Abstract
Epstein-Barr virus (EBV) can infect human B cells and is associated with various types of B cell lymphomas. Studies on the global alterations of the cellular pathways mediated by EBV-induced B cell transformation are limited. In the present study, microarray analysis was performed following generation of two EBV-infected B-lymphoblastoid cell lines (BLCL), in which normal B cells obtained from two healthy Thai individuals and transcriptomic profiles were compared with their respective normal B cells. The two EBV-transformed BLCL datasets exhibited a high degree of similarity between their RNA expression profiles, whereas the two normal B-cell datasets did not exhibit the same degree of similarity in their RNA expression profiles. Differential gene expression analysis was performed, and the results showed that EBV infection was able to dysregulate several cellular pathways in the human B-cell genes involved in cancer and cell activation, such as the MAPK, WNT and PI3K-Akt signaling pathways, which were upregulated in the BLCL and were associated with increased cellular proliferation and immortalization of EBV-infected B cells. Expression of proteins located in the plasma membrane, which initiate a biological response to ligand binding, were also notably upregulated. Expression of genes involved in cell cycle control, the p53 signaling pathway and cellular senescence were downregulated. In conclusion, genes that were markedly upregulated by EBV included those involved in the acquisition of a tumorigenic phenotype of BLCL, which was positively correlated with several hallmarks of cancer.
Keywords: B cells, Epstein-Barr virus, B-lymphoblastoid cell lines, microarray, gene expression
Introduction
The majority of the global population have been infected by the Epstein-Barr virus (EBV). However, EBV subtype distribution in EBV-induced diseases and the EBV antibody levels varies among the infected population, according to their ethnicity and geographic characteristics (1-3). EBV favorably infects human B cells and is found to be associated with several types of human B cell lymphoma as a consequence of disrupted cellular pathways by EBV proteins, such as latent membrane proteins (4-7). EBV causes certain types of B cell lymphomas, such as diffuse large B cell lymphoma (DLBCL), Burkitt lymphoma, Hodgkin lymphoma and EBV-associated lymphoproliferative disorder that can cause mortality in patients with immune deficiency (4). Furthermore, EBV can infect and replicate in epithelial cells, and cause several types of cancer, including nasopharyngeal carcinoma (NPC) and gastric carcinoma in the infected cells (8-10).
EBV can transform human B cells, and alters their normal phenotype properties into a cell with a more transformative phenotype, which includes dysregulated proliferation and immortalization of EBV-transformed B-lymphoblastoid cell lines (BLCL), resulting in increased cell growth and survival (4,7,11). Host transcription factors, including NF-κB, Jun/Fos and AP-1, are induced by EBV proteins, which may be involved in the process of cellular transformation (4,12,13).
EBV can transform B cells into cells with tumorigenic phenotypes, including immortalization and sustained proliferative signaling, which are hallmarks of cancer (14). However, there are limited reports on the cellular gene expression profiles in human BLCL, particularly when compared with normal B cells (15,16). In the present study, microarray expression analysis, a high throughput method, was used to investigate the differential gene expression profiles of human BLCL and cellular pathway alterations associated with tumorigenic phenotypes of B cells. A more in-depth understanding of the specific molecular mechanisms and cellular pathways involved in EBV-induced cellular transformation may highlight the identification of potential biomarkers and targets for treatment of EBV-related diseases.
Materials and methods
Study participants
In the present study, 2 healthy male volunteers (aged 38 and 40 years) were enrolled at The King Chulalongkorn Memorial Hospital, Faculty of Medicine, Chulalongkorn University (Bangkok, Thailand). The present study was approved by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University (Bangkok, Thailand) (approval no. 358/46) and both subjects provided written informed consent for participation.
Peripheral blood mononuclear cell (PBMC) isolation and BLCL preparation
PBMCs were isolated from healthy donor blood according to the manufacturer's protocol using density gradient separation with Isoprep (Robbins Scientific Corporation). PBMCs were adjusted to 1x106 cells/ml with R10 medium; RPMI-1640 supplemented with 100 U/ml penicillin and 100 U/ml streptomycin (all from Gibco; Thermo Fisher Scientific, Inc.) and 10% heat-inactivated FBS (BioWhittaker, Inc.). For the EBV-containing culture supernatant, the marmoset B-lymphoblastoid cell line (B95-8) were maintained in R10 media at an exponentially growing rate in a humidified incubator at 37˚C with 5% CO2. After 3 days, the supernatant was collected and centrifuged at 300 x g for 10 min at 4˚C to separate the EBV-containing culture supernatant from the cells. The supernatant was filtered through a 0.45 µm filter, aliquoted and stored at -80˚C. Typically, the culture supernatant contains >100-1,000 transforming units per ml (17). EBV-transformed BLCL were generated by incubating 1x106 cells/ml PBMCs with 1 ml EBV supernatant from the B95-8 cell line for 1 h at 37˚C with 5% CO2. BLCL were maintained in R20 medium; RPMI-1640 supplemented with 100 U/ml penicillin, 100 U/ml streptomycin and 20% heat-inactivated FBS, with 1 mg/ml cyclosporin A. The B95-8 cell line was kindly provided by The National Institute of Health, Thailand. Following B cell transformation, the BLCL were used for RNA extraction or cryopreserved for future use.
RNA sample preparation
Total RNA was extracted from the cells mentioned above using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RNA purity and integrity qualification were evaluated on a ND-1000 Spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.) and an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). RNA samples were used for RNA labeling and hybridization using an Agilent One-Color Microarray-Based Gene Expression Analysis protocol (version 6.5; Agilent Technologies, Inc.). Briefly, 100 ng total RNA from each sample was linearly amplified using the oligo dT-promoter primer included in the kit, and labeled with Cy3-dCTP and Cy5-dCTP at 40˚C for 2 h (Agilent Technologies, Inc.) for BLCL and normal B cells, respectively. Following purification, the concentration and specific activity of the labeled cRNAs (pmol Cy3/µg cRNA) were measured using a NanoDrop ND-1000.
