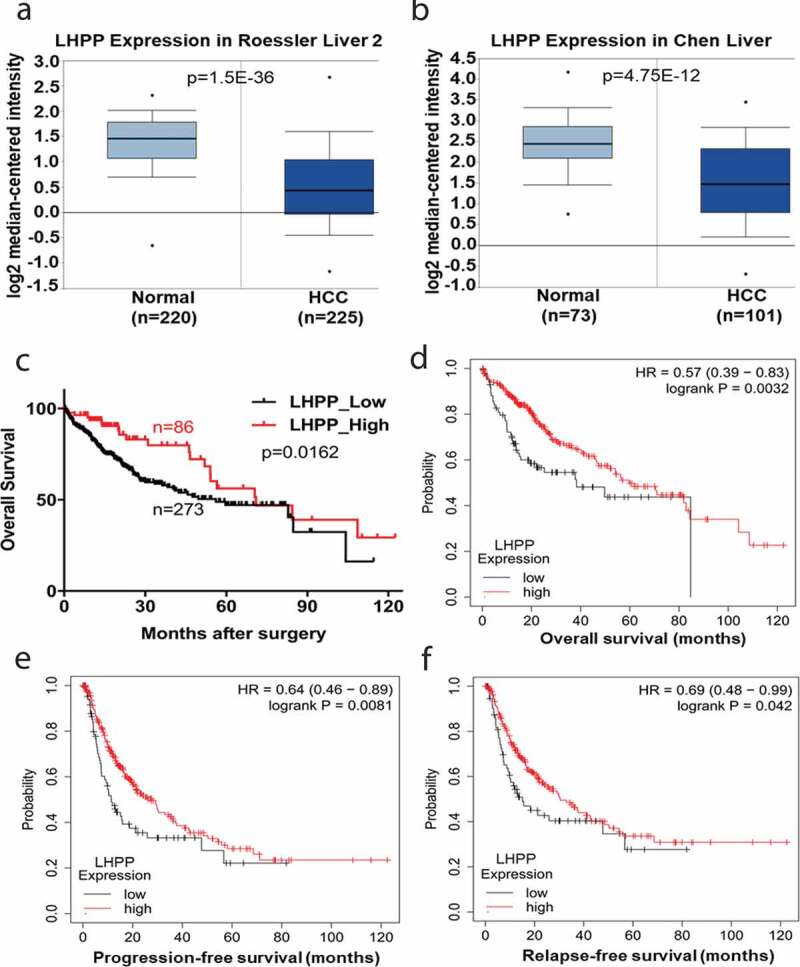
High expression of LHPP is associated with better clinical outcomes in HCC. (a-b) LHPP Expression in Roessler Liver 2 Statistics and LHPP Expression in Chen Liver Statistics. These data come from the Oncomine database (https://www.oncomine.org/resource/main.html). Gene expression of LHPP was down-regulated in the HCC tissues comparing with the normal tissues. (c) Kaplan-Meier curves of overall survival according to LHPP expression in HCC tissues. The overall survival was significantly reduced in the LHPP low expression group (LHPP-Low, n = 273) compared with the LHPP high expression group (LHPP-High, n = 86) (Log-Rank test, p = 0.0162). The HCC patients date comes from the TCGA database. (https://tcga-data.nci.nih.gov/publications/tcga). (d-f) The overall survival, progression-free survival and relapse-free survival probability were compared between LHPP high and LHPP low expression in the HCC patients from the Kaplan-Meier Plotter database (https://kmplot.com/analysis/). Log-rank p values were shown.
LHPP inhibits HCC cell viability and colony formation
In order to evaluate the function ofLHPP in HCCcell growth and metastasis in vitro, we firstly examined the expression levels of LHPP in normal liver cell line (LO2) and HCC cell lines (SMCC-7721, HepG2, Huh7, MHCC-97 H, and LM3), the Real-time PCR assay showed that the expression level of LHPP in the normal liver cell was significantly increased compared with the HCC cell lines (Figure 2(a)). In addition, the Western Blot assay showed a similar result in the above cells (Figure 2(b)), suggesting LHPP’s tumor suppressor activity in HCC progression. Lentivirus-mediatedoverexpressionof LHPPin the Huh7 and LM3 cells which were performed to assess the function of LHPP in the HCC cell viability and colony formation. The efficiency of LHPPoverexpression in the Huh7 and LM3 cells was assessed by Western Blot assay (Figure 2(c)). Next, assessed the LHPP-high expression HCC cell viability ability via the CCK8 assay. Overexpression of LHPP in the Huh7 and LM3 cells significantly inhibited the cell viability (Figure 2(d,e)). Also, overexpression of LHPP inhibited colony formation ability of the above cells (Figure 2(f,g)). Taken together, these results indicate that LHPP inhibits HCC cell growth in vitro.
