ABSTRACT
In oral squamous cell carcinoma (OSCC), abnormal expression of microRNAs has been extensively reported. MiR-let-7a has been validated as a critical regulator of multiple cancers, but the biological process involved and its potential role in OSCC remain unknown.We first analyzed the differential expression of miR-let-7a in cancer tissues, adjacent noncancerous tissues and cell lines. The functional role of miR-let-7a in OSCC cell lines was evaluated by using colony formation assays, cell proliferation and transwell invasion assays in vitro. In addition, subcutaneous xenotransplantation of miR-let-7a transfected cells into nude mouse model was carried out to explore the potential function of miR-let-7a in vivo.miR-let-7a levels were found to be significantly downregulated in OSCC tissues compared with matched normal tissues (n = 60), and lower expression of miR-let-7a was related to poor prognosis in OSCC patients. Overexpression of MiR-let-7a induced a suppression in proliferation, invasion and migration and inhibited tumourigenesis in the nude mouse model. We also determined that c-Myc may serve as a direct target of miR-let-7a; furthermore, upregulated c-Myc expression could partially rescue the effects caused by miR-let-7a overexpression. miR-let-7a is low expression in OSCC, and promotes tumor development by directly targeting c-Myc. Our results may provide a potential therapeutic role for miR-let-7a in human OSCC.
KEYWORDS: miR-let-7a, proliferation, migration, invasion, OSCC
Introduction
As an aggressive neoplasm, oral squamous cell carcinoma (OSCC) is responsible for 3–10% of cancer mortality worldwide, and commonly displays lymph node metastasis [1,2]. Although surgical techniques, molecular-targeted therapy, chemotherapy and radiation therapy have been performed in treating OSCC patients, the five-year survival rate of patients with advanced OSCC remains less than 50% because of high metastasis and recurrence rate [3]. Accumulating evidence has revealed that OSCC is a multifactorial malignancy related to human papillomavirus (HPV) infection, alcohol consumption, betel nut chewing and genetic susceptibility [4]. However, the precise mechanisms underlying OSCC development remain largely unknown. In recent decades, little improvement has been achieved to better understand the aggressive behavior of OSCC. Hence, exploring the mechanism of OSCC is urgently needed for new therapeutic strategies.
MicroRNAs are conserved endogenous non-coding small RNAs of 17–25 nucleotides in length, that have been proven to be important modulators of target messenger RNA expression and promote mRNA degradation by binding to the 3ʹ-UTR (untranslated region) [5,6]. miRNAs play vital roles in various biological processes, such as cell growth, angiogenesis, DNA injury repair, differentiation, cell cycle progression and immunity [7,8]. In recent years, emerging evidence has indicated that aberrantly expressed miRNAs are involved in many types of human disease, and they could serve either as oncogenes or as tumor-suppressors in the tumourigenesis of diverse tumor types [9–12]. So far, an increasing number of miRNAs have been reported to serve as oncogenes in OSCC [13,14]. miR-let-7a has been found to be differentially expressed in various human cancers. Moreover, miR-let-7a has been reported to be downregulated in clinical human samples compared with matched normal tissues [15]; however, the precise functions of miR-let-7a in OSCC remain to be elucidated.
Known as proto-oncogenes, the Myc gene family were comprised of membersl-Myc, n-Myc and c-Myc [16,17]. Overexpression of Myc genes has been reported in many malignancies. c-Myc could act as a transcription factor that affects multidrug resistance, cell cycle progression, apoptosis and metabolism [18–20]. Nearly 15% of genes are regulated by myc-transcription factors. In human carcinomas, a change in the expression level of c-Myc is one of the most common gene abnormalities, and overexpression of the c-Myc protein was found in OSCC [21,22]. Recently, there are some research reports that c-Myc is a direct target of miR-let-7a [23,24], and miR-let-7a overexpression markedly suppresses c-Myc expression level, suggesting that microRNAs may be involved in modulating c-Myc expression.
In the current work, we aimed to elucidate the potential roles of miR-let-7a in OSCC and evaluate its relationship with c-Myc. We showed that miR-let-7a expression was downregulated in OSCC cell lines and clinical human samples, and the low expression of miR-let-7a was correlated with the developments of OSCC. These data suggest that miR-let-7a might play a regulatory role in c-Myc expression. These results reveal let-7a as a novel prognostic indicator and a potential therapeutic target in OSCC.
Materials and methods
OSCC cell lines and transfection
The human OSCC cell lines SCC4, SCC25, HN4, HN6 and Cal27 and the HOK cell line (human normal oral keratinocytes) were purchased from the Cell Bank of Chinese Academy of Science (Shanghai, China). All cells were maintained in DMEM (Dulbecco’s modified Eagle’s medium) supplemented with 10% FBS (fetal bovine serum) and 100 μL/mL penicillin/streptomycin in a humidified atmosphere of 5% CO2 at 37°C.
A total of 2.0 × 105 cells/mL were seeded into 6-well plates for 24 h prior to transfection. OSCC cell lines were transfected with chemically synthesized miR-let-7a mimics, inhibitor or scrambled control by using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). In detail, cells should be 70–90% confluent at the time of transfection. Diluted miR-let-7a mimics, inhibitor or scrambled control and diluted Lipofectamine 3000 were mixed and incubated for 15 min at room temperature. Then, the 100 μl of complexes was added into each well containing cells and medium. Then, incubated OSCC cell lines at 37°C in a CO2 incubator for 24–48 h prior to testing for transgene expression.
Patients and OSCC tissues
Informed consent was obtained from all patients. Tumor tissue or paired adjacent non-tumorous tissue samples were obtained from 60 OSCC patients. The diagnosis of OSCC was independently validated by two experienced pathologists. After surgical resection, tissues were collected, rapidly frozen and stored in liquid nitrogen until further processing. Our study was approved by the Institutional Ethical Board of Nanjing medical university.
