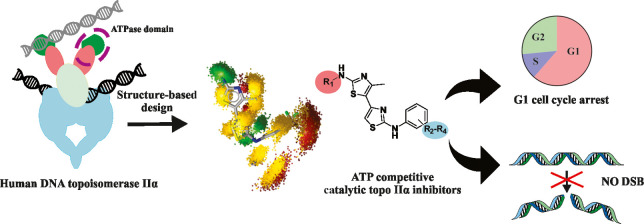
Abstract
Human type II topoisomerases, molecular motors that alter the DNA topology, are a major target of modern chemotherapy. Groups of catalytic inhibitors represent a new approach to overcome the known limitations of topoisomerase II poisons such as cardiotoxicity and induction of secondary tumors. Here, we present a class of substituted 4,5′-bithiazoles as catalytic inhibitors targeting the human DNA topoisomerase IIα. Based on a structural comparison of the ATPase domains of human and bacterial type II topoisomerase, a focused chemical library of 4,5′-bithiazoles was assembled and screened to identify compounds that better fit the topology of the human topo IIα adenosine 5′-triphosphate (ATP) binding site. Selected compounds showed inhibition of human topo IIα comparable to that of the etoposide topo II drug, revealing a new class of inhibitors targeting this molecular motor. Further investigations showed that compounds act as catalytic inhibitors via competitive ATP inhibition. We also confirmed binding to the truncated ATPase domain of topo IIα and modeled the inhibitor molecular recognition with molecular simulations and dynophore models. The compounds also displayed promising cytotoxicity against HepG2 and MCF-7 cell lines comparable to that of etoposide. In a more detailed study with the HepG2 cell line, there was no induction of DNA double-strand breaks (DSBs), and the compounds were able to reduce cell proliferation and stop the cell cycle mainly in the G1 phase. This confirms the mechanism of action of these compounds, which differs from topo II poisons also at the cellular level. Substituted 4,5′-bithiazoles appear to be a promising class for further development toward efficient and potentially safer cancer therapies exploiting the alternative topo II inhibition paradigm.
1. Introduction
Cancer represents one of the most pervasive diseases. The overall mechanisms behind cancer development depend on genetic predispositions and environmental influences and thus represents a major challenge for successful treatment. A solid foundation for understanding the conversion of healthy cells into cancer cells provides seminal work of Hanahan and Weinberg, in which they defined and discussed the “hallmarks of cancer”, common features that control multistage transformation leading to cancer cells.1,2 An established treatment approach in chemotherapy is to affect the mechanism of cell replication. Among the many enzymes involved in this complex process DNA topoisomerases,3 a broad family of molecular motors catalyze and enable various topological changes in the DNA molecule. Thus, they are inseparably linked with cell proliferation and cancer pathogenesis.1,2
Topoisomerases, in both bacterial and eukaryotic organisms, are divdied into two major groups labeled type I and type II topoisomerases. Type I topoisomerases catalyze the topological changes involving transient single-strand breaks of the DNA, while the type II counterparts catalyze the topological changes involving transient double-strand breaks (DSBs).4,5 Mammalian type II DNA topoisomerase can be found in two isoforms: α and β. The isoforms are encoded by different genes, share about 70% of amino acid sequence identity and have different levels of expression in the cells; α is expressed predominantly in proliferating cells, while the β isoform is expressed in equal parts in dormant and proliferating cells.6−9
Type II topoisomerases act through a complex catalytic cycle that starts with the binding of the first DNA segment (G segment) to the enzyme to catalyze its double-strand break (cleavage reaction) so that a second bound DNA segment (T segment) can pass through the break. The cycle ends with the relegation of the G segment and the release of both DNA segments. Topo II uses the chemical energy of adenosine 5′-triphosphate (ATP) hydrolysis, which transformed into molecular movement, further enables the action of this biological molecular motor.10−13
Topoisomerase inhibitors are roughly divided into two groups: topo II poisons and catalytic inhibitors,14−16 the first group being the more established. Topo II poisons stabilize the normally transient covalent complex between topo II and DNA, which leads to the formation and accumulation of DNA strand breaks that cause the cell to enter the mitotic phase of cell division and lead to cell death. Some examples of the popular topo II poisons used in clinical practice are etoposide (nonintercalator),17 doxorubicin (intercalator),18 and amsacrine (intercalator).19 The side effects of this group, in particular cardiotoxicity associated with anthracyclines18 and induction of secondary malignancies, which is more pronounced in the podophyllotoxin group of topo II poisons,20 have stimulated further drug design efforts in this field. Another reason for the development of new cancer drugs is drug resistance; many cancers can over time develop resistance to cancer drugs through DNA mutations, metabolic changes, and other mechanisms.21−24 Thus, catalytic inhibitors of the human topo IIα, which use alternative ways to tackle this enzyme, are now being actively researched.15 They can, for example, prevent the binding between DNA and enzyme (e.g., aclarubicin),25,26 inhibit DNA cleavage (e.g., merbarone),27 or inhibit ATP hydrolysis and trap the topo II in a closed clamp (e.g., ICRF-187 and ICRF-193).28,29 Finally, the catalytic inhibitors also act by inhibiting the binding of the ATP molecule such as various groups of purine-based inhibitors.30−33
Some of our earlier research activities were involved in the investigation of the last-mentioned approach. So far, we have characterized several classes of catalytic inhibitors targeting the ATP binding site, including triazin-2(1H)-ones,34,35 1,3,5-triazines,36 1H-pyrazolo[3,4]pyrimidines,37 9H-purines,37 and 1H-indazoles.38 In addition, we also investigated inhibitors of bacterial DNA gyrase. Starting from the binding mode of the natural product clorobiocin, we identified a series of 4′-methyl-N2-phenyl-[4,5′-bithiazole]-2,2′-diamines as inhibitors of DNA gyrase and determined for the representative compound 13 its binding mode in the ATP binding site using protein crystallography.39
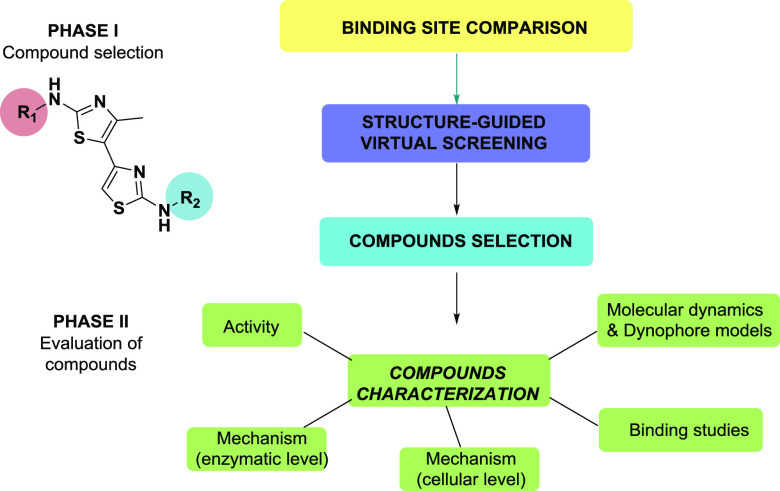
Building on our previous research, we report here on a new class of substituted 4,5′-bithiazoles as catalytic inhibitors of the human topo IIα with promising anticancer activity and thoroughly investigated inhibition mechanism. The outline of our work, which combines computational and experimental methods in a synergetic way, is presented in Figure 1.
Figure 1.
Workflow used in the identification and characterization of novel substituted 4,5′-bithiazoles as catalytic inhibitors of the human DNA topoisomerase IIα.
Compounds were designed by structural analysis and comparison of the ATPase domains of human and bacterial type II topoisomerases, followed by virtual screening of a compiled focused library of compounds. In this step, we looked for substitutions of the 4,5′-bithiazole scaffold, which would allow an optimal interaction with the ATP binding site of human topo IIα. The inhibition was first evaluated with a high-throughput screening (HTS) relaxation assay, and for several active compounds, the catalytic mechanism of topo IIα inhibition was investigated with functional assays and biophysical binding studies. Then, to study the dynamic properties that guide the inhibitor molecular recognition process, molecular dynamics (MD) simulations were coupled with dynamic pharmacophore (dynophore) calculations. We also performed cytotoxicity measurements on HepG2 and MCF-7 cancer cell lines, which was followed by an investigation of the mechanism of action at the cellular level.
2. Results and Discussion
2.1. Comparison of the ATP Binding Sites of Human and Bacterial Type II Topoisomerases and Virtual Screening of a Focused Chemical Library of Substituted 4,5′-Bithiazoles
In the first phase of our design, we aligned the ATPase domains of the human topo IIα (PDB: 1ZXM)40 and the bacterial DNA gyrase (PDB: 1EI1),41 both with bound AMP-PNP ligand, to determine the differences and similarities between the two ATP binding sites. Figure 2B shows the result of the alignment of the compared ATP binding sites and outlines some of the most important amino acids.
Figure 2.
(A) Experimental binding mode of the substituted 4,5′-bithiazole (13) in the DNA gyrase ATP binding site (PDB: 4DUH). (B) Structural alignment of the ATP binding sites of human topo IIα (PDB: 1ZXM) and bacterial DNA gyrase (PDB: 1EI1) located on their corresponding ATPase domains with AMP-PNP ligand. Identical or similar residues are depicted in orange, while residues dissimilar site between the enzymes are shown in red and italics. The Pro126/Arg136 residue pair located outside of the ATP pocket is shown in blue. (C) Structure-based design of the substituted 4,5′-bithiazoles analogues by virtual screening of a focused library of compounds. (Left) Predicted binding mode of the initial 4,5′-bithiazole 13 in the human topo IIα ATP binding site (PBD: 1ZXM). (Right) Docked binding mode of a favorable 4,5′-bithiazole compound 1 from a focused chemical library selected for experimental evaluation.