Hybridization and scanning
A transcriptome database of each RNA sample was created using Agilent SurePrint G3 Human GE 8x60 K Microarray kit (Agilent Technologies Inc.). Briefly, 600 ng of each labeled cRNA was fragmented by adding 5 µl 10x blocking agent and 1 µl 25x fragmentation buffers, and then heated at 60˚C for 30 min. Finally, 25 µl 2x GE hybridization buffer was added to dilute the labeled cRNA. Hybridization solution (40 µl) was dispensed into the gasket slide and assembled into the Agilent SurePrint G3 Human GE 8X60K, V3 Microarrays. The slides were incubated for 17 h at 65˚C in an Agilent hybridization oven and then washed at room temperature using the Agilent One-Color Microarray-Based Gene Expression Analysis protocol. The hybridized array was immediately scanned with an Agilent Microarray Scanner D (Agilent Technologies, Inc.)
Data processing and bioinformatics analyses
Raw data were extracted using Agilent Feature Extraction Software (version 11.0.1.1; Agilent technologies, Inc.). The raw data for the same genes were then summarized automatically using the Agilent feature extraction protocol to generate a raw data text file, providing expression data for each gene probed on the array. Array probes with Flag A in the samples were filtered out. The selected gProcessedSignal value was log-transformed and normalized using the Quantile method (18). Gene-Enrichment and Functional Annotation analysis for the significant probe list was performed using g: Profiler, a web server for functional enrichment analysis (19) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (20).
Statistical analysis
R version 2.15.1 software (21) was used for image and statistical analysis. Statistical significance of the expression data was determined using the fold-change and local-pooled-error test for identifying significantly differentially expressed genes in microarray experiments in which the null hypothesis was that no difference existed between 2 groups. Hierarchical cluster analysis was performed using complete linkage and Euclidean distance as a measure of similarity. Pearson's correlation (scatter plot) and multidimensional scaling plot (2-D graphics) were used to analyze the degree of reproducibility and similarity between samples, respectively. Log2 fold-change values >4.0 and <-4.0 were set as the cut off for significantly upregulated and downregulated genes, respectively. False discovery rate adjusted P<0.05 was considered to indicate a statistically significant Gene Ontology (22,23) and KEGG pathway.
Results
Morphology of EBV-transformed BLCL
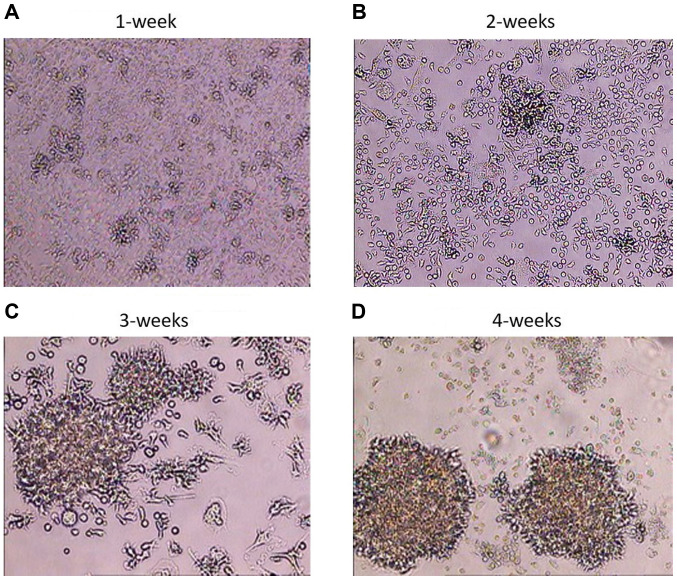
Following the infection of PBMCs and the transformation of B cells into BLCL, the BLCL grew as small cell clusters that were visible by light microscopy within a week (Fig. 1A). With continued cell culturing, microscopic clusters became larger and single cells exhibited large nuclei and numerous vacuoles on weeks 2 and 3 (Fig. 1B and C). The highest degree of cell clumping, cell elongation, cell trajectory and formation of rosette shapes was visible macroscopically on week 4 (Fig. 1D). The infection rate was ~100%, at this time point, and the B cell transformation was considered successful (17,24-26); BLCL were considered ready for RNA extraction.
Figure 1.
Visualization of cell clusters in EBV-infected cells. PBMCs were infected with EBV and examined during transformation using phase-contrast microscopy. Magnification, x100. (A) Cells after 1 week of EBV infection; the BLCL grew as small cell clusters. (B) Cells after 2 weeks of EBV infection; the BLCL grew as cell clusters. (C) Cells after 3 weeks of EBV infection; the BLCL grew as big clusters and single cells exhibited large nuclei. (D) Cells after 4 weeks of EBV infection; increased cell clumping and formation of rosette shapes were observed in BLCL. EBV, Epstein-Barr virus; PBMC, peripheral blood mononuclear cell; BLCL, B-lymphoblastoid cell lines.
Quality control check of gene expression analysis by microarray analysis
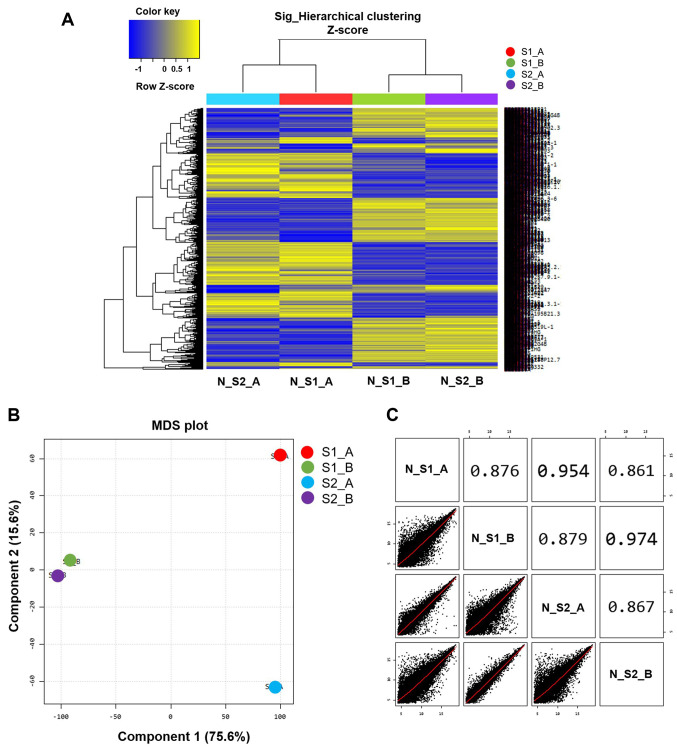
Microarray analysis was used to differentiate gene expression profiles between normal B cells (S1_A and S2_A) and EBV-transformed BLCL (S1_B and S2_B). A multidimensional scaling plot was used to investigate the similarities amongst samples. Of note, the two EBV-transformed BLCL datasets exhibited similar RNA expression profiles that were represented by two points (S1_B and S2_B; Fig. 2B), whereas the two normal B cell datasets exhibited a long distance of RNA expression profiles as represented by two separated points (S1_A and S2_A; Fig. 2B). The scatter plots and correlation analysis showed a close correlation between S1_B and S2_B and S1_A and S2_A (Fig. 2C).
