ABSTRACT
Osteoarthritis (OA) is a very common chronic and degenerative joint disease characterized by persistent destruction of articular cartilage. Recently, increasing evidence showed that circular RNAs (circRNAs) play critical roles in OA progression. However, the functions of circRNAs in OA and their underlying mechanisms of action remain unclear. In the present study, the expression levels of circRNA-UBE2G1 and HIF-1a were significantly increased in OA tissues, whereas miR‑373 expression was downregulated. Function assays showed that circRNA-UBE2G1 inhibition reduced the effects of LPS on C28/I2 cells viability and apoptosis. In terms of mechanism, we revealed that circRNA-UBE2G1 binds to miR‑373 as competing endogenous RNAs (ceRNAs). HIF-1a might act as a target of miR‑373. Moreover, miR‑373 suppression or HIF-1a overexpression restored the effects of circRNA-UBE2G1 downregulation on LPS-induced chondrocytes injury. Collectively, our data suggest that circRNA-UBE2G1 facilitates the progression in the LPS-induced OA cell model via regulating the miR‑373/HIF-1a axis.
Abbreviations
OA: Osteoarthritis; Circular RNAs; miRNAs: MicroRNAs; Mut: Mutant; WT: Wild type; UTR: Untranslated region.
KEYWORDS: Osteoarthritis, circRNA-UBE2G1, miR‑373, HIF-1a
Introduction
Osteoarthritis (OA) is a chronic degenerative disease of the articular cartilage, characterized by synovitis and osteophyte formation at the joint edges [1,2]. Currently, drug treatment can only relieve the clinical symptoms of OA, and surgery is still the only treatment [3,4]. Abnormal metabolism of chondrocytes is the main factor leading to articular cartilage degeneration [5]. Therefore, it is of critical significance to explore the molecular mechanism of chondrocyte proliferation for OA treatment.
Circular RNAs (circRNAs) are a novel type of ncRNAs derived from exons, introns, or intergenic regions that have a covalently closed continuous loop, display cell or tissue specific expression, and are conserved across species due to resistance to RNase R [6,7]. Recently, increasing evidence has shown that circRNAs play key regulatory roles in diverse cellular processes, such as cell proliferation, apoptosis, differentiation, and invasion [8,9]. For example, Ge et al. found that circMTO1 reduced colorectal cancer cell proliferation and invasion through the Wnt/beta-catenin axis [10]. Wei et al. established that circLMO7 regulated myoblasts differentiation and survival by sponging miR-378a-3p [11]. Xu et al suggested that hsa_circ_0081143 promoted cisplatin resistance in gastric cancer by targeting miR-646/CDK6 axis [12]. However, the roles and underlying mechanisms of circRNA remain largely unknown.
In the present study, we found that the expression levels of circRNA-UBE2G1 and HIF-1a in OA tissues were upregulated, whereas miR‑373 expression was downregulated. Subsequently, our function experiments showed that circRNA-UBE2G1 facilitated the cell proliferation and apoptosis in LPS-induced chondrocyte cells through regulating the miR‑373/HIF-1a axis. These findings suggest that circRNA-UBE2G1 might be considered a promising therapeutic target for OA treatment.
Materials and methods
Clinical specimens
This study was performed with the approval of the Ethics Committee of the Henan Provincial People’s Hospital (Zhengzhou, Henan Province, China). A number of 53 patients diagnosed with OA and undergoing total knee arthroplasty were identified, along with 13 healthy tissue donors who were deceased or had suffered a trauma. After preoperative signing of informed consent by all patients, OA and normal tissues were acquired and immediately saved at −80°C until use.
Cell culture and transfections
A human normal chondrocyte C28/I2 cell line was purchased from the Cell Bank of the Chinese Academy of Science (Shanghai, China). The C28/I2 cells were incubated in RPMI-1640 containing 10% FBS at 37°C with 5% CO2.
Small interfering RNA (siRNA) targeting circRNA-UBE2G1 (si-circRNA), miR‑373 mimics, HIF-1a overexpression vector (HIF-1a), and negative controls were bought from GenePharma (Shanghai, China). All plasmids were transfected into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Quantitative real-time reverse transcription PCR (qRT-PCR)
Total RNA was purified from the tissue and cells by TRIzol reagents (Invitrogen). Then, RNA was retrieved into cDNA by the PrimeScript RT reagent kit (Takara, Dalian, China) as a template of the following amplification experiment by using the SYBR Premix ExTaqII kit (Takara). Amplification conditions: 95°C for 10 min (45 cycles), 95°C for 15 s, 60°C for 20 s, and 72°C for 20 s. GAPDH was used as an internal reference of circRNA-UBE2G1 and mRNA and U6 as an internal reference of miRNAs. The gene expression levels were calculated by the 2 −ΔΔCT method.