Both ATP active sites contain an amino acid that interacts with the purine ring of the AMP-PNP ligand. This corresponds to Asn120 in the case of topo IIα and Asp73 for the DNA gyrase, both acting as H-bond acceptors. The interaction of these residues with ATP is also partly mediated by water. Residue pairs with similar roles in ATP interactions include Asn91/Asn46, Lys378/Lys337, and the Gln367/Gln335 pairs. The latter two pairs of residues could help to ensure the correct position of the γ-phosphate group during ATP hydrolysis. The structural analysis also showed that the human topo IIα contains two serine residues—Ser149 and Ser148—which anchor the sugar moiety, while only Gly102 plays this role in its bacterial counterpart. There is also a difference between Asn150 and Lys103 residue pair interacting in the phosphate part of the ATP site. Another critical difference, which should be outlined as it will become important in our design steps, was found between the Arg136 found in the DNA gyrase, while the human topo IIα has a rigid Pro126 at this position (Figure 2B in blue).40,41 In Table S2, the similarities and differences in the interaction patterns between the AMP-PNP ligand in the ATP binding sites in both type II topoisomerase are also presented in the two-dimensional (2D) representation.
At the beginning of the design activities, we took the substituted 4,5′-bithiazole compound 13 from its DNA gyrase co-crystal structure (PDB: 4DUH) depicted in Figure 2A and docked it to the ATP active site of the human topo II ATPase domain (PDB: 1ZXM) (Figure 2C). In addition, we also docked compound 13 to PDB 1EI1 (DNA gyrase with bound AMP-PNP) and PDB 1KZN (DNA gyrase structure used in the original virtual screening that led to the discovery of compound 13) to determine whether our docking protocol could replicate the experimental determined conformation as well as to detect any differences of the ligand conformation in different DNA gyrase structures (Figure S2). The obtained binding positions were also analyzed to determine the ligand–target interaction patterns.
In the human topo IIα, the predicted binding mode of 4,5′-bithiazole 13 resembled the conformation of a native ATP-based ligand (Figure 2C). The N-terminal group on the first thiazole ring interacted with Asn120 acting as a H-bond acceptor, analogous to the Asp73 interaction in DNA gyrase, while the far end of the molecule positioned itself around the position of the first AMP-PNP phosphate group. In the DNA gyrase co-crystal structure with compound 13 (PDB: 4DUH), this compound retained the conformation of the aminothiazole head by interacting with the Asn120 analogue of the DNA gyrase, Asp73 (Figure 2A). Here, however, the remainder of the molecule carrying the R2 carboxyl group was oriented outside the binding pocket that normally interacts with the phosphates of ATP, and it was preferred to form H-bonds with the Arg136 moiety (Figure 2A), though an ionic interaction is also a possible interaction between these two interacting partners. When we docked this compound into the bacterial DNA gyrase ATP binding site, where the flexible loop Gly97-Ser108 was not visible (PDB: 1EI1), no major differences in overall placement were observed, and the docked pose was fully comparable to the experimental pose (Figure S2).
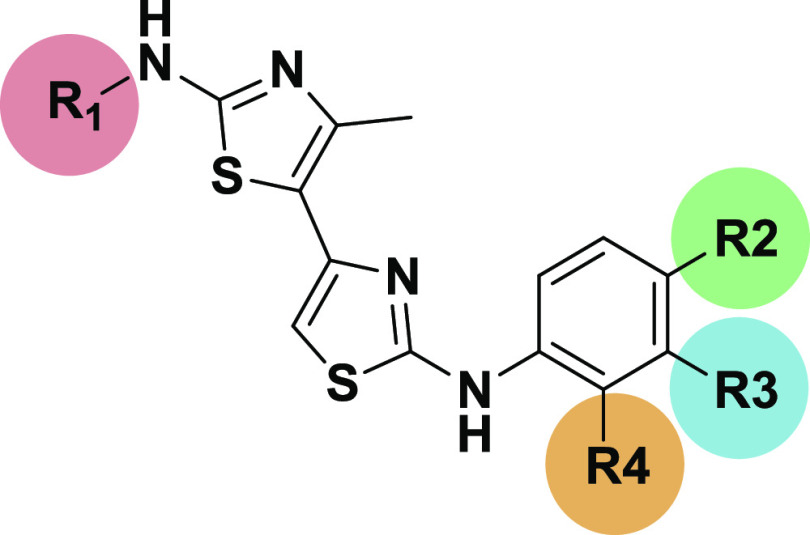
The results of the docking thus indicated a potential difference in the binding of the bithiazole class to the bacterial vs the human type II topoisomerase. To search for compounds that better fit the ATP binding site of human topo IIα, we compiled a focused chemical library of available 4,5′-substituted bithiazoles using an E-molecule database.42 First, we started a substructure search with N-(thiazol-2-yl)acetamide as search query and retrieved 3349 compounds. We then filtered out and visually scanned the bithiazoles, removed fragments and oversized compounds, and then further narrowed down the selection using our chemical intuition. We focused primarily on the modifications of its R1 2′amine group and the R2–R4 substitution of the phenyl ring, which would allow additional interactions of these compounds with the amino acids in the phosphate part of the topo IIα ATP binding site (Figure 2C). Finally, 275 bithiazole compounds from the focused library were docked to the active site of the human ATPase domain.
The predicted poses were manually analyzed using LigandScout to verify the predicted interactions of these compounds with the ATP binding site of topo IIα. We focused on the compounds that showed a favorable hydrogen-bonding interaction of the R1 nitrogen with Asn120. The presence of this hydrogen bond was evaluated by deriving three-dimensional (3D) structure-based pharmacophores for the docking poses of the focused bithaizole library. The second interaction criterion was the presence of detectable H-bonds between the R2 and R4 substituents on the benzene moiety and the “ribose sugar part” (Ser148, Ser149, and Asn150 binding site residues) and/or the “triphosphate part” (residues Asn91, Ala167, and Lys168) of the ATP binding site. In addition, we checked for the presence of hydrophobic interactions with the binding site residues Ile125, Ile141, and Phe142 to outline a few. An example of a docking mode for a hit compound 1 from the focused library is shown in Figure 2C. After the analysis and selection procedure, substituted 4,5′-bithiazole analogues 1–14 (see Table S1, Supporting Information), which showed the most promising interactions, were selected for the experimental evaluation of topo IIα inhibition.
2.2. In Vitro HTS Relaxation Assay of the Selected Compounds and Initial Structure–Activity Relationship (SAR) Data
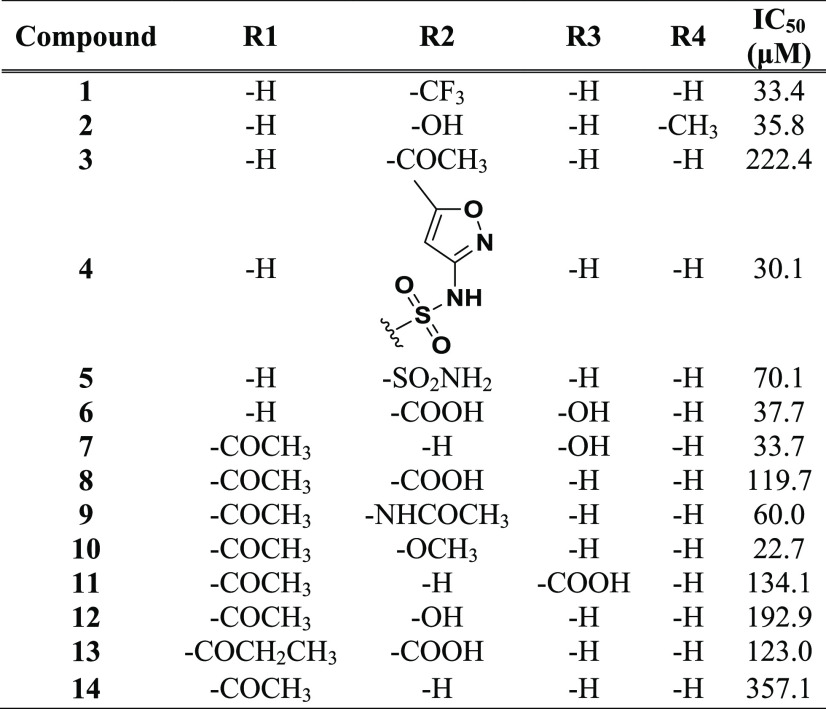
Selected 14-substituted 4,5′-bithiazoles 1–14 were experimentally assayed in a standard high-throughput screening (HTS) relaxation assay.43 Etoposide was used as a control compound to validate the assay; our experimentally determined half-maximal inhibitory concentration (IC50) value of 41.6 μM compared well with the value of 60.3 μM given in the literature.44 The results of the initial HTS relaxation assay are presented in Table 1. It was gratifying to observe that many of our compounds exhibited comparable topo IIα inhibitory activity to the reference etoposide drug, establishing the substituted 4,5′-bithiazoles, to the best of our knowledge, as a newly discovered class of human topo IIα inhibitors. In particular, compounds 1, 2, 4, 6, 7, and 10 showed comparable inhibition to etoposide, with IC50 values in the activity range between 30 and 50 μM, which corresponds to the potency of many topo IIα compound classes reported in the literature.14,15,45
Table 1. IC50 Values of the Selected Substituted 4,5′-Bithiazoles 1–14 Determined in the HTS Screening Relaxation Assay.
The results of the HTS inhibition test assay provided further insight into the structure–activity relationship (SAR) data of the substituted 4,5′-bithiazole class. Both, compounds 1–6 with nonacetylated free amine and 7–14 with included R1-acetylated amino group, exhibited comparable inhibitory activity (Table 1), indicating some flexibility of these compounds when binding to the target. This is probably due to the fact that the ATP binding pocket of human topo IIα provides enough space around the Asn120 residue. An additional increase in R1 substituent size such as the ethyl substituent in compound 13 did not lead to the improved topo IIα inhibitory activity. Molecular docking also indicated a water-mediated H-binding interaction between Asn91 and the first thiazole heterocycle as an important aspect of the molecular recognition of the ligand (Figure 2C).
Selected R2–R4 substituents on the terminal benzene moiety, which is bound to the 4,5′-bithiazoles via an amino linker, showed a certain chemical diversity. This is due to the rather spacious lower part of the ATP pocket, which accommodates three phosphate groups and contains various interaction points with different properties. The identification of different inhibitors at this point provides a beneficial basis for further optimization both from a chemical and physical point of view, since we can choose a substituent that ensures better absorption or that can be more easily incorporated into a future pharmaceutical formulation. The compounds also possessed favorable druglike properties that made them applicable for further development: calculated log P values below 5 and topological surface areas (TPSAs) below 120.
