ABSTRACT
Neoadjuvant intense androgen deprivation therapy for high-risk localized prostate cancer is an emerging but unproven treatment paradigm that is hoped to delay or prevent disease recurrence. We found that a patient enrolled in a clinical trial harbored two completely independent prostate cancers that responded differently to this therapy.
KEYWORDS: Prostate cancer, response, non-response, enzalutamide, intense neoadjuvant therapy
Androgen deprivation therapy (ADT) is a mainstay of treatment for patients diagnosed with recurrent or metastatic prostate cancer, and although newer agents such as abiraterone and enzalutamide intensify ADT and prolong survival, metastatic disease is ultimately lethal.1 Men who present with high-risk but localized prostate cancer are routinely treated with radical prostatectomy (RP) and/or external beam radiation therapy, which serves to abrogate the risk of recurrence in 10-50% of individuals. In patients who undergo surgery with curative intent but ultimately relapse, the drop of prostate-specific antigen (PSA) levels to undetectable and lack of visible lesions on imaging coincides with an extended period of undetectable tumor outgrowth with potential for selection of aggressive prostate cancer subclones. Identifying which patients are at the highest risk of recurrence under this standard of care remains immensely challenging due to the decade-long interval between RP and PSA recurrence, emphasizing the importance of considering intensified treatment options for high-risk disease that is potentially in the curative window.
Rather than waiting for PSA levels to rise, neoadjuvant intense ADT moves the treatment earlier – potentially by years – with the premise that occult micrometastatic or locoregional subclones that existed at the time of surgery would be as equivalently sensitive as the primary tumor. The evidence supporting this rationale comes in part from three recent neoadjuvant intense ADT trials2-4 in which men whose prostates showed either a pathologic complete response (pCR) or minimal residual disease (MRD) did not experience recurrence during the period of follow-up.5 Ideally, a precision-guided clinical workflow could identify more of these exceptional responders prior to treatment, but extensive tumor heterogeneity remains a confounding variable for predicting outcome.6
In our recent case report,7 we described a patient with a large heterogeneous prostate cancer who received neoadjuvant intense ADT as part of a clinical trial but demonstrated substantial resistance to therapy. With access to multiple pre-treatment biopsies targeted by MRI-ultrasound fusion guidance to different regions of the tumor, we extensively sampled this heterogeneity to reveal that this patient harbored a polytumor, defined as two clonally-independent tumor systems that evolved separately. These two distinct tumors (Figure 1) maintained unique genomic features that ultimately drove different responses to neoadjuvant intense ADT in this patient. Of the two lesions identified at baseline on multiparametric magnetic resonance imaging (mpMRI) and characterized using whole exome sequencing, the larger lesion exhibited a more aggressive phenotype, including a substantial population of intraductal carcinoma, the transmembrane protein serine 2-ETS related gene (TMPRSS2-ERG) fusion, and deletions to chromosome arms 10q and 17p involving the tumor suppressors phosphatase and tensin homolog (PTEN) and tumor protein p53 (TP53). The smaller lesion did not harbor any of these events but instead carried a set of mutations and somatic copy number alterations that did not encompass known cancer genes despite showing a focal reduction in PTEN immunostaining. After 6 months of therapy, the patient underwent an additional mpMRI, which showed a reduction in volume of the larger lesion (from 6.26 cc to 2.28 cc), while failing to identify the smaller lesion, suggesting a complete response of the other tumor to treatment. Although the responding focus resolved on imaging, examination of the surgical specimen revealed PTEN-reduced MRD of the smaller lesion only, which was spatially distinct from the large residual tumor. Meanwhile, the large lesion maintained both invasive and intraductal components, with the substantial intraductal component showing a deep deletion of PTEN and a TP53 nonsense mutation by whole exome sequencing.
Figure 1.