Cell proliferation assay
Cell proliferation was detected by CCK-8 assay. In brief, 2 × 103 cells were seeded into a 96-well plate and cultured for different time. Subsequently, CCK-8 solution (Dojindo, Japan) was added to each well, followed by incubation at 37°C for another 2 h. The absorbance was measured at a wavelength of 450 nm using a microplate reader (Bio-Rad, Benicia, CA, USA).
Alternatively, cell proliferation was assessed using 5-ethynyl-2ʹ-deoxyuridine (EdU) assay (Beyotime, Shanghai, China). In brief, cells were treated with EdU (50 μM) for 2 h and fixed in 4% formaldehyde for 15 min. After rinsing with PBS, cells were permeated 143 with P0097 solution, stained with Hoechst 33,342, and visualized by fluorescence microscopy.
Cell apoptosis assay
Cell apoptosis was analyzed using a FITC Annexin V apoptosis detection kit (BD Biosciences, Franklin, NJ, USA). Cells (1 x 106) were collected and incubated with Annexin V-FITC and PI for 15 min at 4°C. Then, the cells were washed by precooled PBS three times and suspended in buffer for the next analysis. The Caliber system (Beckman) was utilized to detect the percentage of apoptosis.
Luciferase reporter assay
Fragment sequences of circRNA-UBE2G1 and HIF-1a 3ʹUTR containing the wide-type or mutant-type miR‑373 binding sites were cloned into the pGL3-Basic luciferase vector (Promega, Madison, WI, USA) to generate circRNA-UBE2G1-WT, circRNA-UBE2G1-Mut, HIF-1a-WT, and HIF-1a-Mut. Further, the luciferase vectors were transfected into cells along with miR‑373 mimics or miR-NC. The relative luciferase activity was assessed 48 h after the transfection using the dual-luciferase assay system (Promega) [13].
RIP assay
Magna RIP kit (Millipore) was used to perform the RIP assay, with specific steps in strict accordance with the instructions of the kit and of previous studies [14,15].
Western blot
Cells were harvested and extracted using lysis buffer. Protein concentration was then determined by the BCA method. Protein (50 ug) was separated by 10% of SDS-PAGE gels, which were transferred to the PVDF membranes. The membranes were blocked in 5% milk for 1 h and then incubated with primary antibodies overnight at 4°C. Next, the membranes were washed with TBST three times and incubated with secondary antibody at room temperature for 1 h. Finally, an enhanced chemiluminescence solution (Pierce; Thermo Fisher Scientific) was used for protein signal detection.
Statistical analysis
Data are expressed as mean ± standard deviation (SD). Statistical analysis was performed by SPSS version 21.0 (Chicago, IL, USA). Student’s t-test and one-way ANOVA analyzes were employed. P < 0.05 was considered statistically significant.
Results
CircRNA-UBE2G1 and HIF-1a expression was upregulated in OA tissues, whereas miR‑373 expression was downregulated
The microarray data of a previous study showed that lots ofmore circRNAs were upregulated in OA tissues than in normal tissues [16]. To explore the roles of circRNAs in OA, we determined the five most upregulated circRNAs expression in OA tissues, of which hsa _circ_0008956 was chosen for a further study (Figure 1(a–e)). Hsa_circ_0008956 (CircRNA-UBE2G1) is spliced from the UBE2G1 gene and has an ultimate length of 396 nt (Figure S1).
Figure 1.