Figure 2.
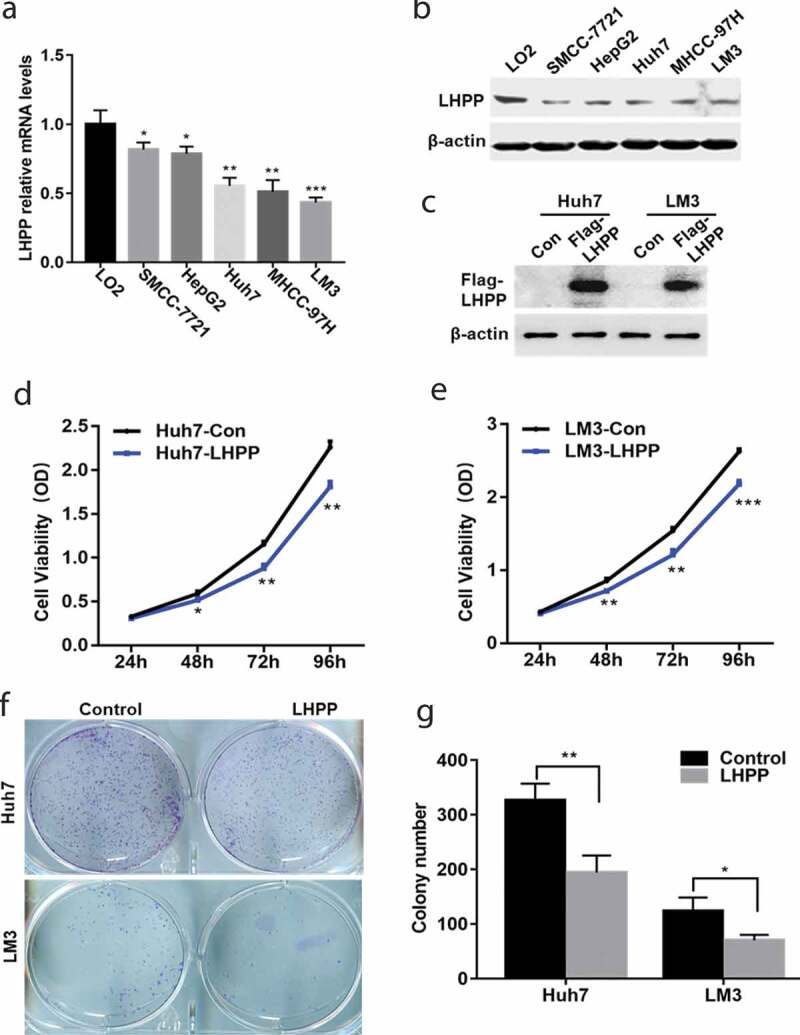
LHPP inhibits HCC cell viability and colony formation. (a-b) The expression level of LHPP was detected in a normal liver cell line (LO2) and HCC cell lines (SMCC-7721, HepG2, Huh7, MHCC-97 H, and LM3) using Real-time PCR assay and Western Blot. Means±SD from three independent experiments were presented as a relative ratios to the LO2 whose value was taken as 1.0. (c) Overexpression of LHPP in the Huh7 and LM3 cells infected with lentivirus. The efficiency of overexpression LHPP in the above cells were assessed by Western Blot. (d-e) Overexpression of LHPP down-regulated the cell viability in the Huh7 and LM3 cells using the CCK8 assay. (f-g) Overexpression of LHPP in the Huh7 and LM3 cells significantly inhibited cell colony formation assay in colony formation assay. The permeable cells were stained with Giemsa. Significant differences were determined using a one-way analysis of variance, *P < 0.05, **P < 0.01, ***P < 0.001.