RNA extraction and qRT-PCR assay
Total RNA from fresh-frozen tissues and cells was extracted by using the TRIzol reagent. For mRNA profiling detection, PrimeScript RT Reagent was used to convert total RNA into cDNA, and the Step One Plus Real-Time PCR System was used to perform the qRT-PCR amplification. mRNA expression was normalized to β-actin expression. To examine the miR-let-7a expression level, qRT-PCR was carried out by using a miRNA qPCR detection kit on the Step One Plus Real-Time PCR System. The U6 gene was chosen as an internal reference. All assays mentioned were conducted in triplicate.
Western blot assay
Whole tissues or cell protein extracts were prepared with RIPA buffer supplemented with a protease inhibitor. Then, 30 μg protein samples were electrophoresed in SDS-PAGE gels (10% or 12%) and transferred to PVDF membranes. The PVDF membranes were then incubated with primary antibodies against c-Myc (1:1000, Abcam), Vimentin (1:1000, Abcam), p38 (1:1000, Abcam), p-ERK (1:1000, Abcam), E‑cadherin (1:1000, Abcam), p-p38 (1:1000, Abcam), ERK (1:1000, Abcam), or β-actin (1:1000, Abcam) overnight at 4°C. After washing with PBST and incubating with secondary antibodies for 2 h at 37°C, the membranes were washed another three times with TBST buffer, and protein expression was visualized with a peroxide LumiGLO reagent.
Cell proliferation analysis
To detect the biological effect of miR-let-7a on cell proliferation was evaluated by CCK-8 assay. First, cells at equal density (2 × 103) were plated in a 96-well plate for each group. After 24 h, when cells were well adhered, 10 μL of CCK-8 reagents mixed with 100 μL of medium was added to each well after the transfection (0, 24, 48, 72 and 96 h) and incubated for 2 h. Viable cells were then measured by a microplate reader at 450 nm. Cell proliferation was also evaluated by colony formation assay, whereby OSCC cell lines were placed in 6-well microplates at a density of 600 cells/well and cultured for 2 wk, with the complete medium replaced with new culture medium every 2–3 d. Colonies were stained with methylene blue, and number of viable clones were calculated and imaged to evaluate cell proliferation.
Cell migration assays
Cell migration and invasion were assayed by a Millicell chamber (Merck Millipore). Approximately 2 × 105 transfected cells were placed into the upper part of the transwell chamber with or without Matrigel (BD Bioscience Pharmingen) coating. Complete medium was added to the lower compartment of the chamber. After incubation for 24 h at 37°C, cells that migrated to the bottom of the filter were fixed with 70% ice-cold methanol (MeOH) and stained with crystal violet (0.1% crystal violet (w/v)) for 30 min. Photographs of five randomly selected fields of the fixed cells were taken and counted under a light microscope at the magnification of 100 ×.
Target analysis and luciferase reporter gene assay
The putative targets of miR-let-7a were predicted by using bioinformatic analysis. The 3ʹ-UTR of the c-Myc gene containing wild-type (WT) or mutated (MT) miR-let-7a binding site was chemically synthesized and cloned into the pGL3-promoter vector containing the firefly luciferase reporter. The OSCC cell lines were placed into 96-well microplates (3 × 103 cells) and incubated in 5% CO2 at 37°C before transfection. Cultured cells were co-transfected with WT or MT c-Myc 3ʹ-UTR together with 40 nM of let-7a mimic or mimic NC. Cells were harvested after incubation for 48 h, and the firefly and Renilla luciferase activities were measured independently by using a dual-luciferase reporter assay kit. The relative normalized luciferase activity of each construct was detected according to the luciferase signal ratio (Rluc/luc).
Tumor growth in nude mice
Five-week-old BALB/c nude mice (Shanghai Experimental Animal Center, PR China), weighing 15–20 g, were randomly allocated into 2 equal groups with 5 animals per group. Nude mice were implanted subcutaneously with 1 × 106 stably transfected cells (miR-let-7a sponge or control sponge vector) into the neck. Tumor sizes were measured with a caliper every week for 5 wk and evaluated by using the following formula: volume = (width2 × length)/2 [25]. The mice were sacrificed by carbon dioxide overdose, and tumors were excised and weighed after 28 d.
Statistical analysis
Numerical data are expressed as the mean ± s.d. Student’s t-tests were used to analyze the differences between two groups. Comparison of measurement data was analyzed using one-way ANOVA. Survival curves were obtained by the Kaplan–Meier method, and the distribution between patient subsets was compared using the log-rank test. Statistically significant differences were considered when P < 0.05.
Results
The expression of miR-let-7a is reduced in OSCC cell lines and OSCC tissues
The previous expression profiling of miRNAs in 61 primary OSCC tissue specimens showed that miR-let-7a was found to be significantly downregulated [15]. To confirm and extend the previous results, an analysis of miR-let-7a expression in 60 OSCC tissues and paired adjacent noncancerous tissue samples was performed using qRT-PCR, and significant low expression of miR-let-7a was observed in OSCC tissues compared to paired adjacent noncancerous tissue samples (Figure 1(a)). Furthermore, the expression of miR-let-7a was dramatically decreased in SCC4, SCC25, HN4, HN6 and Cal27 cells compared with human normal oral keratinocytes (HOK). These results suggest that reduced miR-let-7a expression occurs frequently in OSCC tissues and OSCC cell lines (Figure 1(b)).
Figure 1.