Figure 2.
Gene expression microarray analysis. (A) Heat map analysis of differential gene expression profiles amongst the 4 samples tested. The rows display genes and the columns show the different samples. (B) Degree of similarity amongst the 4 samples was tested, and determined using an MDS plot. Component 1 and 2 are 22,628 normalized and filtered cellular genes expressed in each sample. (C) Scatter plot between 2 samples; the correlation was analyzed. MDS, Multidimensional scaling plot. EBV, Epstein-Barr virus; BLCL, B-lymphoblastoid cell lines; normal B cells, S1_A and S2_A; EBV-transformed BLCL, S1_B and S2_B.
Gene expression and molecular pathway analysis
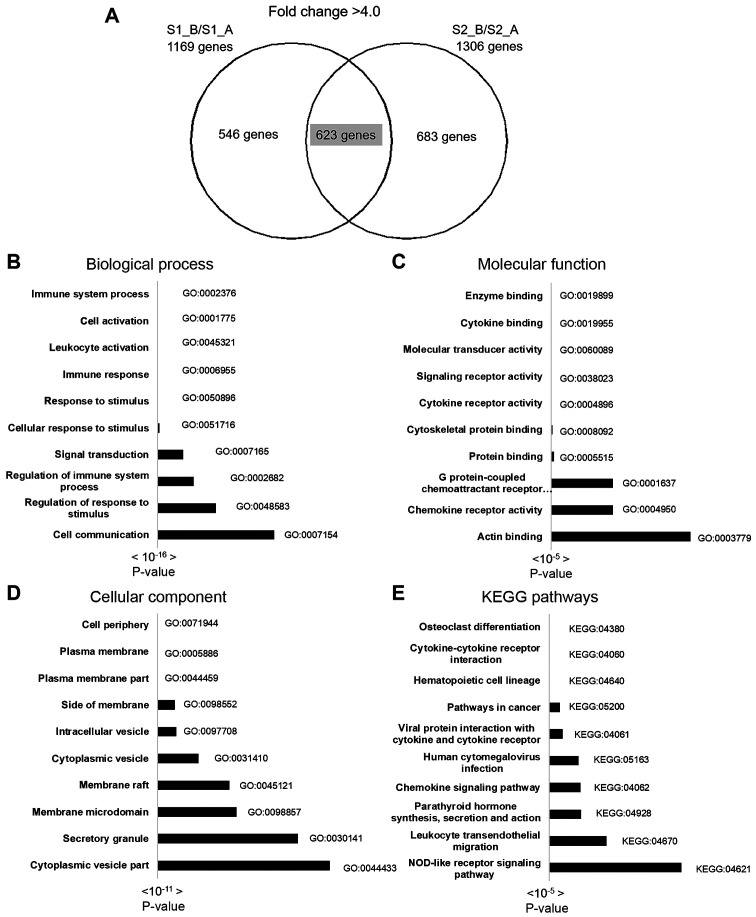
Following data normalization, heat map clustering was plotted to express the distance similarity between normal B cells and EBV-transformed BLCL (Fig. 2A). A log2 fold-change of >4.0 and <4.0 were set as the cut off points for the differential gene expression analysis of up and downregulated genes, respectively. A total of 1,018 and 1,061 upregulated, and 1,169 and 1,306 downregulated genes were identified in samples S1_B and S2_B, respectively. There were 623 overlapping upregulated genes with a >4.0 fold-change and 503 overlapping downregulated genes with a <4.0 fold-change in the two datasets (S1_B and S2_B) (Table SI). The top 30 most up and downregulated genes are shown in Table I.
Table I.
The 30 most significantly up- and downregulated genes in EBV-transformed BLCL samples (S1_B and S2_B), as compared with normal B cell samples (S1_A and S2_A).