LHPP prevents HCC cell migration and invasiveness
We next assessed whether LHPP had an inhibitory role in HCC cell migration and invasiveness ability via transwell assay. Overexpression of LHPP in the Huh7 and LM3 cells significantly inhibited cell migration ability. LHPPoverexpression displayed sharp declines in cell migration compared with the control group (Figure 3(a,b)). Moreover, LHPP had a similar role in the HCC cells invasiveness ability (Figure 3(c,d)). Collectively, the data showed that LHPPcan prevent HCC cell metastasis.
Figure 3.
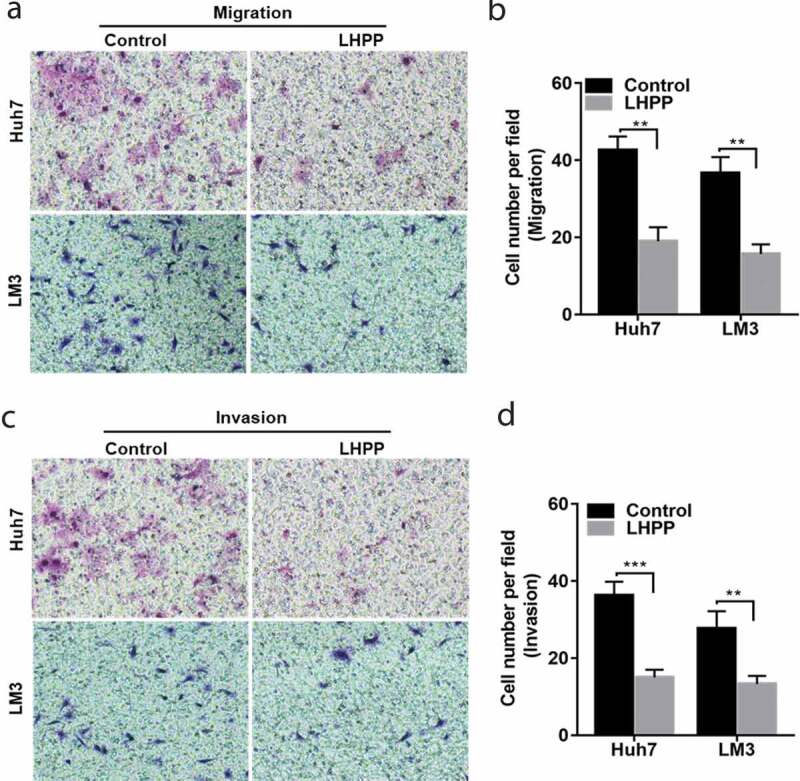
LHPP prevents HCC cell migration and invasiveness. (a-b) Overexpression of LHPP in the Huh7 and LM3 cells significantly inhibited cell migration ability. (c-d) Used the matrigel transwell assay to investigate LHPP’s role in cell invasiveness. The permeable cells were stained with Giemsa. Significant differences were determined using Student t-test, **P < 0.01, ***P < 0.001.
LHPP expression levels negatively correlated with cell cycle and metastasis in humanHCC tissues
To explore the mechanism underlying LHPP’s suppression of HCC cell growth and metastasis, we firstly analyzed the RNA-seq data of HCC patients from TCGA-LIHC (https://xenabrowser.net/datapages/?dataset=TCGA.LIHC; dataset ID: TCGA.LIHC. -sampleMap/HiSeqV2), and then divided into two groups, LHPP low expression, and LHPP high expression. Next, we performed the gene set enrichment analysis (GSEA) for the above data (Figure 4(a)). Using GSEA, we showed that cell cycle, mitotic spindle, and G2 M checkpoint pathways were found significantly enriched in HCC patients with LHPP low expression group (Figure 4(b–d)). Moreover, Liaometastasis and Roesslerliver cancer metastasis were also highly enriched in the LHPP low expression group (Figure 4(e,f)), suggesting LHPP’s suppression in HCC metastasis. However, GSEA analysis showed that metabolic pathways such as oxidative phosphorylation and fatty acid metabolism were remarkably compromised in the LHPP low expression group (Figure 4(g,h)). Thus, these findings indicated that LHPP suppresses cell cycle and metastasis in the HCC tissues.
Figure 4.