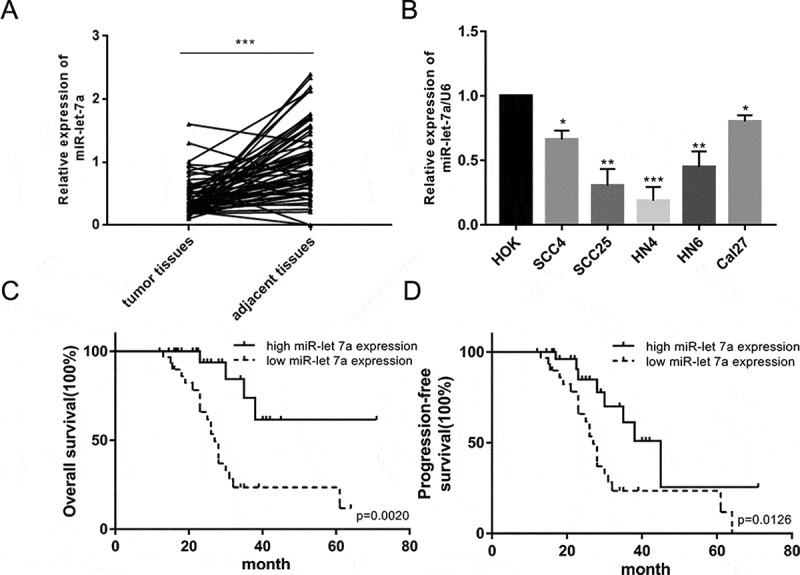
Down-regulated expression of miR-let-7a in OSCC tissues and OSCC cell lines. (a) The relative expression of miR-let-7a in 60 paired OSCC tissues and adjacent normal tissues were detected by miRNA RT-PCR. (b) Expression of miR-let-7a in OSCC cell lines and the human normal oral keratinocytes (HOK) cell line were detected by miRNA RT-PCR. (*P < 0.05, **P < 0.01, ***P < 0.001) (c, d) Overall survival and progression-free survival curves were compared between OSCC patients with low expression level of miR-let-7a and those with high level of miR-let-7a. All assays in this part were performed in triplicates.
Clinicopathological characteristics of patients with reduced miR-let-7a expression
We sought to analyze the clinical significance of miR-let-7a expression in OSCC. The median value of the relative miR-let-7a expression in all 60 OSCC cases was chosen as the cutoff point for separating tumors with lower expression of miR-let-7a from those with higher expression miR-let-7a. The correlation of downregulated miR-let-7a and clinicopathological features was investigated and shown in Table 1. Lower expression of miR-let-7a was correlated significantly with larger tumor size (P = 0.04). However, the expression level of miR-let-7a had no significant relationship with other clinical factors. Kaplan-Meier survival analyses also determined that people with lower miR-let-7a levels had a poorer prognosis than those with higher miR-let-7a expression both in terms of overall survival (OS) and progression-free survival (PFS) (Figure 1(c,d)).
Table 1.
Correlation between clinical pathologic features and the miR-let-7a and c-myc expressions in patients with oral squamous cell carcinoma.
| miR-let-7a expression |
c-myc expression |
||||||
|---|---|---|---|---|---|---|---|
| Characteristics | Number | High group |
Low group |
P value |
High group |
Low group | P value |
| Age(years) | |||||||
| <60 | 8 | 4 | 4 | 0.83 | 6 | 2 | 0.22 |
| >60 | 52 | 28 | 24 | 27 | 25 | ||
| Gender | |||||||
| Male | 29 | 15 | 14 | 0.79 | 16 | 13 | 0.44 |
| Female | 31 | 15 | 16 | 14 | 17 | ||
| Clinical stage | |||||||
| I–II | 34 | 14 | 20 | 0.12 | 17 | 17 | 0.76 |
| II–IV | 26 | 16 | 10 | 12 | 14 | ||
| Differentiated | |||||||
| Well/moderate | 28 | 13 | 15 | 0.22 | 11 | 17 | 0.07 |
| Poor | 32 | 10 | 22 | 20 | 12 | ||
| Lymph node metastasis |
|||||||
| No | 20 | 9 | 11 | 0.06 | 8 | 12 | 0.01 |
| Yes | 40 | 28 | 12 | 29 | 11 | ||
| Tumor size(cm) |
|||||||
| <3 | 38 | 21 | 17 | 0.04 | 12 | 26 | 0.02 |
| ≥3 | 22 | 6 | 16 | 14 | 8 | ||
miR-let-7a overexpression inhibits the proliferation, migration and invasion of OSCC cell lines
We performed loss-of-function experiments by transfecting miR-let-7a mimics or scramble miR as a negative control to evaluate the potential role of miR-let-7a in the development and progression of OSCC. First, the miR-let-7a expression levels were lower in SCC25 and HN4 cells, and both cell lines were transfected with miR-let-7a mimics or miR-NC. RT-PCR was used to examine miR-let-7a expression levels in the two OSCC cell lines (Figure 2(a)). Furthermore, we transfected Cal27 cells with miR-let-7a inhibitor based on the relatively higher expression of let-7a in this OSCC cell line, and the efficiency of transfection was confirmed by qRT-PCR at 48 h after transfection (Figure 2(b)). The CCK-8 assay results showed that cell proliferation was significantly inhibited in SCC25 and HN4 cells transfected with miR-let-7a mimics compared to the control group, inhibition of miR-let-7a significantly increased the proliferation of Cal27 cells (Figure 2(c)). In addition, transwell migration and invasion assays were performed to investigate the cell migration and invasion capacities in each group. As shown in Figure 2(d,e), the miR-let-7a overexpression group had significantly impaired invasion and migration compared with the control groups in SCC25 and HN4 cells. Meanwhile, miR-let-7a inhibition markedly enhanced cell migration and invasion in Cal27 cells. Collectively, the above results proved that overexpression of miR-let-7a may inhibit the proliferation, migration and invasion of OSCC cell lines.
