Figure 2.
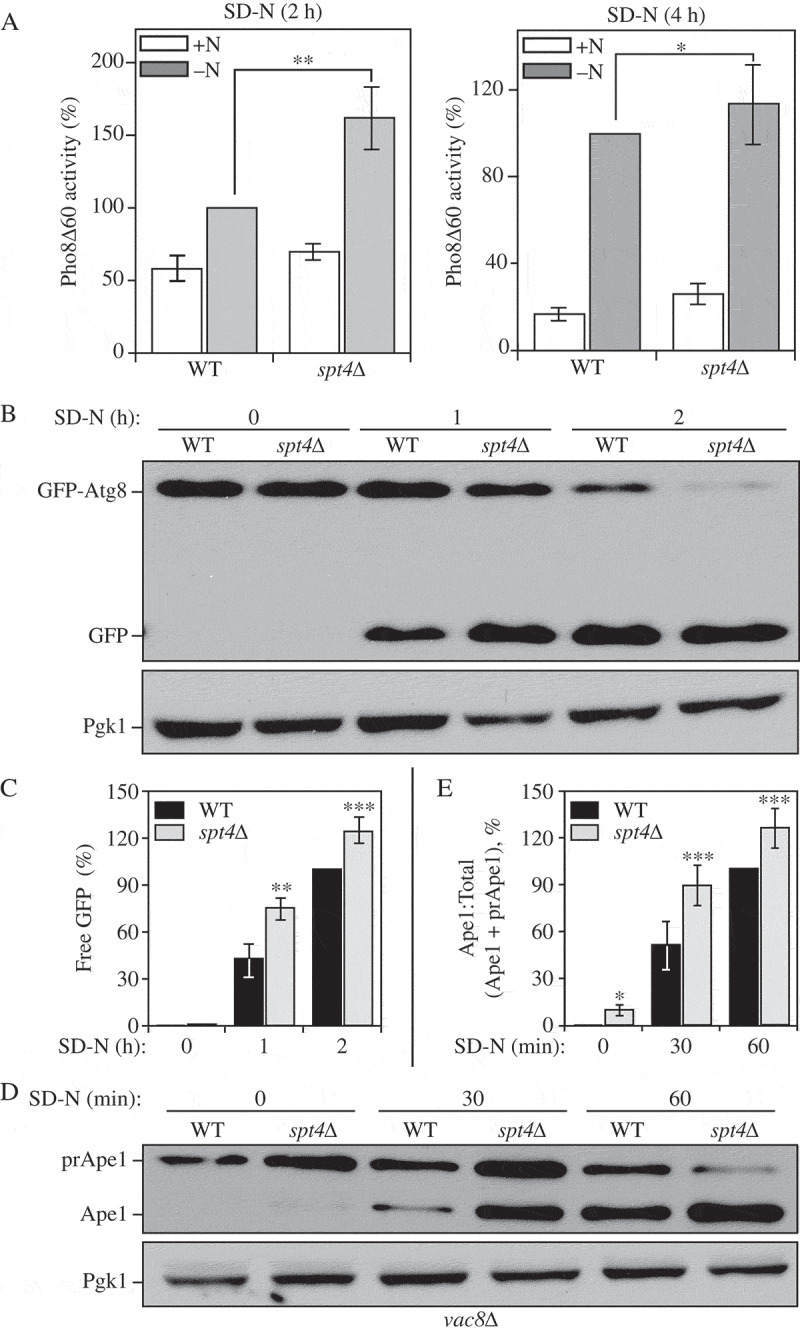
Spt4 negatively regulates autophagy activity. (A) Autophagy activity was measured with the quantitative Pho8∆60 assay in WT (JMY347) and spt4∆ (WXY131) cells under growing conditions (+N) and after 2 or 4 h of nitrogen starvation (-N). Error bars represent the SEM of at least 3 independent experiments. *, p < 0.05; **, p < 0.01. (B-C) Autophagy activity was measured using the GFP-Atg8 processing assay. Wild-type (WXY112) and spt4∆ cells (WXY113) with CUP1 promoter-driven GFP-ATG8 were grown to mid-log phase in YPD and then starved (SD-N) for the indicated times. Samples from growing (YPD, 0 h) and starvation (SD-N, t = 1–2 h) conditions were collected. Proteins were analyzed by western blot with anti-YFP antibody and anti-Pgk1 (loading control) antiserum. The quantitative analysis of processed GFP is shown in (C), and the error bar represents the SEM of 3 independent experiments. The processed GFP after 2 h of starvation was set as 1, and other values were normalized. *, p < 0.05; **, p < 0.01. (D-E) Wild-type (WXY121) and spt4∆ (WXY122) cells with a deletion of VAC8 were collected in both growing (YPD) and starvation (SD-N, t = 30–60 min) conditions. The precursor (pr) and mature forms of Ape1 were separated by SDS-PAGE and detected with anti-Ape1 antiserum by western blot. Pgk1 was used as a loading control. The ratio of processed Ape1 was quantified in (E), and the error bar represents the SEM of 3 independent experiments. The maturation of prApe1 after 60 min of starvation has been set as 1, and other values were normalized. *, p < 0.05; ***, p < 0.005.