In further analysis of the docking results, most of the additional interactions of the R2–R4 substituents of the active compounds were associated with the formation of new interactions with Ser148, Ser149, and Asn150 residues in the “ribose sugar” part of the ATP binding site and with Ala167 and Lys168 residues in its “triphosphate” part. In addition, the bithiazole backbone as well as the phenyl ring showed hydrophobic interactions with Ile125, Ile141, and Phe142, as displayed by the yellow dotted lines in Figure 2C for compound 1 (see also the predicted mode of compound 9; Figure S3, Supporting Information). We also compared the inhibition results for the tested bithiazoles that were favored in our DNA gyrase design campaign.39 The compounds generally showed comparable micromolar inhibition of both topoisomerases (Table S3).
2.3. Investigation of the Inhibitory Mechanism
The promising results of the HTS relaxation assay have encouraged us to further investigate the inhibition mechanism of the discovered 4,5′-bithiazoles. Due to the complex catalytic cycle associated with the function of the topo IIα molecular motor,10 only additional functional and biophysical assays can provide the necessary data for a deeper understanding of the specific inhibition mechanism of these compounds. For further assays, we selected the active R2 CF3-substituted compound 1 with free amino group and the N-acetylated compound 9 with R2 carboxyl group from the second subgroup of active compounds. In addition to the measured inhibition activities, favorable physicochemical properties, especially solubility, played a decisive role in the selection of these compounds for further analysis.
2.3.1. Substituted 4,5′-Bithiazoles Act as Catalytic Inhibitors of the Human DNA Topoisomerase IIα
To investigate whether 4,5′-bithiazoles can inhibit DNA decatenation catalyzed by human topo IIα, we performed the kinetoplast (kDNA) decatenation assay for selected compounds 1, 9, and etoposide as the control compound. The results of the decatenation assay are shown in Figure 3A.
Figure 3.
Substituted 4,5′-bithiazoles act as catalytic inhibitors of human DNA topoisomerase IIα. (A) Compounds 1 and 9 inhibited topo IIα-mediated DNA decatenation. Control reactions had no compounds and were done in the presence of topo IIα (+) and in the absence of topo IIα (−). Etoposide was used as the standard compound. (B) Same as (A), except topo IIβ was used instead of topo IIα. (C) Results of the topo IIα-mediated cleavage assay for etoposide and compounds 1 and 9 at four concentrations. Etoposide was again used as the standard compound.
Both compounds significantly inhibited the decatenation of kDNA in a concentration-dependent manner. Etoposide showed inhibition of human topo IIα decatenation activity, comparable to the literature data,46 with significant inhibition levels in the concentration range of 500 and 125 μM, with inhibition of 95.6 and 67.2%, respectively. The free R1-amine bithaizole 1 was shown to be more effective in inhibiting decatenation, with complete inhibition observed at 125 and 500 μM and 19.1% of inhibition at 31.5 μM. Compound 9 was also found to be an active inhibitor, although to some extent less active than compound 1. These results confirmed the significant influence of the 4,5′-bithiazole class on the catalytic activity of human topo IIα, with compound 1 showing a higher inhibitory activity of human topo IIα-catalyzed decatenation compared to the etoposide standard. In addition, a comparable degree of inhibition was also determined for compound 10, for which the decatenation was performed at a later stage, as it showed strong cytotoxicity to cancer cell lines, as will be shown in the following chapter (see Tables S5 and S6 and Figures S5 and S6 for further details).
We were also interested in whether compounds can act on both isoforms of the human topo II. Thus, we performed human topo IIβ decatenation assays for compounds 1 and 9. The results showed that compound 1 completely inhibited the human topo IIβ at 500 and 125 μM concentrations and showed an inhibition of 20.8% at 31.5 μM. Compound 9 showed 88.6% of inhibition at 500 μM and 73.9% of inhibition at 125 μM (Figure 3B). This was comparable to the inhibition observed with the topo IIα isoform. Etoposide showed a certain selectivity against human topo IIα vs human topo IIβ isoforms, in accordance with the reported experiments47,48 (see also Tables S5 and S6 for more details).
In dealing with the catalytic topo II inhibitors, including those targeting the ATP binding site, it has been found that inhibition of both α and β isoforms could be a desirable property of the compounds.30 In this respect, the catalytic inhibitors differ from the topo II poisons where selectivity for the topo IIα isoform is preferred.30,35,49 Recent experiments with mice have demonstrated that the topo IIα but not IIβ is essential for cell proliferation,50,51 but further experiments with siRNA showed that the topo IIβ isoform can compensate for the depletion of topo IIα in certain cell lines.52 Therefore, partial compensation should be considered in the development of catalytic inhibitors, and inhibition of both topo II isoforms could be beneficial.30 Some researchers also suggested that topoisomerase IIβ could be used as a cancer target all by itself when targeting both nonproliferative cells and cancer stem cells, and it has been proposed as a target to counteract glioblastoma cell resistance in glioblastoma therapy.53
To determine whether compounds act as catalytic inhibitors, a cleavage assay was next performed for compounds 1 and 9. After treatment with human topo IIα, the negatively supercoiled plasmid was incubated with four different concentrations of the investigated compounds 1 and 9 and the etoposide control. The results obtained (Figure 3C) clearly show the poison activity of etoposide, with the amount of linear DNA increasing as the concentration of the drug increases. In contrast, the same titration with compounds 1 and 9 did not reveal a significant amount of linear DNA above the background level, indicating that they act as catalytic inhibitors (see Table S7 and Figure S7, Supporting Information).
2.3.3. Substituted 4,5′-Bithiazoles Act as ATP Competitive Catalytic Inhibitors of Human DNA Topoisomerase IIα
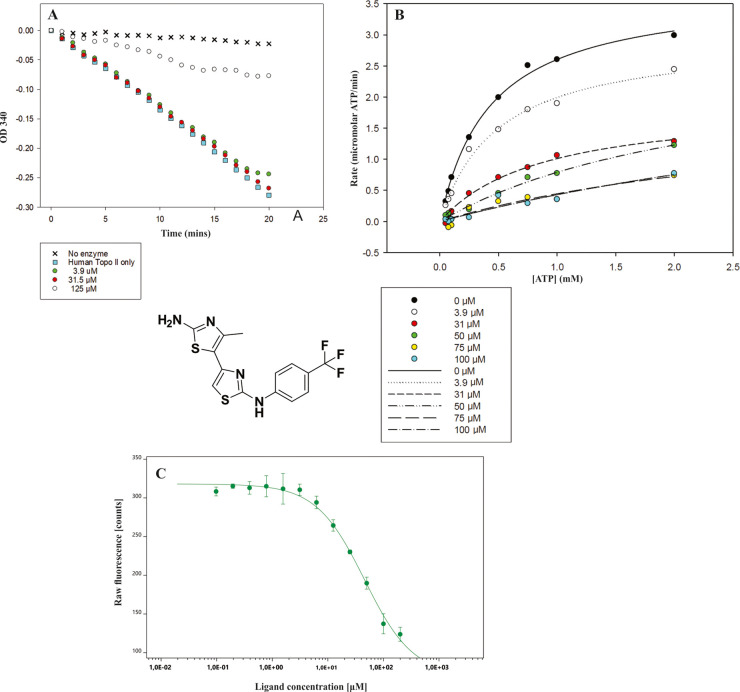
To investigate whether our compounds can inhibit ATP hydrolysis catalyzed by human topo IIα, ATP hydrolysis assay was performed for compound 1. This assay is coupled to the oxidation of reduced nicotinamide adenine dinucleotide (NADH) monitored by the decrease in optical density at 340 nm (OD340). Etoposide was used as a control compound. As presented in Figure 4A, compound 1 successfully inhibited 75% (first parallel) or 85% (second parallel) of the ATP hydrolysis at 125 μM. For comparison, etoposide inhibited ATP hydrolysis by 70% (first parallel) or 63% (second parallel) at this concentration. Further data on this assay are provided in the Supporting Information (Figures S8 and S9 and Table S8).
Figure 4.
Substituted 4,5′-bithiazoles act as ATP competitive catalytic inhibitors of human DNA topoisomerase IIα. (A) Results of the topo IIα ATPase assay. Compound 1 almost fully inhibited the human topo IIα-mediated ATP hydrolysis at 125 μM. The graph shown is representative of two independent experiments. (B) Results of the competitive topo IIα ATPase assay. (C) Microscale thermophoresis (MST) experimental binding curve of compound 1. In the MST experiment, we kept the concentration of the human topo IIα ATPase domain labeled molecule constant, while the concentration of unlabeled compound 1 was varied between 0.03 and 1000 μM.
We then performed the competitive ATPase assay for compound 1 to investigate how our compound class affects ATP hydrolysis as a function of ATP concentration. Figure 4B depicts the observed rates of ATP hydrolysis plotted against the increased ATP concentration for different concentrations of compound 1. From the graph obtained, it can be seen that the rate of ATP hydrolysis was significantly faster at lower concentrations of inhibitor 1 and was then slowed down as the concentration of the compound increased. This showed that compound 1 has a significant concentration-dependent effect on the ATP hydrolysis rate. In addition, we also calculated the IC50 values of compound 1 compared to different concentrations of ATP. These results are presented in Table 2, and a significant decrease in IC50 values is observed with decreasing ATP concentrations. This observed behavior corresponds to the targeted ATP-competing inhibition mode of the substituted 4,5-bithiazole class. Additional data on this assay are provided in the Supporting Information (Table S9).
Table 2. IC50 Values for Compound 1 at Different Concentrations of ATP with the R2 Values for the Generated Fitsa.
| c(ATP) | 2 | 1 | 0.75 | 0.5 | 0.25 | 0.1 | 0.075 | 0.05 | 0.025 |
|---|---|---|---|---|---|---|---|---|---|
| IC50 (μM) | 25.4 | 21.6 | 17.8 | 15.2 | 18.3 | 9.4 | 14.8 | 8.8 | 3.8 |
| R2 | 0.981 | 0.971 | 0.972 | 0.985 | 0.992 | 0.974 | 0.958 | 0.929 | 0.474 |
The percent activity values at each ATP concentration were calculated by dividing the activity at each concentration of inhibitor by the activity in the absence of inhibitor then multiplied by 100. This was done for each of the different ATP concentrations. These percent activities were then plotted against inhibitor concentration. Curves were fitted using the equation y = y0 – (a e(−bx)), and then IC50 values were calculated from the generated values of yo, a and b.