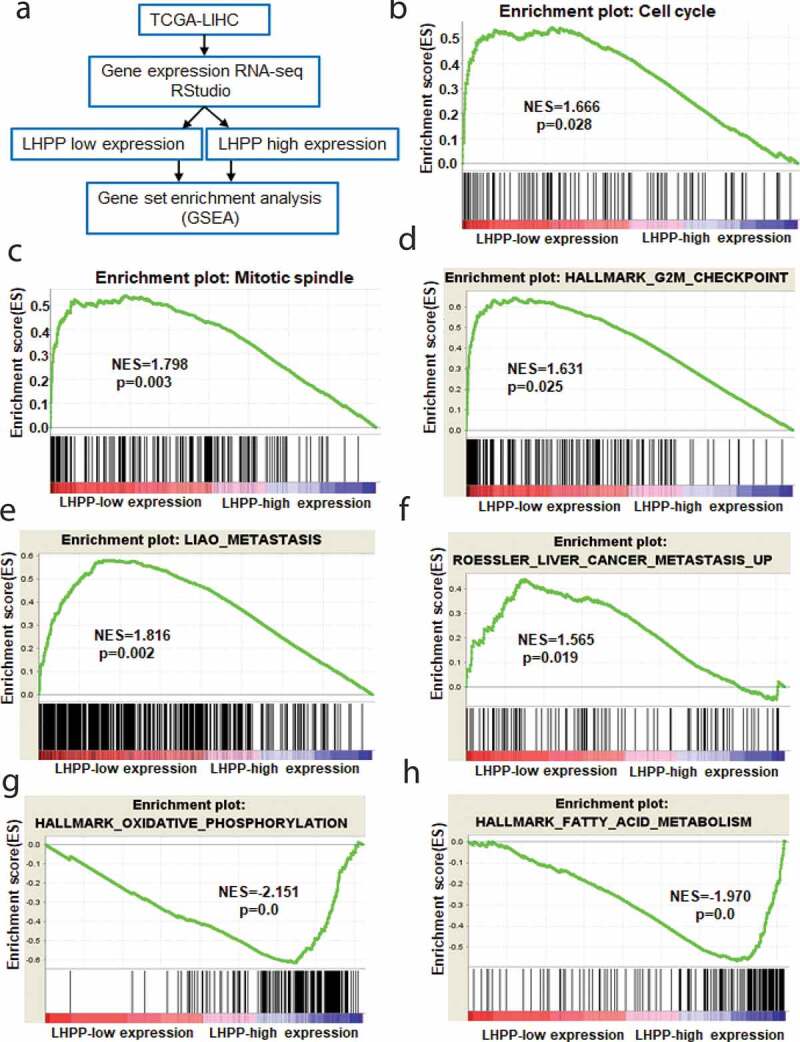
LHPP expression levels negatively correlated with cell cycle and metastasis in HCC tissues. (a) Analysis of the RNA-seq data of HCC patients from TCGA, and then divide into LHPP low expression and LHPP high expression group, followed using GSEA analysis. (b-f) GSEA analysis showed that pathways including cell cycle, mitotic spindle, G2 M checkpoint, metastasis and liver cancer metastasis were highly enriched in HCC patients with LHPP low expression. (g-h) The metabolic pathways such as oxidative phosphorylation and fatty acid metabolism were remarkably enriched in the LHPP high expression group.
LHPPdecreases the expression of CCNB1, PKM2, MMP7 and MMP9 in HCC cells
Next, we wanted to investigate any possible antitumor mechanismsaboutLHPPin humanHCC cells. We firstly detected the several oncogenes (including CCNB1, PKM2, MMP7, and MMP9) expression levels in the above LHPPoverexpression HCC cellsviaReal-time PCR assay, and found that overexpressionLHPPdecreasedCCNB1, PKM2, MMP7, and MMP9mRNA levels (Figure 5(a,b)). Moreover, a negative correlation was found between the expression levels of LHPP and CCNB1 in humanHCC tissues (r = −0.34, p < 0.0001) (Figure 5(c)). Also, the similar results could be found in the PKM2, MMP7, and MMP9 genes (Figure 5(d–f)). These results suggested that LHPPconstrained HCC cell growth and metastasis, likely through acting on the expression of CCNB1, PKM2, MMP7, and MMP9.
Figure 5.