| Gene symbol | Upregulated genes Description | Mean Log FC | Gene symbol | Downregulated genes Description | Mean LogFC |
|---|---|---|---|---|---|
| TYROBP | TYRO protein tyrosine kinase-binding protein | 1,429.88 | EBI3 | Epstein-Barr virus-induced gene 3 | -385.97 |
| CX3CR1 | CX3C chemokine receptor 1 | 1,177.16 | MIR155HG | MIR155 Host Gene | -263.21 |
| HBA2 | Hemoglobin subunit alpha | 1,114.79 | LAMP3 | Lysosome-associated membrane glycoprotein 3 | -210.37 |
| S100A9 | Protein S100-A9 | 912.38 | lnc-EFR3B-4 | EFR3 Homolog B, binding | -161.03 |
| CD8A | T-cell surface glycoprotein CD8 alpha chain | 707.23 | CD70 | CD70 antigen | -124.01 |
| CSF1R | Macrophage colony-stimulating factor 1 receptor | 652.57 | PYCR1 | Pyrroline-5-carboxylate reductase 1, mitochondrial | -119.83 |
| CSF1R | Colony stimulating factor 1 receptor | 625.56 | HMMR | Hyaluronan mediated motility receptor | -100.36 |
| GNLY | Granulysin | 548.88 | VWCE | von Willebrand factor C and EGF domain-containing protein | -87.10 |
| S100A10 | Protein S100-A10 | 485.02 | UBE2C | Ubiquitin-conjugating enzyme E2 C | -80.61 |
| CD3D | T-cell surface glycoprotein CD3 delta chain | 477.29 | PHLDA3 | Pleckstrin homology-like domain family A member 3 | -80.53 |
| FOS | Proto-oncogene c-Fos | 462.16 | PIK3CD-AS2 | PIK3CD Antisense RNA 2 | -80.32 |
| CD14 | Monocyte differentiation antigen CD14 | 433.84 | CDCA2 | Cell division cycle-associated protein 2 | -75.40 |
| SERPINA1 | serpin family A member 1 | 423.99 | FSCN1 | Fascin | -72.29 |
| MAFB | Transcription factor MafB | 410.38 | MELK | Maternal embryonic leucine zipper kinase | -68.64 |
| NKG7 | Protein NKG7 | 393.96 | CENPM | Centromere protein M | -67.54 |
| PRAM1 | PRAME family member 1 | 334.75 | PLEKHH3 | Pleckstrin homology domain-containing family H member 3 | -65.46 |
| LILRB3 | Leukocyte immunoglobulin-like receptor subfamily B member 3 | 309.86 | CCNB2 | G2/mitotic-specific cyclin-B2 | -63.19 |
| PTGDS | Prostaglandin-H2 D-isomerase | 305.56 | TYMS | Thymidylate synthase | -61.40 |
| SPOCK2 | Testican-2 | 291.96 | CDK1 | Cyclin-dependent kinase 1 | -59.03 |
| CD2 | T-cell surface antigen CD2 | 283.29 | MND1 | Meiotic nuclear divisions 1 | -54.32 |
| DUSP6 | Dual specificity protein phosphatase 6 | 281.45 | CDCA3 | Cell division cycle-associated protein 3 | -53.49 |
| F13A1 | Coagulation factor XIII A chain | 270.72 | lnc-SPAG1-3 | lincRNA | -49.32 |
| CSF3R | Granulocyte colony-stimulating factor receptor | 226.40 | BIRC5 | Baculoviral IAP repeat-containing protein 5 | -49.25 |
| GP1BB | Platelet glycoprotein Ib beta chain | 214.81 | ASPM | abnormal spindle microtubule assembly | -47.99 |
| CTSW | Cathepsin W | 190.24 | CDCA5 | cell division cycle associated 5 | -47.78 |
| C1orf162 | Transmembrane protein C1orf162 | 173.42 | TROAP | Trophinin-associated protein | -45.83 |
| NRGN | Neurogranin | 172.89 | TPX2 | Targeting protein for Xklp2 | -45.54 |
| LST1 | Leukocyte-specific transcript 1 protein | 157.63 | LOC283710 | Uncharacterized LOC283710 | -42.64 |
| GLIPR2 | Golgi-associated plant pathogenesis-related protein 1 | 143.79 | C16orf59 | Chromosome 16 open reading frame 59 | -41.83 |
| DUSP1 | Dual specificity protein phosphatase 1 | 137.27 | CDC45 | Cell division control protein 45 homolog | -39.36 |
Gene-enrichment and functional annotation analysis were performed using KEGG pathway and gene ontology analysis in g:Profiler. A total of 623 overlapping upregulated genes were used for complete gene ontology analysis; 178 terms in biological process (BP), 37 terms in cellular component (CC), 24 terms in molecular function (MF) and 15 KEGG pathways were identified. The top 10 BP, CC, MF and KEGG pathways are shown in Fig. 3, and the top 3 pathways of the 30 most upregulated genes and gene lists are shown in Table II. The most significant BPs included immune system process, cell activation, response to stimulus and leukocyte activation. The most significant CCs included cell periphery, plasma membrane and plasma membrane part. The most significant MF pathways included enzyme binding, cytokine binding and molecular transducer activity. The most significant KEGG pathways included osteoclast differentiation, cytokine-cytokine receptor interaction, hematopoietic cell lineage and pathways involved in cancer.
Figure 3.
Molecular pathway and functional analysis of upregulated genes using g:Profiler and KEGG pathway enrichment analysis. (A) A total of 623 overlapping differentially upregulated genes with a fold-change of >4.0 were identified. The top 10 pathways in (B) biological process, (C) cellular component, (D) molecular function and (E) KEGG pathways. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table II.
Top 3 pathways and gene lists of enriched GO categories, and KEGG pathways of 30 upregulated genes.