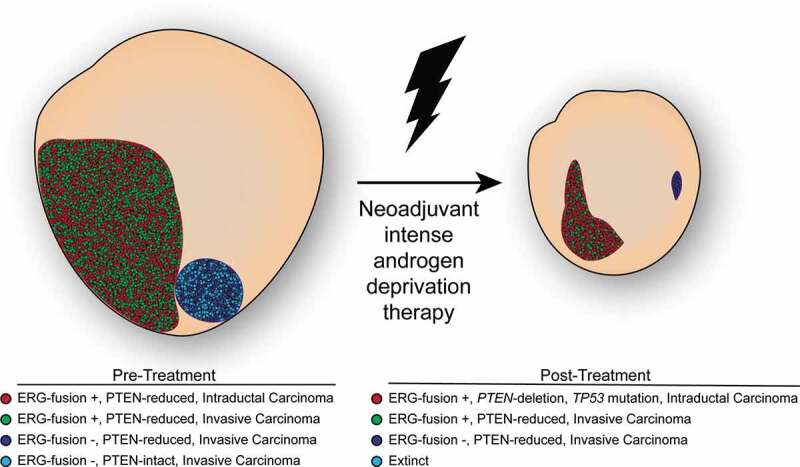
Molecular changes in prostate cancer following neoadjuvant intense androgen deprivation therapy. Scaled graphical depiction of prostate and prostate tumor changes following 6 months of neoadjuvant intense androgen deprivation therapy. The large lesion harbors an admixture of intraductal (red) and invasive (green) tumor cells that are positive for the transmembrane protein serine 2-ETS related gene (TMPRSS2:ERG) fusion protein and deficient for the phosphatase and tensin homolog (PTEN) tumor suppressor. The small lesion is comprised of an ERG-negative tumor which is itself an admixture of PTEN-reduced (dark blue) and PTEN-intact (light blue) tumor cells. Selected by treatment, the intraductal tumor expands substantially and contains an additional nonsense mutation to tumor protein p53 (TP53) although all three PTEN-reduced tumors survive.
While tumor heterogeneity has remained a well-known phenomenon across multiple cancer types, our understanding of the multifocality of prostate cancer and its implications for clinical management is only recently becoming clearer. Prostate cancer diagnosis and management tends to rely upon the status of the index lesion, which is usually the largest and/or more aggressive tumor component identified on imaging, biopsy or final pathology, believed to contribute the most to a patient’s outcome. However, advances in molecular pathology over the last decade have provided evidence to support the evolution of polytumors in prostate cancer, which further aligns with the independent probabilities of a high-risk patient developing prostate cancer twice.8,9
Although we were able to identify independent lesions by integrating advanced imaging and genomic dissection of targeted biopsies, the possibility remains that under routine clinical practice this heterogeneity would have been undersampled. As this patient experienced a two-thirds reduction in index lesion tumor volume, the neoadjuvant intense ADT was mostly effective. In our case, a standard biopsy that had targeted the larger tumor at baseline would most likely have still sampled the lesion exhibiting intrinsic resistance to therapy. The question remains how to further improve reduction in tumor volume, especially as TP53 and PTEN mutations are molecular characteristics of prostate cancer resistance to ADT.10
While it may not be possible to predict the final disposition of the more sensitive, smaller tumor if a precision workflow had directed the patient in our report away from neoadjuvant intense ADT, one can still examine the mechanisms of resistance following treatment. PTEN loss is amongst the most recurrent alterations driving tumorigenesis of aggressive prostate cancers.10 If the effect of neoadjuvant intense ADT is an overall debulking of the tumor to expose subclones driving resistance, future trials may include a precision neoadjuvant or adjuvant approach. In this case, PI 3-kinase pathway targeted agents such as ipatasertib (an inhibitor of AKT, also known as protein kinase B) might effectively eliminate residual tumor with biallelic PTEN deletions.
Nonetheless, work by our group and others examining the multifocality of prostate cancer highlights the importance of considering multiple targets in diagnosing and treating aggressive prostate cancer. While the mere presence of multiple independent tumors may inherently increase a patient’s recurrence risk,9 a more complete assessment can identify potential subclones that might mediate future cancer recurrence. In turn, having a catalog of mutations and matched precision agents offers clinicians a valuable tool for treating aggressive prostate cancer that might otherwise progress to lethal disease.