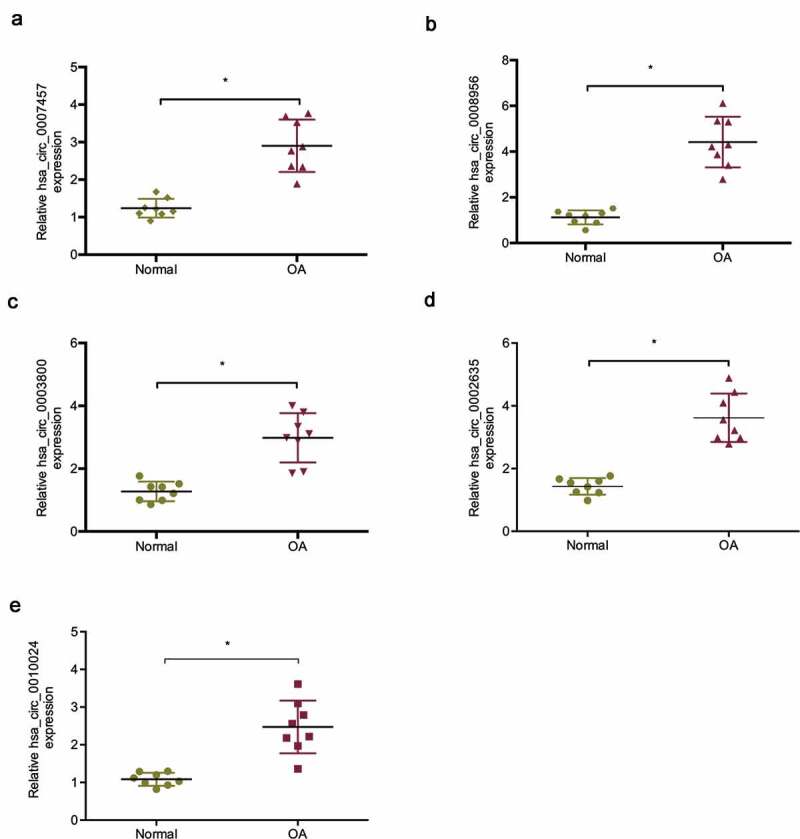
Expression of the five most upregulated circRNAs in OA tissues. (a) Relative hsa_circ_0007457 expression in OA tissues; (b) Relative hsa_circ_0008956 expression in OA tissues; (c) Relative hsa_circ_0003800 expression in OA tissues; (d) Relative hsa_circ_0002635 expression in OA tissues; (e) Relative hsa_circ_0010024 expression in OA tissues. *P < 0.05.
Next, we explored circRNA-UBE2G1 expression in 13 healthy tissue donors and 53 OA patients. QRT-PCR showed that the expression levels of circRNA-UBE2G1 and HIF-1a were significantly more increased in OA tissues than in normal tissues (Figure 2(a,c)). Conversely, miR‑373 expression was decreased in OA tissues (Figure 2(b)). Subsequently, modified Mankin grading was used to evaluate OA severity. Correlation analysis revealed that high circRNA-UBE2G1 and HIF-1a expression was positively associated with modified Mankin scores in OA patients (Figure 2(d,f)), whereas miR‑373 expression was negatively associated with modified Mankin scores (Figure 2(e)).
Figure 2.
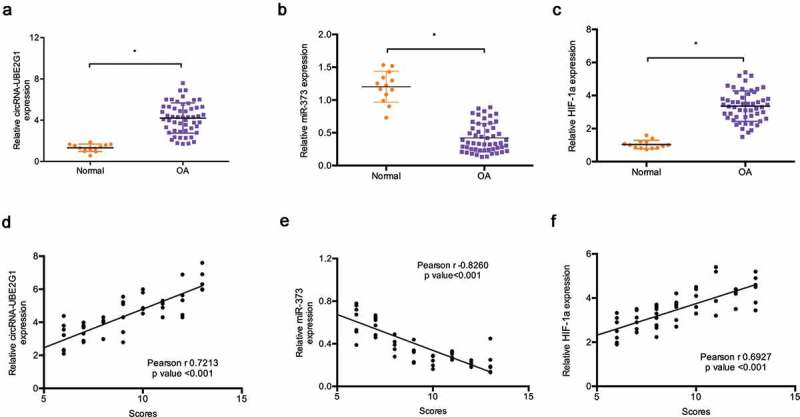
Expression of circRNA-UBE2G1, miR‑373, and HIF-1a in OA. (a–c) Relative circRNA-UBE2G1, miR‑373, and HIF-1a expression in OA tissues and normal tissues; (d, f) CircRNA-UBE2G1 and HIF-1a expression were positively associated with high modified Mankin scores; (e) MiR‑373 expression was negatively associated with modified Mankin scores. *P < 0.05.
CircRNA-UBE2G1 regulates LPS-induced chondrocytes injury
To determine the biological functions of circRNA-UBE2G1 in OA, we used LPS-treated C28/I2 cells as a cellular model of OA [17,18]. QRT-PCR data showed that circRNA-UBE2G1 expression was considerably higher in C28/I2 cells treated with LPS in a concentration-dependent manner; 5 ug/mL LPS was chosen for the following studies (Figure 3(a)). Next, we transfected si-circRNA into C28/I2 cells and determined the transfection efficiency by qRT-PCR (Figure 3(b,c)). CCK-8 and EdU assays showed that LPS significantly reduced C28/I2 cells viability, while circRNA-UBE2G1 suppression abolished the effects (Figure 3(d–f)). Cell apoptosis assay revealed that LPS significantly increased C28/I2 cells apoptosis, while circRNA-UBE2G1 inhibition reversed the effects (Figure 3(g,h)). Moreover, we explored the roles of circRNA-UBE2G1 on pro-inflammatory cytokines levels. Using ELISA assay, we established that circRNA-UBE2G1 inhibition significantly reduced the expression of pro-inflammatory cytokines (IL-1β, IL-6, and TNF-α) in C28/I2 cells treated with LPS (Figure 4(a–c)).