| A, Biological process | ||||
|---|---|---|---|---|
| Pathways | GO term | Gene count | Gene lists | P-value |
| Immune system process | GO:0002376 | 18 | TYROBP, GNLY, FOS, CD14, MAFB, CSF1R, CD8A, SERPINA1, LILRB3, CSF3R, S100A9, PRAM1, CTSW, CD3D, CD2, LST1, CX3CR1, DUSP1 | 7.31533E-06 |
| Regulation of immune system process | GO:0002682 | 14 | TYROBP, FOS, CD14, MAFB, CD8A, LILRB3, CSF3R, S100A9, PRAM1, CD3D, CD2, LST1, CX3CR1, DUSP1 | 8.96541E-06 |
| Cell activation | GO:0001775 | 12 | TYROBP, CD14, MAFB, CD8A, SERPINA1, LILRB3, S100A9, PRAM1, GP1BB CD3D, CD2, LST1 | 0.00026598 |
| B, Molecular function | ||||
| Pathways | GO term | Gene count | Gene lists | P-value |
| Signaling receptor activity | GO:0038023 | 9 | CD14, CSF1R, CD8A, LILRB3, CSF3R, GP1BB, CD3D, CD2, CX3CR1 | 0.01814678 |
| Molecular transducer activity | GO:0060089 | 9 | CD14, CSF1R, CD8A, LILRB3, CSF3R, GP1BB, CD3D, CD2, CX3CR1 | 0.02401768 |
| MAP kinase tyrosine/ serine/threonine phosphatase activity | GO:0017017 | 2 | DUSP6, DUSP1 | 0.03972376 |
| C, Cellular component | ||||
| Pathways | GO term | Gene count | Gene lists | P-value |
| External side of plasma membrane | GO:0009897 | 6 | CD14, CD8A, CSF3R, CD3D, CD2, CX3CR1 | 0.00348039 |
| Cytoplasmic vesicle part | GO:0044433 | 10 | TYROBP, GNLY, CD14, F13A1, SERPINA1, LILRB3, S100A9, CTSW, NRGN, CD3D | 0.00526305 |
| Cell surface | GO:0009986 | 8 | TYROBP, CD14, CSF1R, CD8A, CSF3R, CD3D, CD2, CX3CR1 | 0.00549100 |
| D, KEGG pathway | ||||
| Pathways | GO term | Gene count | Gene lists | P-value |
| Hematopoietic cell lineage | KEGG:04640 | 7 | CD14, CSF1R, CD8A, CSF3R, GP1BB, CD3D, CD2 | 5.73E-08 |
| Osteoclast differentiation | KEGG:04380 | 4 | TYROBP, FOS, CSF1R, LILRB3 | 0.01092597 |
| Acute myeloid leukemia | KEGG:05221 | 3 | CD14, CSF1R, DUSP6 | 0.02843111 |
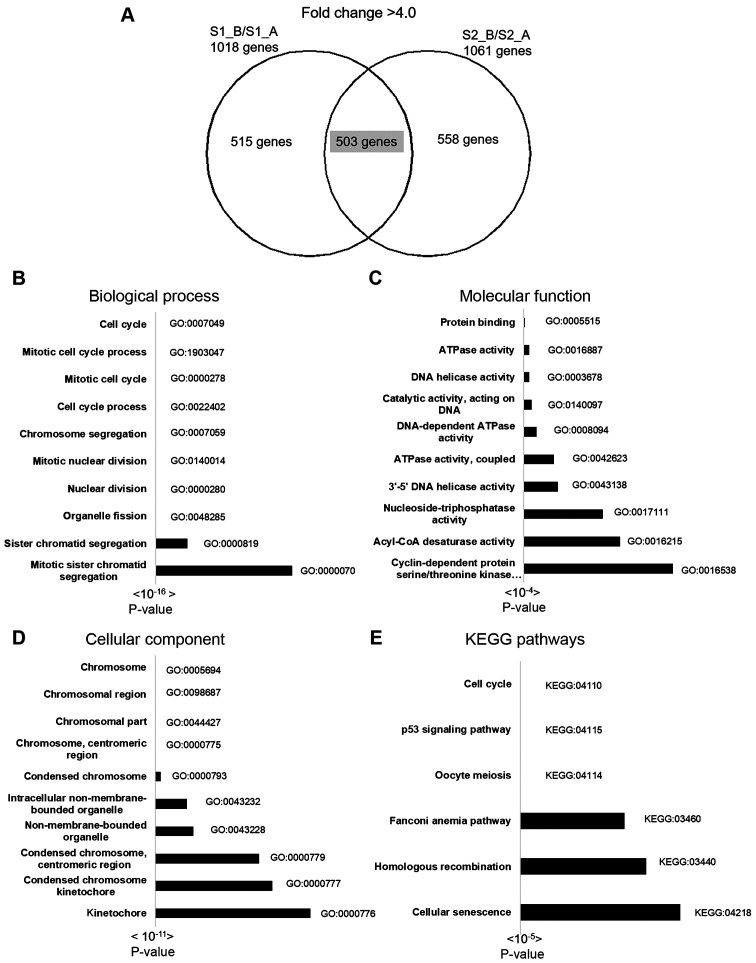
Functional and pathway analysis results of 503 downregulated genes are shown in Fig. 4. There were 250 terms in BP, 112 terms in CC, 28 terms in MF and 6 KEGG pathways identified by g: Profiler. The most significant BPs included cell cycle, mitotic cell cycle process and mitotic cell cycle. The most significant CCs included chromosome, chromosomal region and chromosomal part. The most significant MF pathways included protein binding, ATPase activity and DNA helicase activity. The most significant KEGG pathways included cell cycle, the p53 signaling pathway and oocyte meiosis (Fig. 4). The top 3 pathways of the 30 most downregulated genes with gene lists are displayed in Table III.
Figure 4.
Molecular pathway and functional analysis of downregulated genes using g:Profiler and KEGG pathway enrichment analysis. (A) A total of 503 overlapping differentially downregulated genes with a fold-change of <4.0 were identified. The top 10 pathways in (B) biological process, (C) cellular component, (D) molecular function and (E) KEGG pathways. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table III.
Top 3 pathways and gene lists of enriched GO categories and KEGG pathways of 30 downregulated genes.
| A, Biological process | ||||
|---|---|---|---|---|
| Pathways | GO term | Gene count | Gene lists | P-value |
| Cell cycle | GO:0007049 | 14 | HMMR, UBE2C, CDCA2, MELK, CCNB2, TYMS, CDK1, MND1, CDCA3, BIRC5, ASPM, CDCA5, TPX2, CDC45 | 2.14467E-05 |
| Mitotic cell cycle phase transition | GO:0044772 | 9 | HMMR, UBE2C, MELK, CCNB2, TYMS, CDK1, CDCA5, TPX2, CDC45 | 6.1294E-05 |
| Cell division | GO:0051301 | 9 | UBE2C, CDCA2, CCNB2, CDK1, CDCA3, BIRC5, ASPM, CDCA5, TPX2 | 8.68163E-05 |
| B, Molecular function | ||||
| Pathways | GO term | Gene count | Gene lists | P-value |
| No statistically significant results | ||||
| C, Cellular component | ||||
| Pathways | GO term | Gene count | Gene lists | P-value |
| No statistically significant results | ||||
| D, KEGG pathway | ||||
| Pathways | GO term | Gene count | Gene lists | P-value |
| Cell cycle | KEGG:04110 | 3 | CCNB2, CDK1, CDC45 | 0.04453008 |
EBV infection affected several cellular pathways; according to BP and KEGG pathway analysis, EBV infection induced the expression of cellular genes involved in cancer-related pathways, including the MAPK signaling pathway (ARRB2, CD14, CDC25B, DUSP1, DUSP6, HSPA6, JUND, MAP3K1, MAP3K6, MAPKAPK3, PRKACB, PRKCA, RASGRP2, RASGRP4, TGFBR2 and TNFRSF1A), the WNT signaling pathway (CCND3, CTBP2, FRAT1, FRAT2, LEF1, PLCB1, PLCB2, PRKACB, PRKCA and WNT10B) and the PI3K-Akt signaling pathway (CCND3, CDK4, CDK6, CSF1R, CSF3R, F2R, GNG11, GNG2, IL4R, IL6R, ITGA5, ITGB1, JAK1, LAMB2, PPP2R2B, PRKCA, SGK3 and TLR2). These genes promote proliferation of BLCL and their transformation to immortalized cells. Cellular genes that function and are located in the plasma membrane and cell surface, such as CX3CR1, CD8A, CD3D, CD14, CD2, CDH23, CSF3R, TYROBP, CSF1R, S100A10, NKG7, LILRB3, CD2, GP1BB, NRGN, ITGAM, ITGB and ITGAX were highly expressed, and these genes were associated with the MF pathways; protein binding and cell adhesion. As shown in Fig. 1D, EBV-transformed BLCL exhibited cell clumping and rosette shape formation.