Funding Statement
This work was supported by the Prostate Cancer Foundation (Young Investigator Awards to S.W. and A.G.S.), the Department of Defense CDMRP Prostate Cancer Research Program (Early Investigator Research Award [W81XWH-19-1-0712] to S.W. and Impact Award [W81XWH-15-1-0710] to A.G.S.) and the Intramural Research Program of the NIH, National Cancer Institute.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
Manuscript preparation: S.W., A.G.S.
References
- 1.Wedge DC, Gundem G, Mitchell T, Woodcock DJ, Martincorena I, Ghori M, Zamora J, Butler A, Whitaker H, Kote-Jarai Z, et al. Sequencing of prostate cancers identifies new cancer genes, routes of progression and drug targets. Nat Genet. 2018;50(5):1–3. doi: 10.1038/s41588-018-0086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taplin M-E, Montgomery B, Logothetis CJ, Bubley GJ, Richie JP, Dalkin BL, Sanda MG, Davis JW, Loda M, True LD, et al. Intense androgen-deprivation therapy with abiraterone acetate plus leuprolide acetate in patients with localized high-risk prostate cancer: results of a randomized phase II neoadjuvant study. J Clin Oncol. 2014;32(33):3705–3715. doi: 10.1200/JCO.2013.53.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mostaghel EA, Nelson PS, Lange P, Lin DW, Taplin ME, Balk S, Ellis W, Kantoff P, Marck B, Tamae D, et al. Targeted androgen pathway suppression in localized prostate cancer: a pilot study. J Clin Oncol. 2014;32(3):229–237. doi: 10.1200/JCO.2012.48.6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montgomery B, Tretiakova MS, Joshua AM, Gleave ME, Fleshner N, Bubley GJ, Mostaghel EA, Chi KN, Lin DW, Sanda M, et al. Neoadjuvant enzalutamide prior to prostatectomy. Clin Cancer Res. 2017;23(9):2169–2176. doi: 10.1158/1078-0432.CCR-16-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKay RR, Montgomery B, Xie W, Zhang Z, Bubley GJ, Lin DW, Preston MA, Trinh Q-D, Chang P, Wagner AA, et al. Post prostatectomy outcomes of patients with high-risk prostate cancer treated with neoadjuvant androgen blockade. Prostate Cancer Prostatic Dis. 2018;21(3):364–372. doi: 10.1038/s41391-017-0009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salami SS, Hovelson DH, Kaplan JB, Mathieu R, Udager AM, Curci NE, Lee M, Plouffe KR, de la Vega LL, Susani M, et al. Transcriptomic heterogeneity in multifocal prostate cancer. JCI Insight. 2018;3(21). doi: 10.1172/jci.insight.123468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkinson S, Harmon SA, Terrigino NT, Karzai F, Pinto PA, Madan RA, VanderWeele DJ, Lake R, Atway R, Bright JR, et al. A case report of multiple primary prostate tumors with differential drug sensitivity. Nat Commun. 2020;11(1):837. doi: 10.1038/s41467-020-14657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindberg J, Klevebring D, Liu W, Neiman M, Xu J, Wiklund P, Wiklund F, Mills IG, Egevad L, Grönberg H, et al. Exome sequencing of prostate cancer supports the hypothesis of independent tumour origins. Eur Urol. 2013;63(2):347–353. doi: 10.1016/j.eururo.2012.03.050. [DOI] [PubMed] [Google Scholar]
- 9.Espiritu SMG, Liu LY, Rubanova Y, Bhandari V, Holgersen EM, Szyca LM, Fox NS, Chua MLK, Yamaguchi TN, Heisler LE, et al. The evolutionary landscape of localized prostate cancers drives clinical aggression. Cell. 2018;173(4):1003–1013. e15. doi: 10.1016/j.cell.2018.03.029. [DOI] [PubMed] [Google Scholar]
- 10.Robinson D, Van Allen E, Wu Y-M, Schultz N, Lonigro R, Mosquera J-M, Montgomery B, Taplin M-E, Pritchard C, Attard G, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