Figure 3.
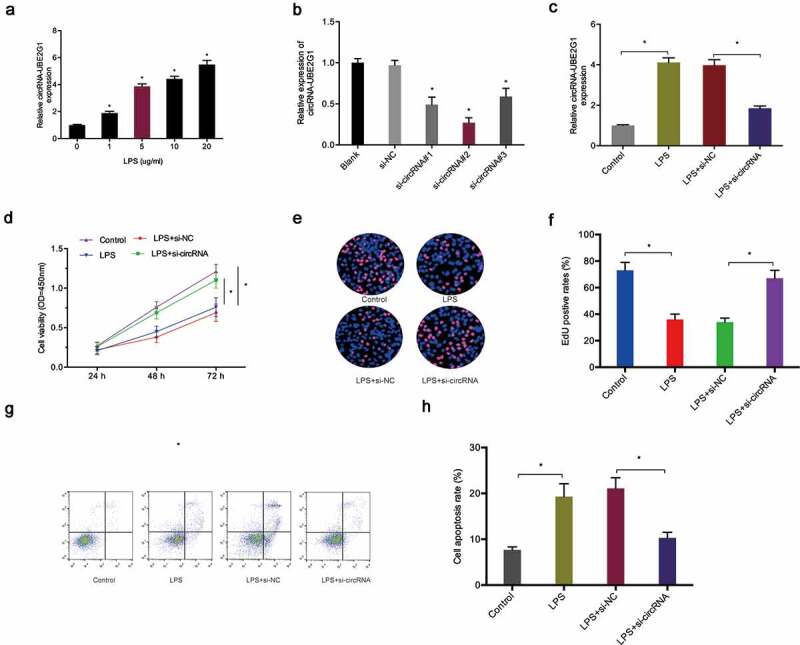
Effects of circRNA-UBE2G1 on LPS‐treated C28/I2 cells. (a) LPS increased circRNA-UBE2G1 expression in C28/I2 cells; (b, c) Transfection effects of si-circRNA in LPS‐treated C28/I2 cells; (d–f) CircRNA-UBE2G1 suppression abolished the effects of LPS on C28/I2 cells viability; (g, h) CircRNA-UBE2G1 suppression reversed the effects of LPS on C28/I2 cells apoptosis. *P < 0.05.
Figure 4.

Effects of circRNA-UBE2G1 on pro-inflammatory cytokines levels in LPS‐treated C28/I2 cells. (a–c) circRNA-UBE2G1 inhibition reduced IL-1β, IL-6, and TNF-α expression in LPS‐treated C28/I2 cells. * P < 0.05.
MiR‑373 is a target of circRNA-UBE2G1
To further uncover the mechanisms of action of circRNA-UBE2G1 in OA, we initially explored the localization of circRNA-UBE2G1 in C28/I2 cells. Subcellular fractionation assay showed circRNA-UBE2G1 was mostly distributed in the cytoplasm in C28/I2 cells (Figure 5(a)). The results implied that circRNA-UBE2G1 might function as a competing endogenous RNA (ceRNA) in OA. Next, through the bioinformatics prediction, miR-373 was discovered to harbor a binding site for circRNA-UBE2G1 (Figure S2A-S2B and Figure 5(b)). Luciferase reporter assay showed that miR‑373 mimics reduced the relative luciferase activity of circRNA-WT group (Figure 5(C)). Additionally, RIP assay revealed that circRNA-UBE2G1 and miR‑373 were preferentially enriched in the Ago2 pellet (Figure 5(d)). QRT-PCR detected markedly increased si-circRNA miR‑373 expression in C28/I2 cells (Figure 5(e)). Next, our correlation analysis showed that the expression of circRNA-UBE2G1 was negatively associated with miR‑373 expression in OA tissues (Figure 5(F)). Moreover, by qRT-PCR, we found that si-circRNA increased miR‑373 expression in LPS‐treated C28/I2 cells, which was restored by miR‑373 inhibitors (Figure 5(g)).
Figure 5.