Downregulated genes were primarily involved in cell cycle (CCNB1, CCNB2, CCNE1, CCNE2, CDC20, CDC45, CDK1, CDKN2A, CHEK1, E2F1, ESPL1, MAD2L1, MCM2, MCM4, MDM2, PCNA, PKMYT1, PTTG1, PTTG2 and SFN), p53 signaling pathway (CCNB1, CCNB2, CCNE1, CCNE2, CDK1, CDKN2A, CHEK1, DDB2, FAS, MDM2, RRM2B and SFN) and cellular senescence (CCNB1, CCNB2, CCNE1, CCNE2, CDK1, CDKN2A, CHEK1, E2F1, MAPK11, MDM2, MYBL2 and SLC25A4).
Discussion
The present study was performed to determine the effects of EBV infection on alterations to cellular signaling pathways in human B cells collected from two healthy Thai individuals. The similarities between the gene expression profiles of the two EBV-transformed BLCL indicated common characteristics of EBV-induced transformation of B cells. A considerable number of processes were upregulated in BLCL, including osteoclast differentiation, cytokine-cytokine receptor interaction, hematopoietic cell lineage, pathways in cancer, the chemokine signaling pathway, leukocyte transendothelial migration and the NOD-like receptor signaling pathway. Significantly downregulated pathways consisted of genes involved in cell cycle regulation, oocyte meiosis, the p53 signaling pathway, metabolic pathways and cellular senescence, which was similar to the results of a previous study using DLBCL samples (27). Carter et al (28) used cDNA array hybridization on nitrocellulose filters and RT-qPCR, similar to the present study, and showed that cells involved in cell adhesion and structure were highly expressed in EBV-infected-lymphoblastoid cells. Previous studies using RNA sequencing (RNAseq) to investigate cellular gene expression patterns in EBV-transformed B cells at different time points found similar alterations in the expression of genes and thus biological pathways to those reported in the present study, such as immune system process, response to stimulus, lymphocyte migration, cellular activation, defense response, tumor necrosis factor-mediated signaling pathway, G1/S transition of mitotic cell cycle, DNA replication and cell cycle arrest (16,29). This similarity suggests that the results of cellular transcriptomic analysis of EBV-transformed B-cells obtained from either microarray analysis or RNAseq analysis are similar.
Cellular genes that function and are located in the external side of the plasma membrane or cytoplasmic vesicle that were upregulated, may have been involved in cell-cell fusion. The LMP-1 protein of EBV has the ability to induce the expression of surface proteins, such as integrins and cadherins, which are involved in cell adhesion (30). It has been reported that integrin and EBV glycoprotein interactions induce epithelial cell fusion (31). The molecular pathways that may be involved in the expression of cell surface proteins in the present study included Rho and GTPase-mediated signal transduction, and these proteins have been reported to result in cytoskeletal rearrangement and induce cell-cell adhesion (32).
The other cellular genes with biological functions in the MAPK, WNT and PI3K-Akt signaling pathways were upregulated and involved in the acquisition of a tumorigenic phenotype in EBV-infected B cells, included B-lymphoproliferative disease, Hodgkin's lymphoma and Burkitt's lymphoma (4). The EBV encoded latent membrane protein 1 (LMP-1) viral oncoprotein functions similar to those of the active receptor CD40/TNFR family, and has an oncogenic potential that results in a long period of cell survival (9). LMP1 has the ability to activate several cellular signaling pathways, including the MAPK signaling pathway, which ultimately increases B cell proliferation (4). The WNT pathway also increases the invasiveness of EBV-positive cells (33). Previous studies have shown that, LEF1, a gene involved in the WNT pathway is also upregulated in EBV-infected epithelial cells and B cells (33,34). In addition, other studies have reported that LMP1 has the ability to activate the PI3K/Akt pathway in NPC and nasal natural killer/T-cell lymphoma, resulting in the inhibition of cell apoptosis (35,36).
Overexpression of JAK1 and STAT4 in BLCL was observed in the present study, which may be involved in the JAK/STAT signaling pathway that has been reported to be activated in DLBCL (37). MYC oncogene has been reported to be overexpressed in Burkitt's lymphoma, and the present study identified increased MYCL proto-oncogene transcription factor in BLCL, which may be involved in cell proliferation (37). CDK4 and CDK6 gene expression was also increased in BLCL, consistent with results of previous studies that used RNAseq (16,29). CDK4/6 proteins induce the phosphorylation of retinoblastoma protein (pRb), as well as release free E2F from the pRb/E2F complex to drive the cell cycle into the S phase (38). In addition, a CDK4/6 inhibitor has been shown to suppress NPC cell growth and induce cell cycle arrest (39).