Since DNA topoisomerase IIα is a complex molecular motor, we also investigated the binding of inhibitor 1 to the isolated human topo IIα ATPase domain using a novel microscale thermophoresis (MST) technique. MST is a versatile technique for the characterization of intermolecular interactions between, among others, biomolecules and small molecules. It quantifies biomolecular interactions based on the physical principle of thermophoresis, the direct movement of molecules in the temperature gradient. The thermophoretic movement of the labeled protein in complex with a selected inhibitor was measured by monitoring the fluorescence distribution within capillary.54−56 MST experiments performed in three independent runs showed concentration-dependent binding to the ATPase domain and yielded Kd = 50.6 ± 7.6 μM for compound 1. The binding curve of compound 1 is presented in Figure 4C. These results confirmed that substituted 4,5′-bithiazoles bind to the truncated human topo IIα ATPase domain, where the ATP binding site is located. This observation coupled with the results of the competitive ATPase assay provides ample evidence that the mode of inhibition occurs via binding to the ATP binding site.
2.3.4. Analysis of the Proposed 4,5-Bithiazoles Interactions in the ATP Active Site Using Molecular Dynamics (MD) and Dynophore Analysis
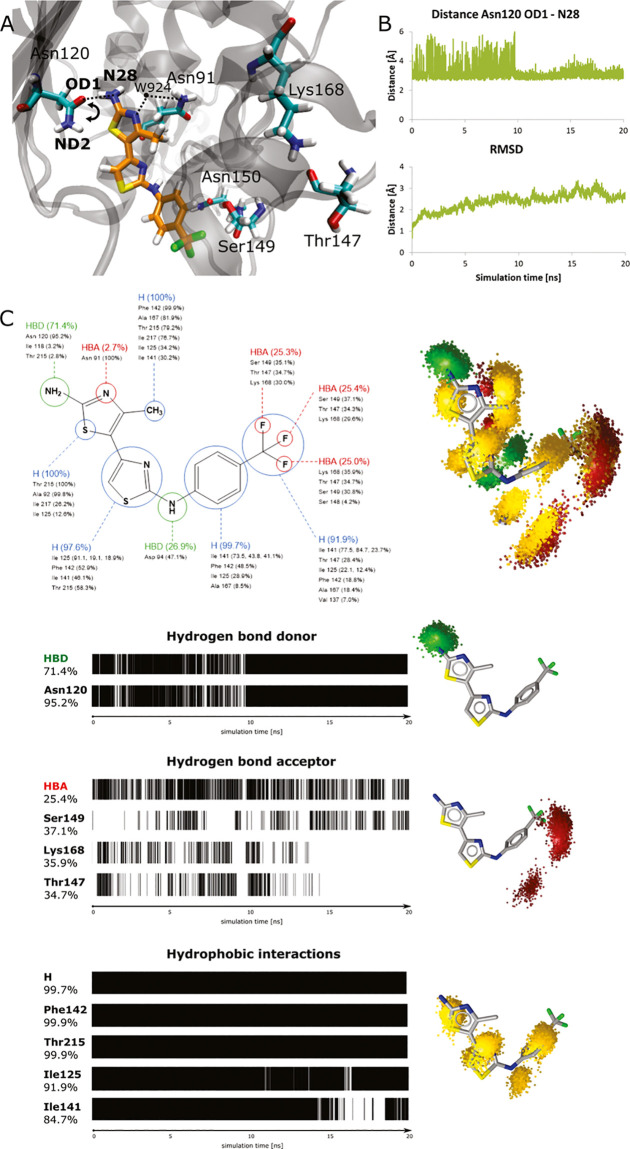
Molecular docking experiments can only provide a static binding pose prediction of the target–ligand complex. Therefore, the application of molecular dynamics (MD) simulations is necessary to obtain further information about the dynamic behavior and properties of a bound compound that guide molecular recognition. As our performed experiments indicated that the ATP binding site located on the human topo IIα ATPase domain serves as the binding site of these compounds, we have further initiated MD simulations57 using the docking binding mode of the active 4,5-bithiazole compound 1. It is important to mention that no complex structure of a small-molecule inhibitor bound to the human topo IIα ATP binding site has been reported to date.
Using the CHARMM-GUI platform,58 we constructed a solvated topo IIα—compound 1 system, which was then equilibrated and simulated in a molecular dynamics simulation. The animations of the MD simulation are available in the Supporting Information, and a representative MD snapshot is depicted in Figure 5A. The bithazole and the amino group of compound 1 were modeled in their deprotonated states, taking into account the available pKa experimental data.59 It should be also noted that ligands’ pKa values can significantly change when compounds bind to the protein, and this can influence the compound protonation pattern.60 In a first step, we evaluated the stability of the docked binding modes for each compound in the topo IIα ATP binding site. The generated conformations proved to be stable overall, with a root-mean-square deviation (RMSD) value of 2.4 ± 0.4 Å (see Figure 5B for the RMSD time-dependent graph).
Figure 5.
(A) Representative conformation of compound 1 in the ATP binding site of the human topo IIα during MD simulation. The arrow indicates an observed rotation of the Asn120 side chain during the simulation. (B) (Top) Time-dependent graph for the distance between the OD1 atom of Asn120 and the N28 nitrogen of compound 1; (bottom) time-dependent RMSD graph for compound 1. (C) Dynophore interaction pattern for compound 1: (left) persistence of pharmacophore elements throughout the MD with listed interacting amino acids; (right) generated dynophore model represented with pharmacophoric features. Hydrogen-bond acceptors, HBAs (red); hydrogen-bond donor, HBD (green); and hydrophobic region, H (yellow).
Next, we analyzed the interactions proposed by the docking of compound 1. The main hydrogen bonding interaction between the amide oxygen of Asn120 and the 4,5-bithiazole core nitrogen N28 was maintained throughout the MD simulation with an average distance of 3.1 ± 0.5 Å. Although we observed the rotation of the side chain during the first part of the MD simulation, the Asn120-mediated H-bond interaction stabilized later and acted as a main anchor of compound 1. This was as often observed in our previous studies and MD simulations of other chemical classes we developed (Figure 5B).34 Then, we investigated several residues that were considered important in the molecular docking experiments. An average distance between compound 1 and Asn91 of 5.5 ± 0.9 Å confirmed that this interaction is primarily water-mediated, as suggested by the docking with two crystal waters taken into account. Next, the average values of ligand interactions with Ser149 5.3 ± 1.3 Å and Asn150 6.7 ± 1.4 Å from the “triphosphate” and “ribose sugar” portions of the ATP pocket were found to be more dynamic (Figure S4). These interactions primarily reflected the interaction of the CF3 group of the docked compound 1 with the ATP binding site.
Since we wanted to further rationalize this observation, we upgraded the geometric analysis of the MD trajectory by generating a dynophore model. This is a powerful method for the analysis of MD trajectories using structure-based pharmacophore models developed at Freie Universität Berlin.61−63 Dynophores should complement the information of the classical pharmacophores, since they contain information of all pharmacophores generated for each frame of the MD simulation.
The calculated dynophore pattern shown in Figure 5C confirmed all predicted interactions. Hydrogen bonding with the bithiazole scaffold was maintained for 71.4% of the MD simulation time, 95.2% of which was with the residue Asn120, which is consistent with our design hypothesis that this residue serves as an anchor in the ATP pocket. Hydrophobic interactions between the core bithiazole scaffold, its methyl substituent, and the phenyl ring of compound 1 on one side, and the ATP binding pocket (e.g. residues Ile125, Phe142, Ile141, and Thr215) on the other, were present practically 100% of the simulation time. Since such interactions cannot be geometrically analyzed as easily as hydrogen bonds, dynophore analysis proved here to be a very valuable tool. In this respect, the obtained binding mode used in virtual screening was additionally validated.
Finally, the dynophore model also provided a rational interpretation of the influence of the CF3 substituent on the phenyl ring of compound 1 for the topo II binding, as shown in Figure 5C. Each fluorine atom comprising the CF3 group forms interactions about one-third of the MD simulation time with residues Thr147, Ser149, and Lys168, as suggested by the initial docking. The CF3 substituent is not static; it rotates and interacts with various amino acids in the ATP binding site, which corresponds to the dynamic properties of this group. Although the simulation MD run has been relatively long, it still does not provide comprehensive coverage of the conformational space for a clear quantitative ligand stability assessment. In the absence of a crystal structure, however, it provides valuable initial information on ligand dynamics, which can be also employed in the ligand optimization.64
2.4. Activity of Substituted 4,5′-Bithiazoles on Human Cancer Cell Lines
2.4.1. In Vitro Cytotoxicity on MCF-7 and HepG2 Cancer Cell Lines
We have determined the cytotoxicity of the compounds using the human breast cancer MCF-7 and human hepatocellular carcinoma HepG2 cancer cell lines by standard 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay.65 The two selected human cancer cell lines are representative and well-established systems for the cell-based evaluation of potential cancer drugs. Etoposide was again used as a positive control (PC). We performed initial screening of compounds 1–14 at one concentration. Depending on the solubility of the compounds in the cell growth media, 200 μM concentration was used for compounds 1, 4, 5, 7, 8, 11, 13, and 14; 100 μM for compounds 2, 6, 9, 10, and 12; and 50 μM for compound 3. Exponentially growing cells were exposed to all compounds for 24 h, and the results are shown in Figure 6A. The most effective compounds reduced cell viability by more than 80% compared to the untreated control in both cell lines.
Figure 6.
Cytotoxic activity of substituted 4,5′-bithiazoles in HepG2 and MCF-7 cells. (A) Results of the initial cytotoxicity screening using the MTS assay at compound concentrations of 200 μM for compounds 1, 4, 5, 7, 8, 11, 13, and 14; 100 μM for compounds 2, 6, 9, 10, and 12; and 50 μM for compound 3 for 24 h. We used dimethyl sulfoxide (DMSO) (0.5%) as solvent control along with etoposide (200 μM) as the positive control (PC). Analysis of variance (ANOVA) method was used to evaluated significant differences between treated cells and solvent control (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.001). (B) Dose–response curves for active substituted 4,5′-bithiazoles at 24 or 72 h treatment on the HepG2 and MCF-7 cells. Experiments were performed in five parallels and repeated three times independently, and standard deviation (SD) values were calculated.