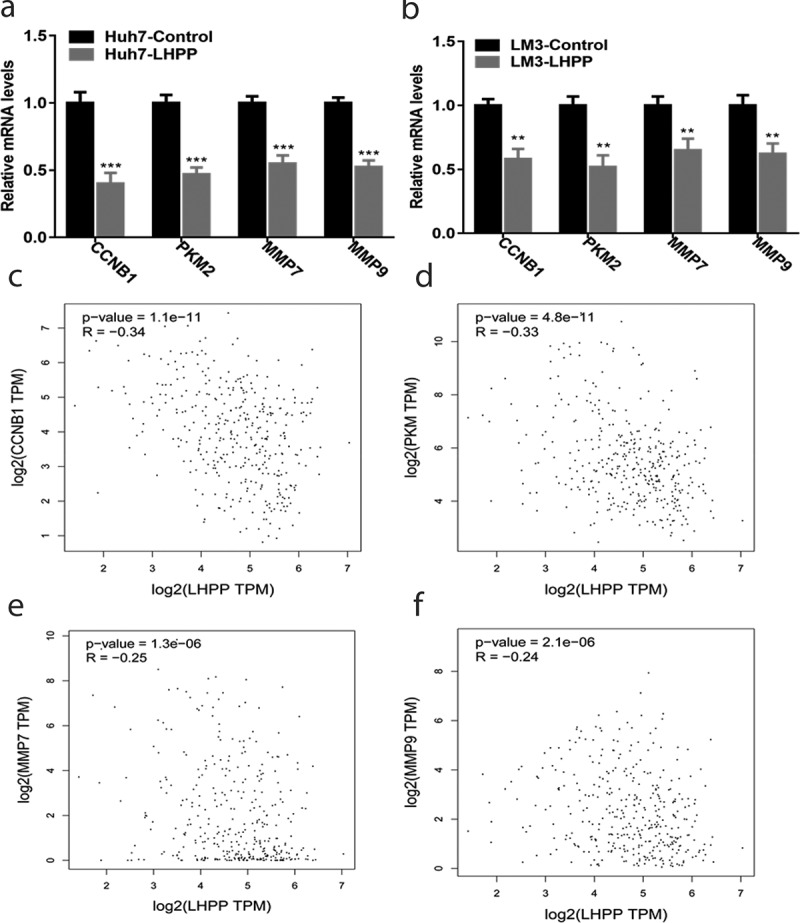
LHPP decreases the expression of CCNB1, PKM2, MMP7, and MMP9 in the HCC cells. (a-b) Overexpression of LHPP in the Huh7 and LM3 cells decreased CCNB1, PKM2, MMP7, and MMP9 mRNA levels. CCNB1, PKM2, MMP7, and MMP9 mRNA level was measured using the Real-time PCR. Means±SD from three independent experiments were presented as relative ratios to the control whose value was taken as 1.0. Significant differences were determined using the Student t-test, **P < 0.01, ***P < 0.001. (c-f) A negative correlation was found between the levels of CCNB1, PKM2, MMP7 and MMP9 with LHPP in the HCC tissues. The HCC tissue date comes from the TCGA database.
Discussion
In the present study, the TCGA database and the Oncomine database showed that LHPP was down-regulated in HCC tissues and high expression of LHPPwas associated with better clinical outcomes in HCC. And then, we found that the protein and mRNA levels of LHPP were reduced in human HCC cells. A functional study demonstrated that LHPP reduction promoted the growth and metastasis of human HCC cells. In addition, we further analyzed the RNA-seq data of HCC patients from TCGA using GSEA, and found that LHPP expression levels negatively correlated with cell cycle, mitotic spindle, G2 M checkpoint pathway, and metastasis in human HCC tissues. Furthermore, we showed that LHPP decreased the expression of CCNB1, PKM2, MMP7, and MMP9 in human HCC cells.
Currently, it has been reported that LHPP knockdown can accelerate cell proliferation, metastasis, and apoptosis by modulating AKT in cervical cancer [24]. Consistent with this finding in bladder cancer, knockdown of LHPPcan enhance the cell viability and colony formation via inactivating AKT/p65 signaling [25]. However, there are few studies available reporting on the roles of LHPP in humanHCC cells. The present study showed that LHPP is downregulation in HCCcells and overexpression of LHPP inhibits the growth and metastasis of HCC cells. However, there are still few studies that describe the precise role of LHPP in human HCC and its underlying mechanisms still unclear. In this study, we found that LHPP expression levels negatively correlated with cell cycle, mitotic spindle, G2 M checkpoint pathway and metastasis in human HCC tissues. Since the CCNB1, PKM2, MMP7, and MMP9 are the key genes in involving the above pathways, we next detected these oncogenes expression in the LHPP overexpressing HCC cells. Also, our data showed that overexpression LHPP decreased CCNB1, PKM2, MMP7, and MMP9 expression. Thus, we conclude that there is a certain relationship between LHPP and these oncogenes, which may represent a new therapeutic target for HCC.