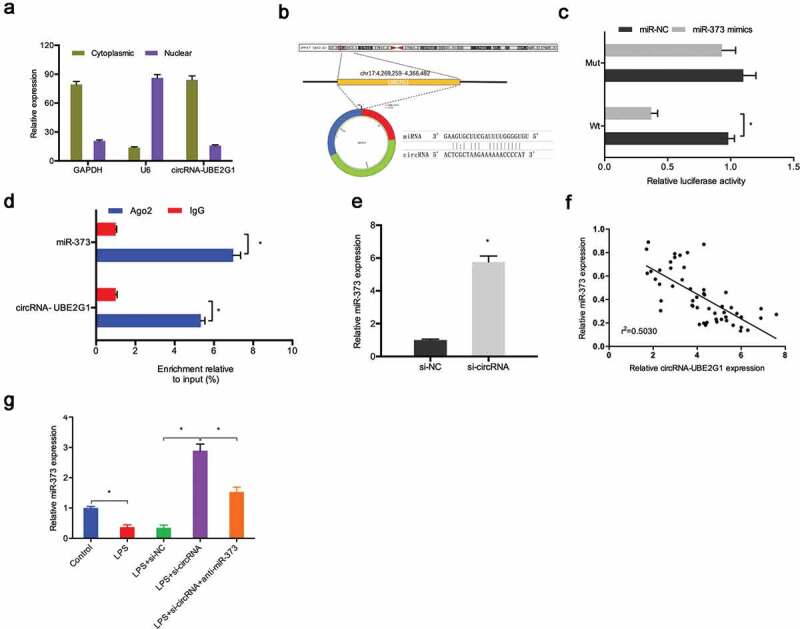
Interaction between circRNA-UBE2G1 and miR‑373. (a) circRNA-UBE2G1 was mostly distributed in the cytoplasm of C28/I2 cells; (b) The predicting binding site of circRNA-UBE2G1 and miR‑373; (c) MiR‑373 mimics reduced the luciferase activity of the circRNA-Wt group; (d) RIP assay showed circRNA-UBE2G1 and miR‑373 was preferentially enriched in the Ago2 pellet; (e) si-circRNA increased miR‑373 expression in C28/I2 cells; (f) CircRNA-UBE2G1 expression was negatively associated with miR‑373 expression in OA tissues; (g) Si-circRNA increased miR‑373 expression in LPS‐treated C28/I2 cells. * P < 0.05.
MiR‑373is targeted by HIF-1a
Next, we explored the potential targets of miR‑373 via bioinformatics analysis. Our results indicated that miR‑373 might be targeted by HIF-1a (Figure 6(a,b)). Luciferase reporter assay results showed that miR‑373 mimics significantly suppressed the luciferase activity of HIF-1a-WT group (Figure 6(c)). Subsequently, qRT-PCR and Western blot assays revealed that miR‑373 mimics significantly reduced HIF-1a expression in C28/I2 cells, whereas miR‑373 inhibitors increased HIF-1a expression (Figure 6(d,e)). Moreover, correlation analysis detected a negative association between the expression of miR‑373 and HIF-1a expression in OA tissues (Figure 6(f)).
Figure 6.
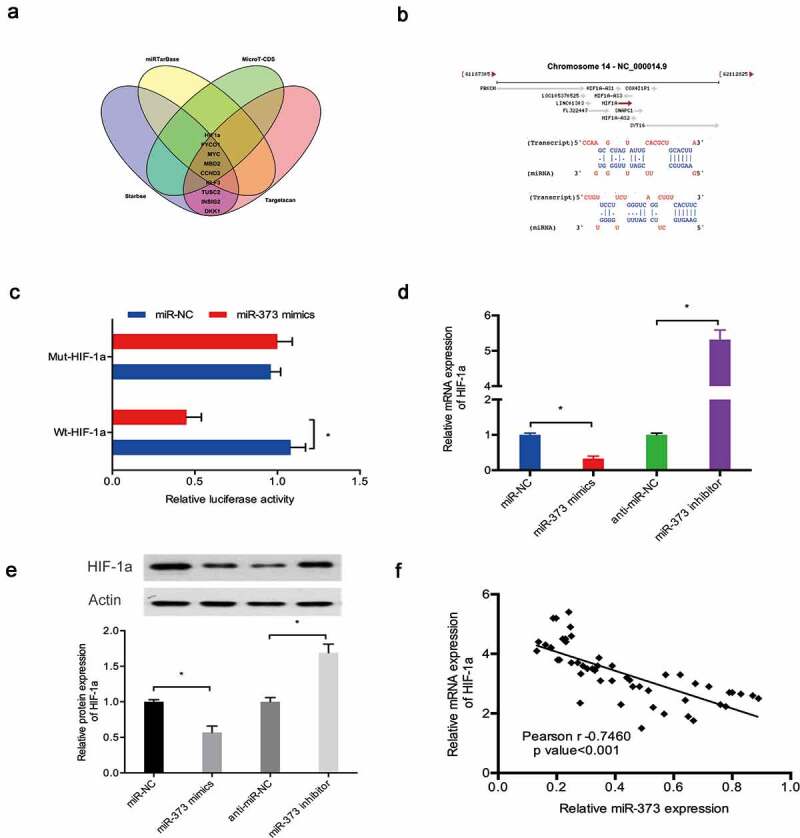
HIF-1a was a target of miR‑373. (a, b) HIF-1a was a potential putative target gene of miR‑373; (c) MiR‑373 mimics suppressed the luciferase activity of the HIF-1a-WT group; (d, e) Effects of miR‑373 on HIF-1a expression in C28/I2 cells; (f) MiR‑373 expression was negatively associated with HIF-1a expression in OA tissues. * P < 0.05.