In addition, EC50 values were determined for the most potent compounds by testing their cytotoxicity at different concentrations to observe the dose–response curves for 24 h (short-term) and 72 h (long-term) exposure treatment. We selected the most potent compounds from the initial screening, namely, compounds 1, 7, 9, 10, and 14 together with etoposide as the positive control. Unfortunately, we could not determine the EC50 value of compound 3 due to its low solubility in the cell growth medium. The obtained dose–response curves are shown in Figure 6B, and the EC50 values obtained after 72 h exposure are listed in Table 3. The EC50 values, which were also determined after 24 h exposure, are listed in Table S10 (Supporting Information). The compounds demonstrated a higher cytotoxic activity after a longer exposure time, which was expected. For comparison, we also determined the EC50 values for etoposide and confirmed that the EC50 values after 72 h were in the same range as for our compounds and in accordance with the reported data.66,67 The compounds were generally more cytotoxic for the MCF-7 cell line compared to HepG2 cells. The most potent compound was compound 10 with an EC50 value of 4.5 μM on MCF-7 cell line after 72 h treatment. For comparison, compound 10 was also the strongest compound most potent in the enzyme relaxation assay as an inhibitor of human topo IIα. Compounds 1, 7, and 9 were also potent topo IIα inhibitors as well as good cytotoxic agents with EC50 values of 59.5, 6.6, and 13.8 μM in MCF-7 cells, respectively. Compound 14 was less potent in the enzymatic assay (IC50 = 357.1 μM) but showed promising cytotoxic activity. The observation could be rationalized by a better cell permeability due to the unsubstituted phenyl substituent. Compounds 6, 8, and 11 with the carboxylic acid present on the phenyl ring had no or only minor effects on the tested cell lines. In addition, in the HepG2 cancer cell line at 72 h exposure, the EC50 values of the tested compounds ranged between 15 and 50 μM, and several compounds had comparable cytotoxic activity compared to etoposide (Table 3).
Table 3. Determined Cytotoxicity of Substituted 4,5′-Bithiazoles Represented by EC50 Values and Value Ranges on HepG2 and MCF-7 Cancer Cell Lines.
| compound | EC50 (μM) MCF-7 (72 h) | EC50 (μM) HepG2 (72 h) |
|---|---|---|
| 1 | 59.5 (58.1–59.3) | 46.8 (47.1–49.3) |
| 7 | 6.6 (4.6–7.6) | 32.3 (29.7–37.2) |
| 9 | 13.8 (9.7–15.9) | 23.5 (17.1–32.1) |
| 10 | 4.5 (3.4–5.9) | 14.6 (12.7–19.5) |
| 14 | 7.5 (5.2–6.7) | 28.2 (20.5–29.9) |
| etoposide | 12.6 (10.1–15.9) | 25.8 (15.1–35.3) |
2.4.2. Investigation of the Effect of 4,5′-Bithiazoles on Cell Cycle, Cell Proliferation, and Induction of DNA Double-Stranded Breaks
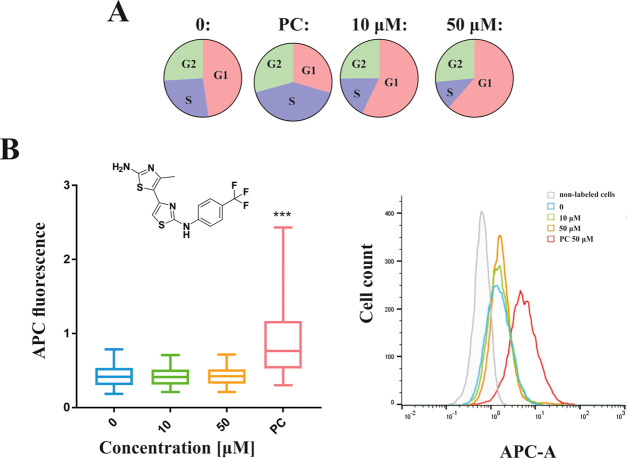
Encouraged by the promising cytotoxicity provided by the MTS assay, we have conducted further studies to gain more insights into the mode of action of selected 4,5′-bithiazoles on cancer cells. To first investigate how our class of compounds affects the cell cycle and the percentage of proliferating cells, we performed a flow cytometry analysis on HepG2 cells after 24 h of treatment. For this step, we selected the in vitro most characterized substituted 4,5′-bithiazole 1. HepG2 cells were selected based on our long experience with this system, plus compound 1 had HepG2 cytotoxicity comparable to the more active compounds, and as well as to etoposide, which was used as the positive control.
While DNA damage can trigger a cell cycle arrest in the G1, S, or G2 phase depending on the time of its occurrence, topo II poisons seem to mainly cause the G2 phase arrest.68,69 On the other hand, catalytic inhibitors do not directly damage DNA, and several such molecules are thought to cause G1 arrest.70−72 In line with this, compound 1 induced the cell cycle arrest in the G1 phase. More precisely, compound 1 at the concentration of 50 μM significantly increased the proportion of cells in the G1 phase of the cell cycle (60.6%) and decreased their proportion in the S phase (17.5%) compared to solvent control (46.7% in the G1 phase and 25.9% in the S phase). At a lower concentration of 10 μM, the changes in the cell cycle were less pronounced—56.5% of the cells were in the G1 phase and 17.5% in the S phase. The results of this experiment for compound 1 are shown in Figure 7A as a pie chart and in the Supporting Information as the percentage of cells accumulated in each phase (Table S11). On the contrary, etoposide (PC) decreased the proportion of cells in the G1 phase (33.1%) and increased the proportion in the S phase (44.0%) of the cell cycle compared to the solvent control. This is in accordance with the literature data that etoposide induces cell cycle arrest in the late S phase and early G2 phase.73 These results confirmed that the presented compounds act at the cellular level via a different mechanism than topo II poisons. Representative histograms for the cell cycle analysis are further described in Figure S10A.
Figure 7.
Cell cycle analysis and induction of the DNA double-stand breaks (DSBs) by the substituted 4,5′-bithiazole compound 1. (A) Percent of HepG2 cells in certain phases of cell cycle after 24 h treatment to compound 1 at 50 and 10 μM in comparison to the solvent control (0). Etoposide (50 μM) was used as a positive control (PC). (B) Results of the analyses of DNA double-strand break induction assessed by γ-H2AX assay. (Left) The results are presented as the distribution of fluorescent signals of individual cells. Data are presented as quantile box plots with the 25th and 75th percentiles (edges of the box), the median value (line through the box), and the bars representing 95% confidence intervals. Ten thousand events were recorded in each sample, and three independent experiments were performed. Significant difference between solvent control (0.5% DMSO; indicated by 0) and treated cells is shown by ***p < 0.001. A positive control (PC; 50 μM etoposide) was included in each parallel. (Right) Representative histograms for nonlabeled cells, vehicle control (0), compound 1 (10 and 50 μM), and etoposide (50 μM).
We also investigated the effect of compound 1 on the proliferation of HepG2 cells labeled with antibodies against the Ki67 cell biomarker and analyzed them by flow cytometry. The expression of the human Ki67 protein is associated with cell proliferation because the protein is present in all active phases of the cell cycle (G1, S, G2, and M) and is not present in resting (G0) cells.74 We treated HepG2 cells with 10 and 50 μM of compound 1 or 50 μM etoposide as a positive control for 24 h treatment. In cells treated with 50 μM of compound 1, we observed significantly reduced cell proliferation (65.1%) comparable to that observed after treatment with 50 μM etoposide (57.6%). The results are additionally shown graphically in Figure S10B.
Finally, we analyzed the induction of DNA double-strand breaks (DSBs) in HepG2 cells after exposure to compound 1 or etoposide. The presence of DSBs was analyzed by flow cytometry by measuring the fluorescence signals of individual cells, indirectly by detecting γH2AX foci. One form of DNA damage that can occur are double-strand breaks (DSBs), which can lead to chromosome breaks and rearrangement.68 DNA-DSB are associated with severe side effects observed by topo II poisons, such as cardiotoxicity and induction of secondary malignancies.18,20,75 The phosphorylated H2AX histones (γH2AX) are used as biomarkers for DSBs and DNA damage while accumulating and forming foci at sites that correlate with DSBs in a 1:1 ratio.76,77 We have performed this assay to confirm, also on the cellular level, besides the previously provided conformation on the in vitro level by the topo IIα-mediated cleavage assay that this class of compounds does not act as topo II poisons, but as catalytic inhibitors. Exposure to compound 1 at concentrations of 10 and 50 μM did not induce an increase in DNA-DSB formation, while etoposide (50 μM) induced a significant increase in DNA-DSBs (Figure 7B). These results demonstrated that different mechanisms of action of topo IIα poisons and catalytic inhibitors are also reflected at the cellular level. While the cytotoxic activity and the inhibition of proliferation are comparable, the differences in the disruption of the cell cycle and the induction of DNA damage are observed. The results represent a first promising indication that the efficacy of the discovered catalytic inhibitors is comparable to that of the well-known topo IIα poisons. Further preclinical studies are, however, necessary to evaluate in more detail the activity of these compounds at the cellular level. Particularly, to fully assess their potential, especially in terms of the safety index, further assays on noncancer cell lines will play an important role.
3. Conclusions
The development of efficient new cancer treatments is essential due to the widespread occurrence of various types of cancer, which represent a significant and ever-increasing health burden due to the aging population and environmental influences. Human type II DNA topoisomerases represent key targets and catalytic inhibitors of these molecular motors that alter DNA topology describe a new paradigm aimed at circumventing the known limitations of topo II poisons such as cardiotoxicity and induction of secondary tumors, along with addressing the emergence of resistance to existing cancer therapies.
Based on our discovered substituted 4,5′-bithiazoles as inhibitors of the bacterial DNA gyrase, we performed a structural comparison and molecular docking to the human topo IIα counterpart, which outlined two different binding modes of these compounds. Based on this observation, we developed a virtual screening campaign of a focused chemical library of substituted 4,5′-bithiazoles to identify compounds more closely matched to the topology of the human topo IIα ATP binding site.
In the inhibition assay, we identified several compounds with an inhibitory activity comparable to that of the etoposide drug, disclosing a new chemical class of topo IIα inhibitors. Further detailed investigations confirmed the catalytic mode of topo IIα inhibition by competitive ATP inhibition, and the MST experiments confirmed compound binding to the isolated ATPase domain. Dynamic properties that guide the inhibitor–topo IIα binding at the targeted topo IIα ATP site were assessed by molecular dynamics and dynamic pharmacophore (dynophore) calculations to model the key determinants that contribute to the bithiazole molecular recognition process. The compounds were also able to inhibit the topo IIα as well as the topo IIβ-catalyzed decatenation reaction, a potentially favorable property of this catalytic inhibitors compared to the topo II poisons.