Figure 2.
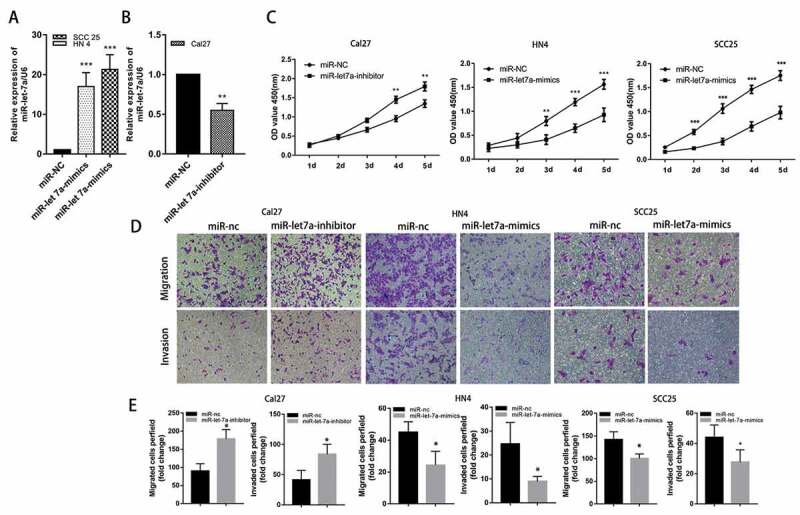
Effects of miR-let-7a on proliferation, migration and invasion of OSCC in vitro. (a, b) MiRNA RT-PCR was used to confirm the expression of miR-let-7a in cells transfected with miR-let-7a-mimics and miR-let-7a-inhibitor. (c) CCK-8 was were used to evaluate the proliferation of in Cal 27, HN4 and HCC-25 cells after transfection with the miR-let-7a inhibitor, miR-let-7a mimics or negative control. (d) Transwell assay was performed to detect the ability of migration and invasion of miR-let-7a inhibitor, miR-let-7a mimics transfected Cal 27, HN4 and HCC-25 cells and their negative control. (e) Quantification of Transwell assay (*P < 0.05, **P < 0.01, ***P < 0.001). All assays in this part were performed in triplicates.
Overexpression of miR-let-7a suppresses tumor growth in vivo
We hypothesized that miR-let-7a overexpression has a growth-attenuating effect on tumors in vivo. A tumor formation assay in nude mice was performed in which 4-6-week-old BALB/c athymic nude mice were injected with SCC25 cells stably transfected with miR-let-7a mimics or negative control. Tumors grown in this animal model of tumourigenicity were observed for 5 wk. As shown in Figure 3(a), in miR-let-7a mimics group, cells transfected with miR-let-7a-mimics generated smaller and lighter tumors. What’ more, we found that tumors in the miR-let-7a overexpression group grew at a slower rate than those in the NC group (Figure 3(b)). Furthermore, the miR-let-7a overexpression group had tumors with lower weights compared to the negative control group (Figure 3(c)). We used miRNA RT-PCR to confirm the expression of miR-let-7a in mice tumor tissues, group transfected with miR-let-7a-mimics owned a higher expression level of miR-let-7a (Figure 3(d)). Our results implied that overexpression of miR-let-7a inhibited tumor growth both in vitro and in vivo.
Figure 3.
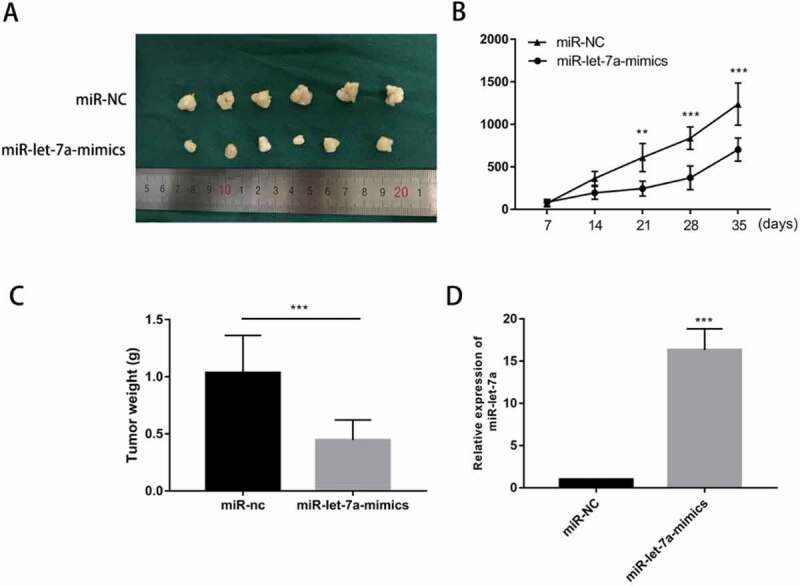
Overexpression of miR-let-7a inhibits OSCC in vivo. (a) Photographs of tumors obtained from the different groups of nude mice transfected with miR-let-7a mimics and miR-NC. (b) Tumor growth curve of nude mice transfected with miR-let-7a-mimics and miR-NC, Tumor diameters were measured every 7 d. (c) The weight of the tumor tissues of nude mice transfected with miR-let-7a-mimics and miR-NC D miR-let-7a expression levels were detected in tumor tissues from miR-let-7a-mimics and miR-NC groups. All assays in this part were performed in triplicates.