In this context, we mainly elucidate the effect of LHPP on the growth and metastasis of human HCC cells. Consequently, we found that LHPPnegatively correlates with cell cycle, mitotic spindle, G2 M checkpoint pathway, and metastasis in human HCC tissues. Importantly, we further demonstrated that LHPP reduces the expression levels of CCNB1, PKM2, MMP7, and MMP9 genes. This provides a new idea for the follow-up study of human HCC.
Materials and methods
LHPP expression analysis from the TCGA database
The transcriptional level of LHPP in HCC tissues was analyzed from the Oncomine database (https://www.oncomine.org/resource/main.html). A total of 329 HCC specimens and 226 normal tissues were available from the LHPP Expression in Roessler Liver 2 Statistics and LHPP Expression in Chen Liver Statistics.
Kaplan-Meier survival analysis of the LHPP
HCC tissues contain patient survival information coming from the TCGA database (https://tcga-data.nci.nih.gov/publications/tcga). According to the expression and median value of the LHPP gene in HCC patients, they were divided into two groups: high expression group and low expression group. The survival curve was drawn from the Kaplan–Meier Plotter database (http://kmplot.com/analysis/).
The gene set enrichment analysis (GSEA) of the LHPP
GSEA was performed using RNA-seq sets from TCGA-LIHC (https://xenabrowser.net/datapages/?dataset=TCGA.LIHC; datasetID:TCGA.LIHC. sampleMap/HiSeqV2). According to the mRNA level of the LHPP gene in HCC patients, they were divided into two groups: high expression group and low expression group.
Cell culture and stable cell lines
Human HCC cell lines (SMCC-7721, Huh7, HepG2, MHCC-97 H, and LM3) and normal liver cells (LO2) were from Cell Bank of Shanghai Institutes of Biological Sciences, Chinese Academy of Sciences (Shanghai, China). All cells were passaged in Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies) supplemented with 10%fetal bovine serum (FBS, HyClone), 1x Pen/Strep, and4 mM L-glutamine and incubated at 37°C in 5% CO2.
For over-expression of LHPP, Flag-tagged full-length LHPP cDNA was cloned in the pCDH-CMV-MCS-EF1-Puro plasmid. For lentivirus production, HEK293 T cells transfected with the lentiviral vectors and helper plasmidsDR8.91 and VSVG. Forty-eight-hour and 72 h after transfection,virus-containing media were collected and concentrated by centrifugation at 10,000 g for 18 h. Cells were transduced using lentivirus with 8 μg/ml polybrene and then selected with the appropriate antibiotics.
RNA isolation and quantitative real-time PCR
Total RNA extraction of the HCC cells was extracted using TRIzol (Invitrogen) and 1.5 μg RNA was reverse transcribed by the First StrandcDNA Synthesis Kit (Marligen Biosciences). Quantitative real-time PCR was performed by SYBR Green PCR Master Mix (Applied Biosystems) in the ABI 7300Detection System (Applied Biosystems). PCR reactions were done in triplicates with following conditions: 95°C/30 s, 40 cycles of 95°C/10 s, 55°C/10 s and 72°C/10 s on Roche cycler and repeated at least three times. Relative mRNA levels were calculated using the – ΔΔCt method using β-actin as control and expressed as 2^(-ΔΔCt). See Table1 for the complete list of qPCR primer sequences.
Table 1.