In the cell-based studies, several compounds showed strong cytotoxicity against HepG2 and MCF-7 cell lines comparable to etoposide. In subsequent assays utilizing HepG2 cancer cell line, no induction of DNA double-strand breaks was detected, along with the significantly reduced cell proliferation and arrest of the cell cycle predominantly in the G1 phase. This confirmed that the mechanism of action differs from the topo II poisons also at the cellular level. Current results clearly demonstrate the potential of the substituted 4,5′-bithiazole class for the development of efficient and potentially safe cancer therapies based on the paradigm of catalytic topo II inhibition.
4. Experimental Section
4.1. Molecular Docking Calculations
Molecular docking was performed using GOLD docking tool78 using human topo IIα ATPase domain (PDB: 1ZXM)40 and DNA gyrase (PDB: 1EI1)41 both with nonhydrolyzable AMP-PNP ligand. Structural alignment of both domains was performed using the Hermes protein alignment tool.
In the first step, the validation of GOLD docking tool was performed79 by redocking the AMP-PNP molecule into the human topo IIα ATP binding site. In our validation docking, the AMP-PNP molecule was repeatedly docked 10 times in the human topo IIα ATPase domain by employing the parameters of the GOLD genetic search algorithm (GA) as they are listed below: population size was 100, the selection pressure was 1.1, the number of operations was 100 000, the number of islands equaled 5, migrate was set to 10, mutate value numbered in 95, the niche size amounted to 2, and the crossover reached 95. The pdb was stripped of all ions and water molecules except for waters W924 and W931. These two molecules are supposed to form important interactions with AMP-PNP and were thus considered during our docking process.31 Spins of W924 and W931 were permitted to vary during docking. The active site was defined as a 10 Å radius around the AMP-PNP ligand, and hydrogen atoms were added to the protein. A docking constraint to Asn120 was added to better preserve the interactions between the purine ring of AMP-PNP and the enzyme.40 The scoring function we selected was GoldScore. The binding pose of AMP-PNP we obtained from our docking calculations is close to the pose from the crystal structure, which confirmed our docking parameters as reliable (Figure S1, Supporting Information). We retrieved the best agreement between the crystallized and docked conformation employing the GoldScore scoring function (RMSD = 0.9 Å). The parameters described above were then utilized in the molecular docking calculations for the generated focused chemical library of the substituted 4,5′-bithiazoles into the topo IIα ATP active site.
In a similar manner, DNA gyrase ATP binding site was prepared for docking of compound 13. The active site comprised a 10 Å radius around the reference ligand AMP-PNP with the water molecule W1601 included in the active site. We used the same GA settings and a GoldScore scoring function. Results of all GOLD docking calculations were subsequently visualized using LigandScout.80
4.2. Molecular Dynamics Simulation and Dynophore Calculations
We employed the CHARMM molecular modeling suite58 for the molecular dynamics (MD) calculations of the complex between a single monomer of the ATPase domain originating from the PDB: 1ZXM and compound 1. We obtained the bound conformations of compound 1 using the protocol described in Section 4.1. The single monomer was preprocessed for MD as described previously.34,36−38 The hydrated protein–compound complex was generated with the CHARMM-GUI tool.81 Parameter and topology files for the monomer were generated with CHARMM—version 36,82,83 while compound 1 was parameterized with the CHARMM general force field (CGenFF).84 The protonation pattern of the bithazole 1 was approximated by considering that the pKa value of the 2-aminothiazole is 5.4 with the first protonation occurring on the ring nitrogen.59 Thus, the bithazole ring and the amino group of 1 were all modeled in their deprotonated forms. See the Supporting Information (Table S4) for the assigned atom types and partial charges of compound 1. Our system was solvated with TIP3 water molecules,85 in an octahedral box, edge distance measuring 10 Å. To obtain an electroneutral system, three chlorine ions were inserted with a standard Monte Carlo method. Both the shape and size of the solvated system were subject to periodic boundary conditions (PBCs). CHARMM-GUI automatically produced this grid on the Particle-mesh Ewald (PME) fast Fourier transform (FFT). The prepared system consisted of 73 253 atoms. To remove bad contacts, short steps of energy minimization were executed. The system was first minimized for 10 000 steps by the steepest descent method and subsequently subjected to a modified adopted basis Newton–Raphson method (also 10 000 steps) and finally an MD equilibration simulation of 1 ns without constraints. The production simulation was 20 ns, in which we employed a SHAKE algorithm (2 fs simulation step) and leapfrog integration. Sampling occurred on every 500th step—10 000 conformations in total. The trajectory was analyzed using visual molecular dynamics (VMD) program.86 RMSD calculations of compound 1 were performed by including all atoms against its initial conformation obtained from docking. We provide two movie animations to further illustrate the conformational behavior during the MD simulation in the Supporting Information.
To provide a more detailed look at the interaction pattern of compound 1, we exported 1000 MD frames at equal time intervals and analyzed them with the DynophoreApp from the Molecular Design Lab led by Prof. Wolber at Freie Universität Berlin, Germany, using their hardware.61−63 The obtained model was visualized and analyzed in LigandScout.80 More data are given in the Supporting Information.
4.3. HTS Relaxation Assay of Human Topo IIα
The assay of all compounds 1–14 was performed as described previously.35,87 The assay was performed at four different concentrations of inhibitors: 7.8, 31.25, 125, and 500 μM. The IC50 values were calculated using GraphPad Prism 6.0 software88 and are shown as the concentrations of tested compounds where the residual activity of the enzyme was 50%.43 All tested compounds were characterized with the high-resolution mass spectrometry (HR-MS) technique. For key compounds used in subsequent assays, the purity was examined using microanalysis performed on a PerkinElmer C, H, N, S analyzer (Pregl–Dumas method) as well as high-performance liquid chromatography (HPLC) analysis (see the Supporting Information).
4.4. Human Topo IIα- and Human Topo IIβ-Mediated Decatenation Assay
We made use of the human topo II decatenation assay kit from Inspiralis (Norwich, U.K.) to assess the ability of selected compounds to impede DNA decatenation.89,90 It was performed for topo IIα and topo IIβ isoforms using the protocol as described previously.35 The assay was carried out for compounds 1, 9, 10, and etoposide (reference compound) in duplicate at four investigated concentrations: 7.8, 31.25, 125, and 500 μM.
4.5. Human Topo IIα-Mediated Cleavage Assay
We performed the assay in collaboration with Inspiralis (Norwich, U.K.). We examined selected compounds 1, 9, and reference compound etoposide at concentrations 3.9, 31.5, 125, and 500 μM, as described previously.35,87
4.6. Inhibition of the ATPase Activity
Determination of whether compound 1 from the class of substituted 4,5′-bithiazoles can inhibit the ATPase activity of human topo II enzyme was performed in collaboration with Inspiralis using a pyruvate kinase/lactate dehydrogenase assay.65 The assay measures the reduction of NADH at 340 nm. Conversion of NADH to NAD is caused by ADP, which is formed from ATP hydrolysis. A mixture of linear pBR322 (1.5 μL of 1 mg/mL per assay), assay buffer (composition: 20 mM Tris–HCl, 125 mM potassium acetate, 5 mM magnesium acetate, 2 mM dithiothreitol (DTT), pH 7.9), phosphoenol pyruvate (0.5 μL of 80 mM per assay), pyruvate kinase/lactate dehydrogenase mix (0.75 μL per assay), NADH (1 μL of 20 mM per assay), and water (34.35 μL per assay) was prepared. This mixture (41.1 μL) was put into the wells on a 384-well microtiter plate. DMSO (0.5 μL), etoposide, and compound 1 were added to the wells and mixed. The dilution buffer (5 μL) or human topo IIα (12 nM final concentration) was then added and mixed. Then, a measurement of OD340 change was performed in a plate reader over in a 10 min time period (called the prerun). Then, 3.4 μL of 30 mM ATP was added and the OD340 was monitored for the next 30 min. The assay temperature was 37 °C. The final DMSO concentration in all of the reactions was 1% (v/v). Assays were performed in duplicate at 3.9, 31.5, 125, and 500 μM final concentrations of the investigated compound 1. Serial dilution of compound 1 was performed in DMSO and added to the mixture before the enzyme was added. Etoposide served as a control compound.
4.7. Human Topoisomerase II Competitive ATPase Assay
The human topo competitive ATPase assay was executed at Inspiralis (Norwich, U.K.). The compound was analyzed using a kinase/lactate dehydrogenase assay as described above.
A mixture of the assay buffer (20 mM Tris–HCl, 5 mM magnesium acetate, 125 mM potassium acetate, 2 mM DTT, pH 7.9), linear pBR322 (1.5 μL of 1 mg/mL per assay), phosphoenol pyruvate (0.5 μL of 80 mM per assay), pyruvate kinase/lactate dehydrogenase mix (0.75 μL per assay), NADH (1 μL of 20 mM per assay), DMSO (1.5 μL per assay), and water (32.85 μL per assay) was prepared. This mixture (41.1 μL) was put into the wells of a 384-well microtiter plate. DMSO (0.5 μL) or the diluted investigated compound in the DMSO was added to the wells and mixed. Subsequently, 5 μL of the dilution buffer or human topo IIα (12 nM final concentration) was added and mixed. Before the run, a prerun was done where we added 3.4 μL of the appropriate concentration of ATP and monitored the OD340 for up to 35 min. The assay temperature was 37 °C. Two negative controls (4% DMSO and dilution buffer without enzyme) were used in the presence of 2 mM ATP. The ATP concentrations in this assay were 0.025, 0.05, 0.075, 0.1, 0.25, 0.5, 0.75, and 1 mM. The assays were executed in duplicate at 3.9, 31, 50, 75, and 100 μM final concentrations of the investigated compound 1. DMSO has a final concentration of 4% (v/v) in all of the reactions.