In the present study, downregulation of genes involved in cell cycle control, p53 signaling pathway and cellular senescence was observed in the BLCL. By contrast, analysis of EBV-positive undifferentiated NPC cells showed that genes involved in the cell cycle and p53 signaling pathway, such as CDK1, CCNB1, CCNB2 and MDM2 were upregulated (40).
A study of liver hepatocellular carcinoma (HCC) reported high levels of BIRC5 expression in HCC compared with the present study, which reported the downregulation of BIRC5 in EBV-transformed BLCL (41). TROAP was found to be upregulated in cholangiocarcinoma (42); however, the present study reported a downregulation of TROAP expression in EBV-transformed BLCL. These findings indicate that each pathogen has an independent ability to alter cellular gene expression that transforms normal cells into immortalized cells and causes cancer progression.
In conclusion, EBV-mediated transformation of different cell types, affects the cellular gene expression profiles and activities of cellular pathways. However, infected cells exhibit alterations to common cellular pathways, such as upregulation of the MAPK, WNT and PI3K-Akt signaling pathways, which may highlight potential targets for therapeutic treatment of patients with EBV-related cancer.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This work was supported by the Applied Medical Virology Research Unit of Chulalongkorn University, and Chulalongkorn Academic Advancement into Its 2nd Century Project. (CUAASC). This work was partly supported by the TRF Grant for New Researchers from the Thailand Research Fund Grant (grant no. TRG6080008) and Chulalongkorn University Office of International Affairs Scholarship for Short-term Research.
Availability of data and materials
The datasets used and/or analyzed in the present study are available from the corresponding author on reasonable request.
Authors' contributions
NK and SB designed and performed the experiments. AC and NK organized and analyzed the data. AC wrote the manuscript. RS performed the experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University (Bangkok, Thailand) (approval no. 358/46) and both subjects provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Balfour HH Jr, Sifakis F, Sliman JA, Knight JA, Schmeling DO, Thomas W. Age-specific prevalence of Epstein-Barr virus infection among individuals aged 6-19 years in the United States and factors affecting its acquisition. J Infect Dis. 2013;208:1286–1293. doi: 10.1093/infdis/jit321. [DOI] [PubMed] [Google Scholar]
- 2.Condon LM, Cederberg LE, Rabinovitch MD, Liebo RV, Go JC, Delaney AS, Schmeling DO, Thomas W, Balfour HH Jr. Age-specific prevalence of Epstein-Barr virus infection among Minnesota children: Effects of race/ethnicity and family environment. Clin Infect Dis. 2014;59:501–508. doi: 10.1093/cid/ciu342. [DOI] [PubMed] [Google Scholar]
- 3.Hjalgrim H, Friborg J, Melbye M. doi: 10.1017/CBO9780511545313.054. The epidemiology of EBV and its association with malignant disease. In: Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R and Yamanishi K (eds). Cambridge, 2007. [DOI] [PubMed] [Google Scholar]
- 4.El-Sharkawy A, Al Zaidan L, Malki A. Epstein-Barr virus-associated malignancies: Roles of viral oncoproteins in carcinogenesis. Front Oncol. 2018;8(265) doi: 10.3389/fonc.2018.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esmeray E, Küçük C. Genetic alterations in B cell lymphoma subtypes as potential biomarkers for noninvasive diagnosis, prognosis, therapy, and disease monitoring. Turk J Biol. 2020;44:1–14. doi: 10.3906/biy-1908-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li CW, Jheng BR, Chen BS. Investigating genetic-and-epigenetic networks, and the cellular mechanisms occurring in Epstein-Barr virus-infected human B lymphocytes via big data mining and genome-wide two-sided NGS data identification. PLoS One. 2018;13(e0202537) doi: 10.1371/journal.pone.0202537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saha A, Robertson ES. Mechanisms of B-cell oncogenesis induced by Epstein-Barr Virus. J Virol. 2019;93(93) doi: 10.1128/JVI.00238-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutt-Fletcher LM. Epstein-Barr virus replicating in epithelial cells. Proc Natl Acad Sci USA. 2014;111:16242–16243. doi: 10.1073/pnas.1418974111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowe M, Zuo J. Immune responses to Epstein-Barr virus: Molecular interactions in the virus evasion of CD8+ T cell immunity. Microbes Infect. 2010;12:173–181. doi: 10.1016/j.micinf.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wanvimonsuk S, Thitiwanichpiwong P, Keelawat S, Mutirangura A, Kitkumthorn N. Distribution of the Epstein-Barr virus in the normal stomach and gastric lesions in Thai population. J Med Virol. 2019;91:444–449. doi: 10.1002/jmv.25318. [DOI] [PubMed] [Google Scholar]
- 11.Jha HC, Pei Y, Robertson ES. Epstein-Barr virus: Diseases linked to infection and transformation. Front Microbiol. 2016;7(1,602) doi: 10.3389/fmicb.2016.01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tulalamba W, Janvilisri T. Nasopharyngeal carcinoma signaling pathway: An update on molecular biomarkers. Int J Cell Biol. 2012;2012(594681) doi: 10.1155/2012/594681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atsaves V, Leventaki V, Rassidakis GZ, Claret FX. AP-1 transcription factors as regulators of immune responses in cancer. Cancers (Basel) 2019;11(11) doi: 10.3390/cancers11071037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Park HW, Dahlin A, Qiu W, Tantisira KG. Gene expression changes in lymphoblastoid cell lines and primary B cells by dexamethasone. Pharmacogenet Genomics. 2019;29:58–64. doi: 10.1097/FPC.0000000000000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C, Li D, Zhang L, Jiang S, Liang J, Narita Y, Hou I, Zhong Q, Zheng Z, Xiao H, et al. RNA sequencing analyses of gene expression during Epstein-Barr virus infection of primary B lymphocytes. J Virol. 2019;93(93) doi: 10.1128/JVI.00226-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hui-Yuen J, McAllister S, Koganti S, Hill E, Bhaduri-McIntosh S. Establishment of Epstein-Barr virus growth-transformed lymphoblastoid cell lines. J Vis Exp. 2011;(57)(3,321) doi: 10.3791/3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 19.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47 (W1):W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M. Post-genome informatics. Oxford University Press, Oxford, UK, 2000. [Google Scholar]