Primer sequences for qRT-PCR.
| Resource | Gene name | Primer sequence (5→3′) |
|---|---|---|
| Human | LHPP | ForwardGCTGGACGTTTGTCCCTACA |
| ReverseGTGGGCTTCCACTCCTATCG | ||
| Human | CCNB1 | Forward GCAGCAGGAGCTTTTTGCTT |
| Reverse CCAGGTGCTGCATAACTGGA | ||
| Human | PKM2 | Forward GCCTGCTGTGTCGGAGAAG |
| Reverse CAGATGCCTTGCGGATGAATG | ||
| Human | MMP-7 | Forward TTGATGGGCCAGGAAACACG |
| Reverse AGACTGCTACCATCCGTCCA | ||
| Human | MMP-9 | Forward CTGGAGGTTCGACGTGAAGGC |
| Reverse GGCTTTCTCTCGGTACTGGAAG | ||
| Human | β-actin | Forward TCCCTGGAGAAGAGCTACG |
| Reverse GTAGTTTCGTGGATGCCACA |
Western blot analysis
Cells were lysed in RIPA buffer (Beyotime) containing protease inhibitors (KeyGen Biotech) for 30 min on ice and centrifuged at 4°C at 16,000 × g for 15 min to collect the supernatant. The protein concentrations were measured by Bradford assay (Bio-Rad Labs) and mixed with loading buffer before boiled. Equal amounts of protein samples were subjected to SDS-PAGE and transferred to PVDF membranes (Millipore). The membranes were then blotted with primary antibodies overnight at 4°C, followed by incubation with the secondary antibody (1:5000). The proteins were then developed using enhanced chemiluminescence (ECL, Thermo Fisher Scientific).Antibody used for western blot: LHPP (Proteintech), β-actin (Proteintech).
Cell viability analysis
Cell viability was measured with a CCK-8 kit (Boster). Huh7 and LM3 cells stably expressing vector or LHPP(2,500 cells/well) were seeded into 96-well plates. The above cells were cultured at 37°C in 5% CO2 for 24, 48, 72, and 96 h, the medium of each well was replaced with 10% CCK-8 solution in fresh medium. The above cells were then incubated for 2 h, afterward, absorbance at 450 nm was measured using a microplate reader.
Colony formation assay
The above HCC cells were seeded, respectively, in six-well plates at a density of 500 cells/well at 37°C in a 5% CO2 humidified environment. After incubation about 2 wk, the plates were washed by PBS and fixed with methanol for 1 h, and then stained with Giemsa solution (Solarbio) for 30 min. Subsequently, the plates were washed with clean water and the number of the above cell colonies was counted.
Transwell migration and invasion
For transwell migration and invasion assays, matrigel (BD Biosciences) was just used in the invasion assay. Ten percent matrigel allowed to polymerize at the base of the top chamber of a 24 well transwell plate for 30 min at 37°C. The above HCC cells (2x105 cells/well) were starved in serum-free medium for 16 h and added to the top chambers. The bottom chambers were filled serum-containing medium. After 48 h, we used the cotton swabs to remove the non-invading cells from the upper surface of the membrane. The above cells that invaded were fixed in 4% paraformaldehyde for 30 min and then stained with 0.1% crystal violet for 30 minat room temperature. Excess dye was washed off with water.
Statistical analysis
Statistical analysis was performed with SPSS 24. The Kaplan–Meier survival analysis, Student t-test, the Pearson test, and one-way analysis of variance were used to compare the difference of the variables. P-value <0.05 was considered statistically significant.
Funding Statement
This work was supported by the National Natural Science Foundation of China [31801186, 31701005, 81572832, and 81874174]; SIAT Innovation Program for Excellent Young Researchers [201801]; Shanghai Rising-Star Program [18QA1402600]; Shanghai Municipal Commission of Health and Family Planning [2018YQ12] and School of Medicine, Shanghai Jiao Tong University (Excellent Youth Scholar Initiation) Grant [17XJ11015 and 18XJ11006].
Disclosure statement
The authors declare no conflict of interest.