4.8. Microscale Thermophoresis (MST) Measurements of Compound 1 Binding onto Human Topo IIα ATPase Domain
For MST measurements, a Monolith NT 115 (NanoTemper Technologies, München, Germany) was employed, using MST power at 20% and light-emitting diode (LED) power at 20%. We purchased the protein—the ATPase domain of human topo IIα with 1–453 amino acid residues—from Inspiralis.91 It was labeled with the NT-647 dye using the RED-MALEIMIDE labeling kit from NanoTemper (Cysteine Reactive; no. L004, NanoTemper Technologies). The labeling was performed following the supplier’s protocol in the labeling buffer at 20 μM protein concentration (molar dye/protein = 1:3) at room temperature for 30 min. Then, the unbound dye was eliminated by a gravity flow column and the protein was rebuffered in the MST buffer (50 mM Tris–HCl (pH = 7.4), 10 mM MgCl2, 150 mM NaCl, 0.05% Tween-20).54−56 We kept the concentration of the labeled protein constant at an ∼20 nM concentration using MST buffer. For the unlabeled compound, a twofold dilution series was performed with concentrations ranging from 0.098 up to 200 μM (12 concentrations). Samples were prepared and measured three times to calculate average Kd values and SD. The DMSO concentration was 10% in each sample. Premium capillaries were used to load the samples (MO-K025, NanoTemper Technologies, München, Germany). Thermophoresis was measured at temperature 25 °C with 5/30/5 s laser off/on/off times, respectively. Because of titrant-dependent fluorescence changes in the first step of measuring, SDS-denaturation test was performed to confirm specific, ligand-induced binding and data measurements were analyzed (MO, Affinity Analysis, NanoTemper Technology) using the signal from initial fluorescence.
4.9. Cytotoxic Activity of Studied Compounds in HepG2 and MCF-7 Cell Lines
Cytotoxic activity of studied compounds was determined in two human cancer cell lines: hepatocellular carcinoma (HepG2) and breast cancer (MCF-7) cells. Both cell lines were obtained from ATCC. HepG2 cells were cultured in minimum essential medium Eagle (MEME) (Sigma, M2414), supplemented with 2 mM L-Glutamine, 10% fetal bovine serum (FBS), 2.2 g/L NaHCO3, 1 mM sodium pyruvate, 1% nonessential amino acid (NEAA), and 100 IU/mL penicillin/streptomycin, while MCF-7 were cultured in Eagle’s minimum essential medium (MEM) (Sigma, M5650), supplemented with 2 mM glutamine, 10% FBS, and 100 IU/mL penicillin/streptomycin at 37 °C and 5% CO2. After the exposure of cells to the tested compounds, their viability was determined using the MTS assay.
The cells were seeded at densities of 8000 and 7000 cells/well for HepG2 and MCF-7 cells, respectively, in 200 μL of complete growth medium onto 96-well microtiter plates (Nunc, Thermo Fisher Scientific, Waltham, MA) and were left overnight at 37 °C to attach. Subsequently, the growth medium was replaced with a fresh medium containing graded concentrations of the studied compounds. The cells were further incubated for 24 h at 37 °C. After the incubation, 40 μL of an MTS/PMS (20:1) mixture was added to each well. After 3 h incubation (37 °C, 5% CO2), the absorbance was determined at 490 nm using a spectrofluorometer Synergy MX (BioTek, Winooski, VT). Etoposide (200 μM) was used as a positive control. Assay was performed at five concentrations of each compound: 2, 25, 50, 100, and 200 μM for compounds 1, 7, and 14 and 1, 12.5, 25, 50, and 100 μM for compounds 9 and 10. In addition, etoposide was titrated at 5, 50, 100, 150, and 300 μM. Cell viability was calculated by comparing the optical density (OD) of the wells with exposed cells with the wells of solvent control cells, and the results are shown as percentage of cell viability ± SD. Experiments were performed in three independent repetitions each time in at least three replicates. The EC50 values were determined utilizing nonlinear regression analysis available in GraphPad Prism 7.0 software. Statistically significant difference between control and treated groups was determined by one-way analysis of variance combined with Dunnett’s multiple comparison test.
4.10. Effect of Compound 1 on the Cell Cycle, Cell Proliferation, and Formation of the DNA Double-Stranded Breaks
HepG2 cells were seeded onto 25 cm2 plates (Corning Inc., NY) at a density of 750 000 cells/plate and were left to attach overnight. The next day, the cells were exposed to compound 1 (10 and 50 μM) and a positive control (etoposide, 50 μM) for 24 h. After the treatment, the cells were trypsinized and collected (adherent and floating cells). Subsequently, the cells were centrifuged (800 rpm, 4 °C for 5 min), washed with ice-cold 1× phosphate-buffered saline (PBS) twice, resuspended in cold PBS (0.5 mL), and fixed by adding ethanol (1.5 mL) dropwise into the cell suspension, while mixing. The cells were fixed overnight at 4 °C and stored until analysis at −20 °C. The fixed cells were then centrifuged (1200 rpm, 10 min), washed with ice-cold 1× PBS, and labeled with Anti-H2AX pS139 antibodies (50-fold diluted) for DNA DSB analysis, Ki67 antibodies (50-fold diluted) for proliferation analysis, and Hoechst 33342 dye for cell cycle analysis as described in the manufacturer’s protocol. Flow cytometric analysis was performed on an MACSQuant Analyzer 10 (Miltenyi Biotech, Germany). Fluorescein isothiocyanate (FITC) intensity, corresponding to Ki67+ proliferation marker, was measured on the FITC-A channel, and cell cycle analysis was measured on VioBlueA channel. APC intensity, corresponding to DNA DSBs, was measured on the APC-A channel. Rea-FITC and rea-APC antibodies (Miltenyi Biotec, Germany) were used to determine unspecific binding.
Ten thousand events were recorded in each sample. Three independent experiments were performed. In each experiment, a positive control (etoposide; 50 μM) and a vehicle control (0.5% DMSO) were included. For the analysis of the results, the raw data were exported from MACSQuantify software and was converted to .fcs format and then to .csv format. For the γH2AX positive cells, the statistical analysis between vehicle control and treated groups was done with a linear mixed-effects model. Further calculation was performed with the statistical program R92 and its packages reshape93 and nlme.94
Acknowledgments
This research was funded by grants P1-0012 and P1-0245 of the Slovenian Research Agency ARRS. M.J. is an International Program Associate at RIKEN, Japan. Katja Valjavec helped with the interpretation of performed MD simulations. The authors thank Drs. Alison Howells and Nicolas Burton for performing topoisomerase II-mediated cleavage, decatenation, and ATPase assays at Inspiralis. Dr. Barbara Pogorelčnik, Dr. Matjaž Brvar, and Urška Jug from the National Institute of Chemistry are thanked for technical assistance with the HTS relaxation assay in the initial stage of this work. Rok Petrijan and Dr. Samo Andrenšek from CVTA at NIC in Ljubljana are thanked for the HPLC analysis. The authors also thank Dr. Katja Pirc from the same Institute for technical assistance with MST experiments. Drs. Katarzyna Walkiewicz and Jakub Nowak from NanoTemper Technologies, Munich, Germany, are thanked for discussions regarding the interpretation of the MST measurements. Dr. Ahmed S.A. Mady and Prof. Zaneta Nikolovska Coleska from the University of Michigan, Ann Arbor, are acknowledged and thanked for some initial cytotoxicity testing in the scope of the ARRS bilateral research project. Dynophore calculations were made possible due to collaboration with Prof. Gerhard Wolber at their computer facility at FU Berlin. Klara Hercog is thanked for her help with MTS and flow cytometry experiments. Prof. Janez Mavri from the National Institute of Chemistry in Ljubljana is thanked for helpful discussions of this work.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jcim.0c00202.
The authors declare no competing financial interest.