CircRNA-UBE2G1 regulates LPS-induced chondrocytes injury by the miR‑373/HIF-1a axis
To further confirm the circRNA-UBE2G1/miR‑373/HIF-1a axis in OA, we determined HIF-1a expression in C28/I2 cells transfected with si-circRNA. QRT-PCR showed that si-circRNA reduced HIF-1a expression in LPS-treated C28/I2 cells, whereas miR‑373 suppression (or HIF-1a overexpression) reversed these effects (Figure 7(a)). Correlation analysis revealed the presence of positive correlation between circRNA-UBE2G1 expression and HIF-1a expression in OA tissues (Figure 7(b)). Additionally, using CCK-8 assay, we established that the effects of si-circRNA on LPS-treated C28/I2 cells viability were mitigated by miR-373 inhibitors (or HIF-1a overexpression) (Figure 7(c)). Moreover, si-circRNA reduced the expression of IL-1β, IL-6, and TNF-α in LPS-treated C28/I2 cells, which was abolished by miR-373 inhibitors (or HIF-1a overexpression) (Figure 7(d–f)).
Figure 7.
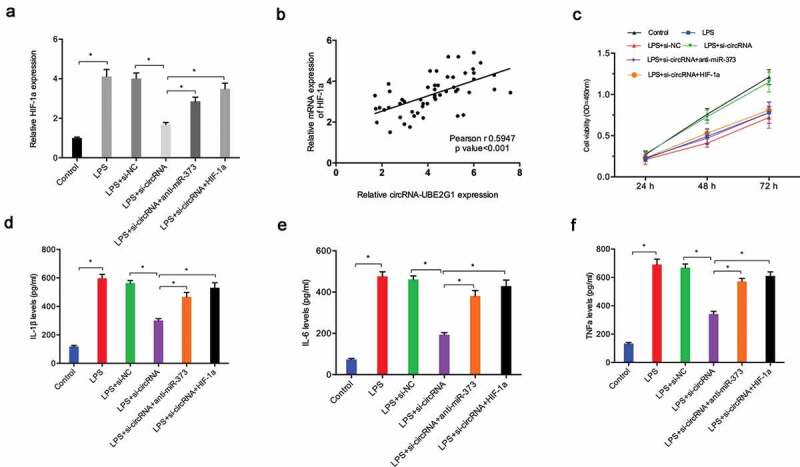
circRNA-UBE2G1/miR‑373/HIF-1a axis in LPS‐treated chondrocytes. (a) MiR‑373 suppression (or HIF-1a overexpression) rescued the effects of si-circRNA on HIF-1a expression in LPS‐treated C28/I2 cells; (b) CircRNA-UBE2G1 expression was positively correlated with HIF-1a expression in OA tissues; (c) MiR‑373 suppression (or HIF-1a overexpression) rescued the effects of si-circRNA on LPS‐treated C28/I2 cells viability; (d–f) MiR‑373 suppression (or HIF-1a overexpression) abolished the effects of si-circRNA on pro-inflammatory cytokines levels in LPS‐treated C28/I2 cells. * P < 0.05.