- 21. R Core Team: (2012). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0. http://www.R-project.org/. [Google Scholar]
- 22.The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47 (D1):330–338. doi: 10.1093/nar/gky1055. The Gene Ontology Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. The Gene Ontology Consortium: Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baik SY, Yun HS, Lee HJ, Lee MH, Jung SE, Kim JW, Jeon JP, Shin YK, Rhee HS, Kimm KC, et al. Identification of stathmin 1 expression induced by Epstein-Barr virus in human B lymphocytes. Cell Prolif. 2007;40:268–281. doi: 10.1111/j.1365-2184.2007.00429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussain T, Mulherkar R. Lymphoblastoid cell lines: A continuous in vitro source of cells to study carcinogen sensitivity and DNA repair. Int J Mol Cell Med. 2012;1:75–87. [PMC free article] [PubMed] [Google Scholar]
- 26.Jeon JP. Human lymphoblastoid cell lines in pharmacogenomics. In: Handbook of pharmacogenomics and stratified medicine. Padmanabhan S (ed). Academic Press, San Diego, pp89-110, 2014. [Google Scholar]
- 27.Kato H, Karube K, Yamamoto K, Takizawa J, Tsuzuki S, Yatabe Y, Kanda T, Katayama M, Ozawa Y, Ishitsuka K, et al. Gene expression profiling of Epstein-Barr virus-positive diffuse large B-cell lymphoma of the elderly reveals alterations of characteristic oncogenetic pathways. Cancer Sci. 2014;105:537–544. doi: 10.1111/cas.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carter KL, Cahir-McFarland E, Kieff E. Epstein-barr virus-induced changes in B-lymphocyte gene expression. J Virol. 2002;76:10427–10436. doi: 10.1128/jvi.76.20.10427-10436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mrozek-Gorska P, Buschle A, Pich D, Schwarzmayr T, Fechtner R, Scialdone A, Hammerschmidt W. Epstein-Barr virus reprograms human B lymphocytes immediately in the prelatent phase of infection. Proc Natl Acad Sci USA. 2019;116:16046–16055. doi: 10.1073/pnas.1901314116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wasil LR, Shair KHY. Epstein-Barr virus LMP1 induces focal adhesions and epithelial cell migration through effects on integrin-α5 and N-cadherin. Oncogenesis. 2015;4:e171–e171. doi: 10.1038/oncsis.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chesnokova LS, Nishimura SL, Hutt-Fletcher LM. Fusion of epithelial cells by Epstein-Barr virus proteins is triggered by binding of viral glycoproteins gHgL to integrins αvβ6 or αvβ8. Proc Natl Acad Sci USA. 2009;106:20464–20469. doi: 10.1073/pnas.0907508106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber GF, Bjerke MA, DeSimone DW. Integrins and cadherins join forces to form adhesive networks. J Cell Sci. 2011;124:1183–1193. doi: 10.1242/jcs.064618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Birdwell CE, Prasai K, Dykes S, Jia Y, Munroe TGC, Bienkowska-Haba M, Scott RS. Epstein-Barr virus stably confers an invasive phenotype to epithelial cells through reprogramming of the WNT pathway. Oncotarget. 2018;9:10417–10435. doi: 10.18632/oncotarget.23824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shackelford J, Maier C, Pagano JS. Epstein-Barr virus activates beta-catenin in type III latently infected B lymphocyte lines: Association with deubiquitinating enzymes. Proc Natl Acad Sci USA. 2003;100:15572–15576. doi: 10.1073/pnas.2636947100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang CF, Yang GD, Huang TJ, Li R, Chu QQ, Xu L, Wang MS, Cai MD, Zhong L, Wei HJ, et al. EB-virus latent membrane protein 1 potentiates the stemness of nasopharyngeal carcinoma via preferential activation of PI3K/AKT pathway by a positive feedback loop. Oncogene. 2016;35:3419–3431. doi: 10.1038/onc.2015.402. [DOI] [PubMed] [Google Scholar]
- 36.Sun L, Zhao Y, Shi H, Ma C, Wei L. LMP-1 induces survivin expression to inhibit cell apoptosis through the NF-κB and PI3K/Akt signaling pathways in nasal NK/T-cell lymphoma. Oncol Rep. 2015;33:2253–2260. doi: 10.3892/or.2015.3847. [DOI] [PubMed] [Google Scholar]
- 37.Pei Y, Lewis AE, Robertson ES. Current progress in EBV-associated B-cell lymphomas. Adv Exp Med Biol. 2017;1018:57–74. doi: 10.1007/978-981-10-5765-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobhani N, D'Angelo A, Pittacolo M, Roviello G, Miccoli A, Corona SP, Bernocchi O, Generali D, Otto T. Updates on the CDK4/6 inhibitory strategy and combinations in breast cancer. Cells. 2019;8(8) doi: 10.3390/cells8040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong CH, Ma BBY, Hui CWC, Lo KW, Hui EP, Chan ATC. Preclinical evaluation of ribociclib and its synergistic effect in combination with alpelisib in non-keratinizing nasopharyngeal carcinoma. Sci Rep. 2018;8(8010) doi: 10.1038/s41598-018-26201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen F, Shen C, Wang X, Wang H, Liu Y, Yu C, Lv J, He J, Wen Z. Identification of genes and pathways in nasopharyngeal carcinoma by bioinformatics analysis. Oncotarget. 2017;8:63738–63749. doi: 10.18632/oncotarget.19478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H, Zhang X, Cai XY, Wen DY, Ye ZH, Liang L, Zhang L, Wang HL, Chen G, Feng ZB. From big data to diagnosis and prognosis: Gene expression signatures in liver hepatocellular carcinoma. PeerJ. 2017;5(e3089) doi: 10.7717/peerj.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chujan S, Suriyo T, Ungtrakul T, Pomyen Y, Satayavivad J. Potential candidate treatment agents for targeting of cholangiocarcinoma identified by gene expression profile analysis. Biomed Rep. 2018;9:42–52. doi: 10.3892/br.2018.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed in the present study are available from the corresponding author on reasonable request.