c-Myc is a potential target gene of miR-let-7a
To gain insight into the mechanisms of miR-let-7a suppressing tumor growth, we conducted bioinformatic analyses in public databases (miRanda, TargetScan) to predict potential miR-let-7a targets. The target prediction program analysis identified a putative binding site for miR-let-7a in the 3ʹ-UTR of c-Myc. Luciferase assays were carried out to validate the prediction. We constructed two types of pGL3 reporter plasmids containing the wild-type or mutant c-Myc 3′-UTR sequence, and reporter plasmids were then co-transfected with miR-let-7a mimics into SCC25 and HN4 cells. miR-let-7a overexpression significantly suppressed the luciferase activity of wild-type c-Myc but not that of the mutant-type c-Myc in SCC25 and HN4 cells (Figure 4(a)). Moreover, higher expression of c-Myc was also significantly correlated with larger tumor size and positive lymph node metastasis (Table 1). These data implied that overexpression of miR-let-7a attenuates c-Myc expression in a posttranscriptional manner.
Figure 4.
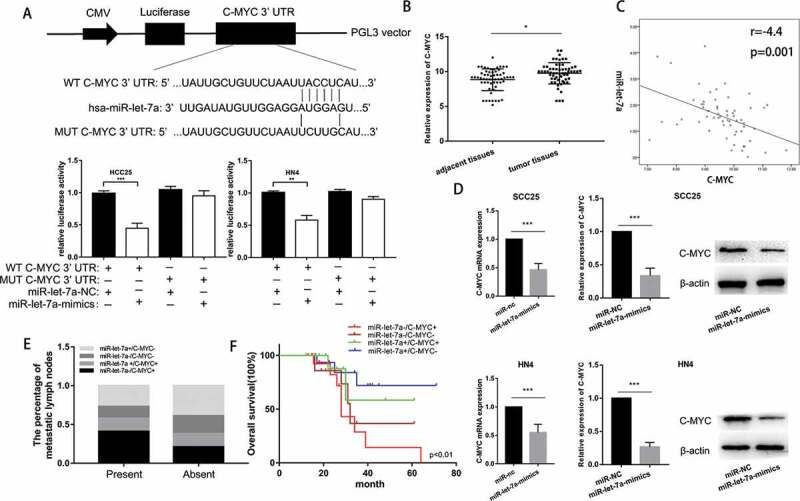
c-Myc is a direct target of miR-let-7a in OSCC cell lines. (a) miR-let-7a and its putative binding sequence in the wild-type and mutant 3′-UTR of c-Myc, Overexpression of miR-let-7a significantly decreased the luciferase activity that carried wild type (WT) but not mutant type (MUT) 3′-UTR of c-Myc in HCC-25, HN4 cells. (b) Expression of c-Myc was upregulated in 60 paired OSCC tissues compared with adjacent normal tissues (c) c-Myc was negatively correlated with miR-let-7a. (d) Expression of c-Myc in HCC-25, HN4 cells transfected with miR-let-7a-mimics or miR-NC was detected by qRT-PCR and western blot, Overexpression of miR-let-7a markedly suppressed the mRNA and protein levels of c-Myc in HCC-25, HN4 cells. (e) The correlation between miR-let-7a/c-Myc co-expression and the percentage of metastatic lymph nodes. (f) Overall survival of OSCC patients. Patient groups were separated based on expression status of miR-let-7a and c-Myc. All assays in this part were performed in triplicates.
To validate the inhibitory effect of overexpression of miR-let-7a on c-Myc, we determined the c-Myc expression level in 60 clinical human samples and paired normal tissues by RT-PCR. Compared with that in the paired non-tumorous tissues, the relative expression of c-Myc was significantly upregulated in OSCC tissues (Figure 4(b)). We also discovered an inverse correlation between miR-let-7a and c-Myc expression in the 60 paired OSCC specimens (Figure 4(c)). Furthermore, we transfected miR-let-7a mimics or NC into SCC25 and HN4 cells, as shown in Figure 4(d), and observed that miR-let-7a negatively regulated c-Myc expression at both the mRNA and protein levels. Then, we divided the 60 OSCC cases into the following four groups according to the miR-let-7a mRNA level and c-Myc protein level: miR-let-7a low expression and c-Myc high expression (miR-let-7a-/c-Myc+), miR-let-7a low expression and c-Myc low expression (miR-let-7a-/c-Myc-), miR-let-7a high expression and c-Myc high expression (miR-let-7a+/c-Myc+), miR-let-7a high expression and c-Myc low expression (miR-let-7a+/c-Myc-). We further analyzed the distribution of lymph node metastasis and overall survival in each group. Our data suggest that the miR-let-7a-/c-Myc+ group had a significantly higher metastasis rate compared to the other groups (Figure 4(e)). Moreover, miR-let-7a-/c-Myc+ levels predicted poor prognosis, while miR-let-7a+/c-Myc- levels indicated relatively favorable prognosis (Figure 4(f)). Taken together, these data may provide a strategy in which low expression of miR-let-7a has a crucial role in promoting the proliferation and metastasis of OSCC through upregulation of c-Myc expression.
c-Myc contributes to the effect of miR-let-7a on OSCC cell lines
To determine whether c-Myc is involved in mediating the biological function of miR-let-7a in OSCC, SCC25 and HN4 cell lines were transfected with miR-let-7a mimics or negative control. We found that the downregulation of miR-let-7a reduced the c-Myc protein and mRNA expression, and the c-Myc protein expression was partially recovered when co-transfection of c-Myc overexpression plasmid (Figure 5(a)). Furthermore, co-transfection of c-Myc overexpression significantly reversed the suppressive effects of miR-let-7a overexpression on cell proliferation, migration and invasion (Figure 5(b,c)). These investigations indicated that overexpression of miR-let-7a inhibited OSCC cell growth, migration and invasion through suppression of c-Myc activity.