Discussion
OA is a joint disease commonly occurring in middle-aged and elderly people, often accompanied by joint pain and bone hyperplasia as the main pathological manifestation [19,20]. Recently, increasing evidence has been accumulated that circRNAs are involved in OA progression. For example, Li et al. revealed that hsa_circ_0045714 regulated chondrocyte proliferation, apoptosis, and extracellular matrix synthesis through the miR-193b/IGF1 R axis [21]. In addition, Shen et al. found that circSERPINE2 protected against OA by targeting miR-1271 and an ETS-related gene [22]. However, the roles and underlying mechanisms of action of circRNA in OA remain unelucidated.
In the present study, we identified circRNA-UBE2G1 as a key upregulated circRNA involved in OA and associated with high modified Mankin scores. Subsequently, we explored the roles of circRNA-UBE2G1 in a cellular model of OA. QRT-PCR showed that LPS increased circRNA-UBE2G1 expression in C28/I2 cells. Loss-of-function assays revealed that circRNA-UBE2G1 suppression increased the viability of LPS‐treated C28/I2 cells and reduced their cell apoptosis. In addition, using ELISA assay, we established that circRNA-UBE2G1 inhibition significantly reduced the expression of pro-inflammatory cytokines (IL-1β, IL-6, and TNF-α) in LPS‐treated C28/I2 cells. Thus, these data suggested that circRNA-UBE2G1 might play critical roles in LPS-induced chondrocytes injury.
A growing number of recent studies have shown that circRNA binds to miRNA as a ceRNA, thus inhibiting the binding of miRNA to target gene mRNA and acting as a sponge of miRNA [23,24]. In this investigation, we revealed that circRNA-UBE2G1 has a binding site of mR-373. Subsequently, the interaction between circRNA-UBE2G1 and miR‑373 was confirmed by luciferase reporter and RIP assays. Correlation analysis established a negative association between circRNA-UBE2G1 expression and miR‑373 expression in OA tissues. Interestingly, rescue experiments showed that miR‑373 suppression reversed the effects (cell proliferation and pro-inflammatory cytokines) of si-circRNA in LPS‐treated C28/I2 cells. These findings suggest that circRNA-UBE2G1 might act as a sponge of miR‑373 in OA.
To further explore the mechanism underlying circRNA-UBE2G1/miR‑373 axis, we identified HIF-1a as a potential target of miR‑373. Besides, we demonstrated that circRNA-UBE2G1 regulated HIF-1a expression through miR‑373 sponging. Previous studies reported that HIF-1a served as an important regular in OA. For example, Yudoh et al and Pfander et al showed that HIF-1a was associated with the progression of articular cartilage degeneration in OA [25,26]. Additionally, Zhang et al. found that increased HIF-1a expression in knee OA aggravated synovial fibrosis via fibroblast-like synoviocyte pyroptosis [27]. In the present study, we established that HIF-1a was overexpressed in OA tissues and HIF-1a, which attenuated the effects of si-circRNA on LPS‐treated C28/I2 cells proliferation. Based on these results, we suggest that circRNA-UBE2G1 mediates OA progression via regulating the miR‑373/HIF-1a axis.
In conclusion, our study for the first time demonstrated the role of circRNA-UBE2G1 in OA. We showed that circRNA-UBE2G1 regulated the injury of LPS-treated chondrocytes through the miR‑373/HIF-1a axis, which provides a potential target for OA treatment.
Supplementary Material
Highlight
circRNA-UBE2G1 was upregulated in OA.
circRNA-UBE2G1 inhibition reduced the effects of LPS on C28/I2 cells viability.
circRNA-UBE2G1/miR‑373/HIF-1a axis in OA.
Authors’ contributions
GC, TL, BFY conceived and designed this study. GC, BYW, QP conducted the experiments and analyzed and checked the data. GC, TL, QP supervised the whole project and revised the manuscript. All authors read and approved the final manuscript.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Disclosure statement
This article has not been published elsewhere in whole or in part. All authors have read and approved the content and agree to submit for consideration for publication in the journal. The authors declared no conflict of interest.