References
- [1].Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- [2].Llovet JM, Ducreux M, Lencioni R, et al.EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908–943. [DOI] [PubMed] [Google Scholar]
- [3].El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. [DOI] [PubMed] [Google Scholar]
- [4].Omer RE, Kuijsten A, Kadaru AM, et al. Population-attributable risk of dietary aflatoxins and hepatitis B virus infection with respect to hepatocellular carcinoma. Nutr Cancer. 2004;48:15–21. [DOI] [PubMed] [Google Scholar]
- [5].Lai SW, Chen PC, Liao KF, et al. Risk of hepatocellular carcinoma in diabetic patients and risk reduction associated with anti-diabetic therapy: a population-based cohort study. Am J Gastroenterol. 2012;107:46–52. [DOI] [PubMed] [Google Scholar]
- [6].Said A, Ghufran A. Epidemic of non-alcoholic fatty liver disease and hepatocellular carcinoma. World J Clin Oncol. 2017;8:429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. [DOI] [PubMed] [Google Scholar]
- [8].Klumpp S, Krieglstein J. Phosphorylation and dephosphorylation of histidine residues in proteins. Eur J Biochem. 2002;269:1067–1071. [DOI] [PubMed] [Google Scholar]
- [9].Klumpp S, Krieglstein J. Reversible phosphorylation of histidine residues in vertebrate proteins. Biochim Biophys Acta. 2005;1754:291–295. [DOI] [PubMed] [Google Scholar]
- [10].Matthews HR. Protein kinases and phosphatases that act on histidine, lysine, or arginine residues in eukaryotic proteins: a possible regulator of the mitogen-activated protein kinase cascade. Pharmacol Ther. 1995;67:323–350. [DOI] [PubMed] [Google Scholar]
- [11].Ek P, Pettersson G, Ek B, et al. Identification and characterization of a mammalian 14-kDa phosphohistidine phosphatase. Eur J Biochem. 2002;269:5016–5023. [DOI] [PubMed] [Google Scholar]
- [12].Klumpp S, Hermesmeier J, Selke D, et al. Protein histidine phosphatase: a novel enzyme with potency for neuronal signaling. J Cereb Blood Flow and Metab. 2002;22:1420–1424. [DOI] [PubMed] [Google Scholar]
- [13].Chaikuad A, Filippakopoulos P, Marcsisin SR, et al. Structures of PGAM5 provide insight into active site plasticity and multimeric assembly. Structure. 2017;25:1089–99.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Koike E, Toda S, Yokoi F, et al. Expression of new human inorganic pyrophosphatase in thyroid diseases: its intimate association with hyperthyroidism. Biochem Biophys Res Commun. 2006;341:691–696. [DOI] [PubMed] [Google Scholar]
- [15].Yokoi F, Hiraishi H, Izuhara K. Molecular cloning of a cDNA for the human phospholysine phosphohistidine inorganic pyrophosphate phosphatase. J Biochem. 2003;133:607–614. [DOI] [PubMed] [Google Scholar]
- [16].Cui L, Gong X, Tang Y, et al. Relationship between the LHPP gene polymorphism and resting-state brain activity in major depressive disorder. Neural Plast. 2016;2016:9162590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Polimanti R, Wang Q, Meda SA, et al. The interplay between risky sexual behaviors and alcohol dependence: genome-wide association and neuroimaging support for LHPP as a risk gene. Neuropsychopharmacol. 2017;42:598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cai N, Bigdeli TB, Kretzschmar W, et al.Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Russell RG. Metabolism of inorganic pyrophosphate (PPi). Arthritis Rheumatism. 1976;19(Suppl 3):465–478. [DOI] [PubMed] [Google Scholar]
- [20].Machens A, Holzhausen HJ, Dralle H. The prognostic value of primary tumor size in papillary and follicular thyroid carcinoma. Cancer. 2005;103:2269–2273. [DOI] [PubMed] [Google Scholar]
- [21].Vijayakrishnan J, Kumar R, Henrion MY, et al. A genome-wide association study identifies risk loci for childhood acute lymphoblastic leukemia at 10q26.13 and 12q23.1. Leukemia. 2017;31:573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lesseur C, Diergaarde B, Olshan AF, et al. Genome-wide association analyses identify new susceptibility loci for oral cavity and pharyngeal cancer. Nat Genet. 2016;48:1544–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hindupur SK, Colombi M, Fuhs SR, et al. The protein histidine phosphatase LHPP is a tumour suppressor. Nature. 2018;555:678–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zheng J, Dai X, Chen H, et al. Down-regulation of LHPP in cervical cancer influences cell proliferation, metastasis and apoptosis by modulating AKT. Biochem Biophys Res Commun. 2018;503:1108–1114. [DOI] [PubMed] [Google Scholar]
- [25].Li Y, Zhang X, Zhou X, et al. LHPP suppresses bladder cancer cell proliferation and growth via inactivating AKT/p65 signaling pathway. Biosci Rep. 2019;39. DOI: 10.1042/BSR20182270 [DOI] [PMC free article] [PubMed] [Google Scholar]