Figure 5.
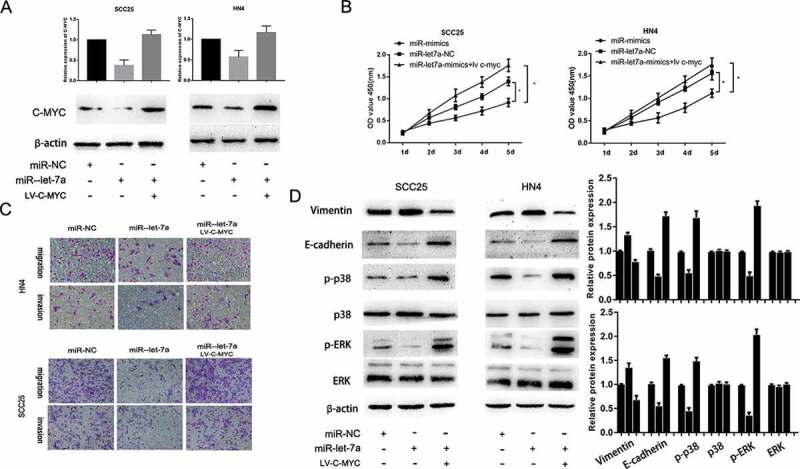
Overexpression of miR-let-7a inhibits proliferation, migration and invasion and regulates MAPK/ERK signaling pathway by targeting c-Myc. (a) qRT-PCR and Western blot analysis showed that over expression of miR-let-7a reduced the c-Myc RNA and protein expression, while co-transfection of c-Myc overexpression plasmids could recover the c-Myc RNA and protein expression. (b, c) Co-transfection of c-Myc plasmid reverted the suppressive effects of miR-let-7a overexpression on the proliferation, migration and invasion. (d) The expression of MAPK/ERK signaling was evaluated by western blot, miR-let-7a overexpression could notably down-regulate the expression of phosphorylated p38 and ERK, while the total amount of p38 and ERK showed no differences. Overexpressed c-Myc could partially counteracted the change of the expression of phosphorylated p38 and ERK. All assays in this part were performed in triplicates.
MiR-let-7a suppresses the MAPK/ERK signaling pathway
To investigate the underlying mechanisms of miR-381 mediated the functional effects, we determined the effect of miR-let-7a overexpression on the expression of MMP (matrix metalloproteinase) and EMT (Epithelial–Mesenchymal Transition) markers, which are well-known factors that mediate the invasiveness of cancer cells. The data showed that the expression of E-cadherin was obviously suppressed and Vimentin expression was significantly increased by miR-let-7a overexpression, while enforced c-Myc expression could partially counteract the changes in these EMT markers compared with miR-NC. A previous study found that c-Myc contributed to migration and invasion through activation of the MAPK/ERK signaling pathway [26,27]. Herein, we explored whether overexpression of miR-let-7a suppressed malignant progression through the MAPK/ERK signaling pathway by targeting c-Myc. Western blot analysis indicated that miR-let-7a overexpression could notably downregulate phosphorylated p38 and ERK expression, while the total protein expression of p38 and ERK was not altered, and c-Myc overexpression could partially counteract the change induced by miR-let-7a overexpression (Figure 5(d)). These data demonstrated that the MAPK/ERK pathway contributed to the role of the miR-let-7a/c-Myc axis in OSCC. However, the results do not rule out the involvement of other signaling pathways in OSCC tumor progression.
Discussion
OSCC is a heterogeneous tumor whose development is widely regulated by multiple molecular mechanisms and signaling pathways [28]. Mounting evidence has suggested that dysregulation of miRNA expression results in progressive and uncontrolled tumor growth. Depending on the specific functions of the targeted mRNAs, miRNAs could be either tumor suppressor genes or cancer-promoting genes by exerting their function on target genes [29]. Inactivation of tumor suppressor genes by increased expression of tumor suppressor miRNAs contributes to the development and biological processes of cancer, and increased expression of oncogenic miRNAs could also cause similar effects [30]. A growing amount of evidence has shown that the abnormal expression of microRNAs plays an important role in the carcinogenesis and metastasis of OSCC [30,31].
miR-let-7a, a member of the let-7 family, was verified to regulate various signaling pathways in tumors by targeting related genes [32]. Let-7a inhibits cell proliferation by regulating E2F2 in osteosarcoma cells and suppresses tumourigenicity in colon cancer [33,34]. miR-let-7a was found to change the expression of MYC and revert MYC-induced growth by targeting caspase-3 in Burkitt lymphoma cells [35]. The tumor-suppressive role of miR-let-7a has been widely reported in previous studies, and downregulation of miR-let-7a could promote proliferation and migration in different tumors [36]. Subsequently, miR-let-7a was identified to down-regulate in 61 primary OSCC tissue specimens [15], but the functional involvement of miR-let-7a in OSCC is still unknown. We performed the expression analysis of miR-let-7a in 60 paired OSCC tumor samples and adjacent normal tissues. Results showed that miR-16 was significantly downregulated in OSCC tumor samples, which was basically consistent with the previous results.
We observed the low expression of miR-let-7a in OSCC tumor samples, and its oncological significance aroused our interest. Here, we show similar results in the OSCC cell lines, and that lower miR-let-7a expression levels are correlated with larger tumor size. Moreover, our data on survival outcomes demonstrated that lower miR-let-7a expression levels in OSCC are associated with shorter OS and PFS. We further suspected that miR-let-7a plays a vital role in biological function changes in OSCC. Our present results show that overexpression of miR-let-7a can suppress tumor growth and invasion ability, while downregulation of miR-let-7a has a tumor-promoting role in OSCC.