Ethics approval and consent to participate
This research was approved by Henan Provincial People’s Hospital and conducted in accordance with the principles of the Declaration of Helsinki.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Arden N, Nevitt MC.. Osteoarthritis: epidemiology. Best Pract Res Clin Rheumatol. 2006;20(1):3–25. [DOI] [PubMed] [Google Scholar]
- [2].Goldring MB, Otero M. Inflammation in osteoarthritis. Curr Opin Rheumatol. 2011;23(5):471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Glyn-Jones S, Palmer AJR, Agricola R, et al. Osteoarthritis. Lancet. 2015;386(9991):376–387. [DOI] [PubMed] [Google Scholar]
- [4].Wang XD, Zhang JN, Gan YH, et al. Current understanding of pathogenesis and treatment of TMJ osteoarthritis. J Dent Res. 2015;94(5):666–673. [DOI] [PubMed] [Google Scholar]
- [5].Mobasheri A, Rayman MP, Gualillo O, et al. The role of metabolism in the pathogenesis of osteoarthritis. Nat Rev Rheumatol. 2017;13(5):302. [DOI] [PubMed] [Google Scholar]
- [6].Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. [DOI] [PubMed] [Google Scholar]
- [7].Haque S, Harries LW. Circular RNAs (circRNAs) in health and disease. Genes (Basel). 2017;8(12):353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lasda E, Parker R. Circular RNAs: diversity of form and function. Rna. 2014;20(12):1829–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: identification, biogenesis and function. Biochimica Et Biophysica Acta (Bba)-gene Regulatory Mechanisms. 2016;1859(1):163–168. [DOI] [PubMed] [Google Scholar]
- [10].Ge Z, Li LF, Wang CY, et al. CircMTO1 inhibits cell proliferation and invasion by regulating Wnt/beta-catenin signaling pathway in colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22(23):8203–8209. [DOI] [PubMed] [Google Scholar]
- [11].Wei X, Li H, Yang J, et al. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 2017;8(10):e3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Xue M, Li G, Fang X, et al. hsa_circ_0081143 promotes cisplatin resistance in gastric cancer by targeting miR-646/CDK6 pathway. Cancer Cell Int. 2019;19(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yang F, Zhang H, Chen SJ, et al. MiR-506 is down-regulated in clear cell renal cell carcinoma and inhibits cell growth and metastasis via targeting FLOT1. PloS One. 2015;10(3):e0120258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yang FQ, Zhang JQ, Jin JJ, et al. HOXA11‐AS promotes the growth and invasion of renal cancer by sponging miR‐146b‐5p to upregulate MMP16 expression. J Cell Physiol. 2018;233(12):9611–9619. [DOI] [PubMed] [Google Scholar]
- [15].Ge Y, Yan X, Jin Y, et al. fMiRNA-192 and miRNA-204 directly suppress lncRNA HOTTIP and interrupt GLS1-mediated glutaminolysis in hepatocellular carcinoma. PLoS Genet. 2015;11(12):e1005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Xiao K, Xia Z, Feng B, et al. Circular RNA expression profile of knee condyle in osteoarthritis by illumina HiSeq platform. J Cell Biochem. 2019. DOI: 10.1002/jcb.29014. [DOI] [PubMed] [Google Scholar]
- [17].Zhao C, Wang Y, Jin H, et al. Knockdown of microRNA-203 alleviates LPS-induced injury by targeting MCL-1 in C28/I2 chondrocytes. Exp Cell Res. 2017;359(1):171–178. [DOI] [PubMed] [Google Scholar]
- [18].Wang Y, Yu T, Jin H, et al. Knockdown mir-302b alleviates lps-induced injury by targeting smad3 in c28/i2 chondrocytic cells. Cell Physiol Biochem. 2018;45(2):733–743. [DOI] [PubMed] [Google Scholar]
- [19].Zhang M, Lygrisse K, Wang J. Role of MicroRNA in osteoarthritis. J arthritis. 2017;6(2). DOI: 10.4172/2167-7921.1000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang M, Wang J. Epigenetic regulation of gene expression in osteoarthritis. Genes Dis. 2015;2(1):69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li B, Zhang Y, Xiao J, et al. Hsa_circ_0045714 regulates chondrocyte proliferation, apoptosis and extracellular matrix synthesis by promoting the expression of miR-193b target gene IGF1R. Hum Cell. 2017;30(4):311–318. [DOI] [PubMed] [Google Scholar]
- [22].Shen S, Wu Y, Chen J, et al. CircSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Ann Rheum Dis. 2019;78(6):826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. [DOI] [PubMed] [Google Scholar]
- [24].Thomas LF, Sætrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics. 2014;30(16):2243–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yudoh K, Nakamura H, Masuko-Hongo K, et al. Catabolic stress induces expression of hypoxia-inducible factor (HIF)-1α in articular chondrocytes: involvement of HIF-1a in the pathogenesis of osteoarthritis. Arthritis Res Ther. 2005;7(4):R904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Pfander D, Cramer T, Swoboda B. Hypoxia and HIF-1a in osteoarthritis. Int Orthop. 2005;29(1):6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang L, Huang Z, Xing R, et al. Increased HIF-1a in knee osteoarthritis aggravate synovial fibrosis via fibroblast-like synoviocyte pyroptosis. Oxid Med Cell Longev. 2019;2019:6326517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.