Growing lines of evidence have demonstrated that the proto‐oncogene c-Myc could promote ribosome biogenesis, transformation, metabolism, cell proliferation and genomic programs in mammalian cells [37–39]. Mechanistically, online predicting database analyses and luciferase reporter assays revealed that miR-let-7a negatively regulated c-Myc mRNA expression by targeting its 3ʹ-UTR and thus mediated the functional effects on cell growth and EMT of OSCC. Moreover, our OSCC tissue study showed that miR-let-7a expression was notably lower and c-Myc expression was significantly higher in OSCC, and an inverse correlation between c-Myc and miR-let-7a expression was found in OSCC patients. Overall, our study provides the first global evidence that downregulation of miR-let-7a promotes OSCC cell proliferation and EMT by negatively regulating c-Myc expression. The function of c-Myc in cell proliferation is well illustrated [40–42], and research has also shown that c-Myc overexpression could be related to the invasion and metastasis of cancer cells [43,44]. c-Myc overexpression showed inhibitory effects on breast cancer cell invasion and migration abilities by silencing the αv and β3 integrin subunits [45]. Overexpression of c-Myc also promoted melanoma metastasis via vasculogenic mimicry [46]. Consistent with the previous report, our findings show that downregulation of miR-let-7a in OSCC induced promotion of OSCC cell proliferation and EMT progression via regulation of c-Myc as well as the MAPK/ERK signaling pathway.
Conclusions
In summary, we found that miR-let-7a is downregulated and may act as a biomarker for predicting poor prognosis in OSCC patients. Our current study revealed that lowering the expression of miR-let-7a promotes cellular proliferation, invasion and migration via the miR-let-7a/c-Myc/MAPK/ERK signaling pathway. The present results demonstrated a novel therapeutic target involved in OSCC progression.
Funding Statement
This work was supported by the National Natural Science Foundation of China (No. 81672169, No. 81974332).
Author contributions
Fan WM designed and supervised this study. Luo CY, Zhang Y, Zhang JY, Zhang X conducted the majority of the experiments and completed the manuscript. Chen YN analyzed the data. All authors read and approved the final manuscript. Data availability Datasets of the current study are available from the corresponding author on reasonable request.
Disclosure statement
The authors do not have any conflict of interest.
References
- [1].Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. Ca A Cancer J Clinicians. 2015;60(5):277–300. [Google Scholar]
- [2].Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. Ca A Cancer J Clinicians. 2016;66(4):271–289. [DOI] [PubMed] [Google Scholar]
- [3].Delaine-Smith RM, Reilly GC.. Chapter twenty – the effects of mechanical loading on mesenchymal stem cell differentiation and matrix production. Vitamins Hormones. 2011;87(10):417. [DOI] [PubMed] [Google Scholar]
- [4].Drummond SN, De MLIA, Barbosa AA, et al. TP53 codon 72 polymorphism in oral squamous cell carcinoma. Anticancer Res. 2002;22(6A):3379–3381. [PubMed] [Google Scholar]
- [5].Farazi TA, Hoell JI, Morozov P, et al. microRNAs in human cancer. Adv Exp Med Biol. 2013;774(2):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Betel D, Wilson M, Gabow A, et al. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(Database issue):149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Song Z, Li G.. Role of specific MicroRNAs in regulation of vascular smooth muscle cell differentiation and the response to injury. J Cardiovasc Transl Res. 2010;3(3):246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sadat AAS, Hossein GSM. The role of microRNAs in cardiovascular disease. Int J Mole Cell Med. 2013;2(2):50–57. [PMC free article] [PubMed] [Google Scholar]
- [9].Ghisi M, Corradin A, Basso K, et al. Modulation of microRNA expression in human T-cell development: targeting of NOTCH3 by miR-150. Blood. 2011;117(26):7053–7062. [DOI] [PubMed] [Google Scholar]
- [10].Kinoshita T, Hanazawa T, Nohata N, et al. The functional significance of microRNA-375 in human squamous cell carcinoma: aberrant expression and effects on cancer pathways. J Hum Genet. 2012;57(9):556–563. [DOI] [PubMed] [Google Scholar]
- [11].Sun T, Wang Q, Balk S, et al. The role of microRNA-221 and −222 in Androgen-independent prostate cancer cell lines. Cancer Res. 2009;69(8):3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Takahashi RU, Miyazaki H, Ochiya T. The roles of MicroRNAs in breast cancer. Cancers (Basel). 2015;7(2):598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tang H, Wu Z, Zhang J, et al. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol Med Rep. 2013;7(3):761–766. [DOI] [PubMed] [Google Scholar]
- [14].Severino P, Oliveira LS, Andreghetto FM, et al. Small RNAs in metastatic and non-metastatic oral squamous cell carcinoma. BMC Med Genomics. 2015;8(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Manikandan M, Rao AKDM, Arunkumar G, et al. Oral squamous cell carcinoma: microRNA expression profiling and integrative analyses for elucidation of tumourigenesis mechanism. Mol Cancer. 2016;15(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lee WM. The myc family of nuclear proto-oncogenes. Cancer Treat Res. 1989;47:37–71. [DOI] [PubMed] [Google Scholar]
- [17].Brenner C, Deplus R, Didelot C, et al. Myc represses transcription through recruitment of DNA methyltransferase corepressor. Embo J. 2014;24(2):336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Koskinen PJ, Alitalo K. Role of myc amplification and overexpression in cell growth, differentiation and death. Semin Cancer Biol. 1993;4(1):3–12. [PubMed] [Google Scholar]
- [19].Ishizaki H, Yano H, Tsuneoka M, et al. Overexpression of the myc target gene Mina53 in advanced renal cell carcinoma. Pathol Int. 2010;57(10):672–680. [DOI] [PubMed] [Google Scholar]
- [20].Mai S. Overexpression of c-myc precedes amplification of the gene encoding dihydrofolate reductase. Gene. 1994;148(2):253–260. [DOI] [PubMed] [Google Scholar]
- [21].Pai R, Pai S, Lalitha R, et al. Over-expression of c-Myc oncoprotein in oral squamous cell carcinoma in the South Indian population. Ecancermedicalscience. 2009;3(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pérez-Sayáns M, Suárez-Peñaranda JM, Pilar GD, et al. What real influence does the proto-oncogene c-myc have in OSCC behavior? Oral Oncol. 2011;47(8):688–692. [DOI] [PubMed] [Google Scholar]
- [23].Wang G, Wang J, Zhao H, et al. The role of Myc and let-7a in glioblastoma, glucose metabolism and response to therapy. Arch Biochem Biophys. 2015;580::84–92. [DOI] [PubMed] [Google Scholar]
- [24].He XY, Chen JX, Zhang Z, et al. The let-7a microRNA protects from growth of lung carcinoma by suppression of k-Ras and c-Myc in nude mice. J Cancer Res Clin Oncol. 2010;136(7):1023–1028. [DOI] [PubMed] [Google Scholar]
- [25].Faustino-Rocha A, Oliveira PA, Pinho-Oliveira J, et al. Estimation of rat mammary tumor volume using caliper and ultrasonography measurements. Lab Anim (NY). 2013;42(6):217–224. [DOI] [PubMed] [Google Scholar]
- [26].Xu XH, Zhang SJ, Hu QB, et al. Effects of microRNA-494 on proliferation, migration, invasion, and apoptosis of medulloblastoma cells by mediating c-myc through the p38 MAPK signaling pathway. J Cell Biochem. 2019;120(2):2594–2606. [DOI] [PubMed] [Google Scholar]
- [27].Qiaoli Z, Assimopoulou AN, Klauck SM, et al. Inhibition of c-MYC with involvement of ERK/JNK/MAPK and AKT pathways as a novel mechanism for shikonin and its derivatives in killing leukemia cells. Oncotarget. 2015;6(36):38934–38951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Saeed NR, Gold JA. Oral squamous cell carcinoma. BMJ. 1994;308(6936):1103. [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang B, Pan X, Cobb GP, et al. microRNAs as oncogenes and tumor suppressors. N Engl J Med. 2007;302(1):1–12. [DOI] [PubMed] [Google Scholar]
- [30].Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25(46):6188–6196. [DOI] [PubMed] [Google Scholar]
- [31].Siow MY, Ng LPK, Abraham MT, et al. P132. MicroRNA profiles of oral squamous cell carcinoma (OSCC) using formalin-fixed, paraffin embedded (FFPE) tissue. Oral Oncol. 2011;47(Suppl 1):S117–S117. [Google Scholar]
- [32].Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–516. [DOI] [PubMed] [Google Scholar]
- [33].Iwasaki T, Tanaka K, Kawano M, et al. Tumor-suppressive microRNA-let-7a inhibits cell proliferation via targeting of E2F2 in osteosarcoma cells. Int J Oncol. 2015;46(4):1543–1550. [DOI] [PubMed] [Google Scholar]
- [34].Yukihiro A, Yoshihito N, Tomoki N. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29(5):903. [DOI] [PubMed] [Google Scholar]
- [35].Sampson VB, Rong NH, Han J, et al. MicroRNA Let-7a Down-regulates MYC and reverts MYC-Induced growth in Burkitt Lymphoma Cells. Cancer Res. 2007;67(20):9762–9770. [DOI] [PubMed] [Google Scholar]
- [36].Wang Y, Cheng N, Luo J. Downregulation of lncRNA ANRIL represses tumorigenicity and enhances cisplatin-induced cytotoxicity via regulating microRNA let-7a in nasopharyngeal carcinoma. J Biochem Mole Toxicolo. 2017;31(7):e21904. [DOI] [PubMed] [Google Scholar]
- [37].Dang C. MYC on the Path to Cancer. Cell. 2012;149(1):22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Grandori C, Cowley SMJames LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior[Review]. Annu Rev Cell Dev Biol. 2000;16(1):653. [DOI] [PubMed] [Google Scholar]
- [39].Stine ZE, Walton ZE, Altman BJ, et al. MYC, metabolism, and cancer. Cancer Discov. 2015;5(10):1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lin C, Lovén J, Rahl P, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nie Z, Hu G, Wei G, et al. c-Myc Is a Universal Amplifier of Expressed Genes in Lymphocytes and Embryonic Stem Cells. Cell. 2012;151(1):68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Susanne W, Francesca L, Jennifer M, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511(7510):483–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Boxer RB, Jang JW, Sintasath L, et al. Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer Cell. 2004;6(6):577–586. [DOI] [PubMed] [Google Scholar]
- [44].Welm AL, Suwon K, Welm BE. MET and MYC cooperate in mammary tumorigenesis. Proc Natl Acad Sci U S A. 2005;102(12):4324–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hong L, Radisky DC, Dun Y, et al. MYC suppresses cancer metastasis by direct transcriptional silencing of αv and β3 integrin subunits. Nat Cell Biol. 2012;14(6):567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lin X, Sun R, Zhao X, et al. C-myc overexpression drives melanoma metastasis by promoting vasculogenic mimicry via c-myc/snail/Bax signaling. J Mol Med (Berl). 2017;95(1):53–67. [DOI] [PubMed] [Google Scholar]