Supplementary Material
References
- Hanahan D.; Weinberg R. A. The hallmarks of cancer. Cell 2000, 100, 57–70. 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D.; Weinberg R. A. Hallmarks of cancer: the next generation. Cell 2011, 144, 646–674. 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Pommier Y.; Sun Y.; Huang S. N.; Nitiss J. L. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016, 17, 703–721. 10.1038/nrm.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates A. D.; Maxwell A. DNA topology: Topoisomerases keep it simple. Curr. Biol. 1997, 7, R778–R781. 10.1016/S0960-9822(06)00403-9. [DOI] [PubMed] [Google Scholar]
- Champoux J. J. DNA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 2001, 70, 369–413. 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Tsutsui K.; Tsutsui K.; Hosoya O.; Sano K.; Tokunaga A. Immunohistochemical analyses of DNA topoisomerase II isoforms in developing rat cerebellum. J. Comp. Neurol. 2001, 431, 228–239. . [DOI] [PubMed] [Google Scholar]
- Watanabe M.; Tsutsui K.; Tsutsui K.; Inoue Y. Differential expressions of the topoisomerase II alpha and II beta mRNAs in developing rat brain. Neurosci. Res. 1994, 19, 51–57. 10.1016/0168-0102(94)90007-8. [DOI] [PubMed] [Google Scholar]
- Capranico G.; Tinelli S.; Austin C. A.; Fisher M. L.; Zunino F. Different patterns of gene expression of topoisomerase II isoforms in differentiated tissues during murine development. Biochim. Biophys. Acta, Gene Struct. Expression 1992, 1132, 43–48. 10.1016/0167-4781(92)90050-A. [DOI] [PubMed] [Google Scholar]
- Bollimpelli V. S.; Dholaniya P. S.; Kondapi A. K. Topoisomerase IIbeta and its role in different biological contexts. Arch. Biochem. Biophys. 2017, 633, 78–84. 10.1016/j.abb.2017.06.021. [DOI] [PubMed] [Google Scholar]
- Berger J. M.; Gamblin S. J.; Harrison S. C.; Wang J. C. Structure and mechanism of DNA topoisomerase II. Nature 1996, 379, 225–232. 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- Berger J. M.; Wang J. C. Recent developments in DNA topoisomerase II structure and mechanism. Curr. Opin. Struct. Biol. 1996, 6, 84–90. 10.1016/S0959-440X(96)80099-6. [DOI] [PubMed] [Google Scholar]
- Schoeffler A. J.; Berger J. M. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008, 41, 41–101. 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- Riccio A. A.; Schellenberg M. J.; Williams R. S. Molecular mechanisms of topoisomerase 2 DNA-protein crosslink resolution. Cell. Mol. Life Sci. 2020, 77, 81–91. 10.1007/s00018-019-03367-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogorelčnik B.; Perdih A.; Solmajer T. Recent developments of DNA poisons - human DNA topoisomerase IIα inhibitors - as anticancer agents. Curr. Pharm. Des. 2013, 19, 2474–2488. 10.2174/1381612811319130016. [DOI] [PubMed] [Google Scholar]
- Pogorelčnik B.; Perdih A.; Solmajer T. Recent advances in the development of catalytic inhibitors of human DNA topoisomerase IIα as novel anticancer agents. Curr. Med. Chem. 2013, 20, 694–709. 10.2174/092986713804999402. [DOI] [PubMed] [Google Scholar]
- Bailly C. Contemporary challenges in the design of topoisomerase II inhibitors for cancer chemotherapy. Chem. Rev. 2012, 112, 3611–3640. 10.1021/cr200325f. [DOI] [PubMed] [Google Scholar]
- Baldwin E. L.; Osheroff N. Etoposide, topoisomerase II and cancer. Curr. Med. Chem.: Anti-Cancer Agents 2005, 5, 363–372. 10.2174/1568011054222364. [DOI] [PubMed] [Google Scholar]
- Minotti G.; Menna P.; Salvatorelli E.; Cairo G.; Gianni L. Anthracyclines: Molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol. Rev. 2004, 56, 185–229. 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- Nelson E. M.; Tewey K. M.; Liu L. F. Mechanism of antitumor drug-action - poisoning of mammalian DNA topoisomerase-II on DNA by 4′-(9-acridinylamino)-methanesulfon-meta-anisidide. Proc. Natl. Acad. Sci. U.S.A. 1984, 81, 1361–1365. 10.1073/pnas.81.5.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix C. A. Secondary leukemias induced by topoisomerase-targeted drugs. Biochim. Biophys. Acta, Gene Struct. Expression 1998, 1400, 233–255. 10.1016/S0167-4781(98)00139-0. [DOI] [PubMed] [Google Scholar]
- Housman G.; Byler S.; Heerboth S.; Lapinska K.; Longacre M.; Snyder N.; Sarkar S. Drug resistance in cancer: an overview. Cancers 2014, 6, 1769–1792. 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilati P.; Nitti D.; Mocellin S. Cancer resistance to type II topoisomerase inhibitors. Curr. Med. Chem. 2012, 19, 3900–3906. 10.2174/092986712802002473. [DOI] [PubMed] [Google Scholar]
- Ganapathi R.; Ganapathi M. Mechanisms regulating resistance to inhibitors of topoisomerase II. Front. Pharmacol. 2013, 4, 89 10.3389/fphar.2013.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakács G.; Paterson J. K.; Ludwig J. A.; Booth-Genthe C.; Gottesman M. M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discovery 2006, 5, 219–234. 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- Case D. C.; Ervin T. J.; Boyd M. A.; Bove L. G.; Sonneborn H. L.; Paul S. D. Phase-II study of aclarubicin in acute myeloblastic-leukemia. Am. J. Clin. Oncol. 1987, 10, 523–526. 10.1097/00000421-198712000-00014. [DOI] [PubMed] [Google Scholar]
- Bojanowski K.; Lelievre S.; Markovits J.; Couprie J.; Jacquemin-Sablon A.; Larsen A. K. Suramin is an inhibitor of DNA topoisomerase II in vitro and in Chinese hamster fibrosarcoma cells. Proc. Natl. Acad. Sci. U.S.A. 1992, 89, 3025–3029. 10.1073/pnas.89.7.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune J. M.; Osheroff N. Merbarone inhibits the catalytic activity of human topoisomerase II alpha by blocking DNA cleavage. J. Biol. Chem. 1998, 273, 17643–17650. 10.1074/jbc.273.28.17643. [DOI] [PubMed] [Google Scholar]
- Tanabe K.; Ikegami Y.; Ishida R.; Andoh T. Inhibition of topoisomerase II by antitumor agents bis(2,6-dioxopiperazine) derivatives. Cancer Res. 1991, 51, 4903–4908. [PubMed] [Google Scholar]
- Chang S.; Hu T.; Hsieh T. S. Analysis of a core domain in Drosophila DNA topoisomerase II. Targeting of an antitumor agent ICRF-159. J. Biol. Chem. 1998, 273, 19822–19828. 10.1074/jbc.273.31.19822. [DOI] [PubMed] [Google Scholar]
- Chène P.; Rudloff J.; Schoepfer J.; Furet P.; Meier P.; Qian Z.; Schlaeppi J. M.; Schmitz R.; Radimerski T. Catalytic inhibition of topoisomerase II by a novel rationally designed ATP-competitive purine analogue. BMC Chem. Biol. 2009, 9, 1 10.1186/1472-6769-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furet P.; Schoepfer J.; Radimerski T.; Chene P. Discovery of a new class of catalytic topoisomerase II inhibitors targeting the ATP-binding site by structure based design. Part I. Bioorg. Med. Chem. Lett. 2009, 19, 4014–4017. 10.1016/j.bmcl.2009.06.034. [DOI] [PubMed] [Google Scholar]
- Jensen L. H.; Thougaard A. V.; Grauslund M.; Sokilde B.; Carstensen E. V.; Dvinge H. K.; Scudiero D. A.; Jensen P. B.; Shoemaker R. H.; Sehested M. Substituted purine analogues define a novel structural class of catalytic topoisomerase II inhibitors. Cancer Res. 2005, 65, 7470–7477. 10.1158/0008-5472.CAN-05-0707. [DOI] [PubMed] [Google Scholar]
- Jensen L. H.; Liang H.; Shoemaker R.; Grauslund M.; Sehested M.; Hasinoff B. B. A three-dimensional quantitative structure-activity relationship study of the inhibition of the ATPase activity and the strand passing catalytic activity of topoisomerase IIalpha by substituted purine analogs. Mol. Pharmacol. 2006, 70, 1503–1513. 10.1124/mol.106.026856. [DOI] [PubMed] [Google Scholar]
- Pogorelčnik B.; Janežič M.; Sosič I.; Gobec S.; Solmajer T.; Perdih A. 4,6-Substituted-1,3,5-triazin-2(1H)-ones as monocyclic catalytic inhibitors of human DNA topoisomerase IIα targeting the ATP binding site. Bioorg. Med. Chem. 2015, 23, 4218–4229. 10.1016/j.bmc.2015.06.049. [DOI] [PubMed] [Google Scholar]
- Bergant K.; Janežic M.; Valjavec K.; Sosič I.; Pajk S.; Štampar M.; Žegura B.; Gobec S.; Filipič M.; Perdih A. Structure-guided optimization of 4,6-substituted-1,3,5-triazin-2(1H)-ones as catalytic inhibitors of human DNA topoisomerase IIα. Eur. J. Med. Chem. 2019, 175, 330–348. 10.1016/j.ejmech.2019.04.055. [DOI] [PubMed] [Google Scholar]
- Pogorelčnik B.; Brvar M.; Zajc I.; Filipič M.; Solmajer T.; Perdih A. Monocyclic 4-amino-6-(phenylamino)-1,3,5-triazines as inhibitors of human DNA topoisomerase IIα. Bioorg. Med. Chem. Lett. 2014, 24, 5762–5768. 10.1016/j.bmcl.2014.10.042. [DOI] [PubMed] [Google Scholar]
- Pogorelčnik B.; Brvar M.; Žegura B.; Filipič M.; Solmajer T.; Perdih A. Discovery of mono- and disubstituted 1h-pyrazolo[3,4]pyrimidines and 9h-purines as catalytic inhibitors of human DNA topoisomerase IIα. ChemMedChem 2015, 10, 345–359. 10.1002/cmdc.201402459. [DOI] [PubMed] [Google Scholar]
- Janežič M.; Pogorelčnik B.; Brvar M.; Solmajer T.; Perdih A. 3-substituted-1 H -indazoles as catalytic inhibitors of the human DNA topoisomerase IIα. ChemistrySelect 2017, 2, 480–488. 10.1002/slct.201601554. [DOI] [Google Scholar]
- Brvar M.; Perdih A.; Renko M.; Anderluh G.; Turk D.; Solmajer T. Structure-based discovery of substituted 4,5′-bithiazoles as novel DNA gyrase inhibitors. J. Med. Chem. 2012, 55, 6413–6426. 10.1021/jm300395d. [DOI] [PubMed] [Google Scholar]
- Wei H.; Ruthenburg A. J.; Bechis S. K.; Verdine G. L. Nucleotide-dependent domain movement in the ATPase domain of a human type IIA DNA topoisomerase. J. Biol. Chem. 2005, 280, 37041–37047. 10.1074/jbc.M506520200. [DOI] [PubMed] [Google Scholar]
- Brino L.; Urzhumtsev A.; Mousli M.; Bronner C.; Mitschler A.; Oudet P.; Moras D. Dimerization of Escherichia coli DNA-gyrase B provides a structural mechanism for activating the ATPase catalytic center. J. Biol. Chem. 2000, 275, 9468–9475. 10.1074/jbc.275.13.9468. [DOI] [PubMed] [Google Scholar]
- Emolecules. www.emolecules.com (accessed Feb 10
- Maxwell A.; Burton N. P.; O’Hagan N. High-throughput assays for DNA gyrase and other topoisomerases. Nucleic Acids Res. 2006, 34, e104 10.1093/nar/gkl504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W. B.; Ou J. B.; Huang Z. H.; Chen S. B.; Ou T. M.; Tan J. H.; Li D.; Shen L. L.; Huang S. L.; Gu L. Q.; Huang Z. S. Synthesis and evaluation of mansonone F derivatives as topoisomerase inhibitors. Eur. J. Med. Chem. 2011, 46, 3339–3347. 10.1016/j.ejmech.2011.04.059. [DOI] [PubMed] [Google Scholar]
- Hu W.; Huang X. S.; Wu J. F.; Yang L.; Zheng Y. T.; Shen Y. M.; Li Z. Y.; Li X. Discovery of novel topoisomerase II inhibitors by medicinal chemistry approaches. J. Med. Chem. 2018, 61, 8947–8980. 10.1021/acs.jmedchem.7b01202. [DOI] [PubMed] [Google Scholar]
- Baviskar A. T.; Madaan C.; Preet R.; Mohapatra P.; Jain V.; Agarwal A.; Guchhait S. K.; Kundu C. N.; Banerjee U. C.; Bharatam P. V. N-Fused imidazoles as novel anticancer agents that inhibit catalytic activity of topoisomerase II alpha and induce apoptosis in G1/S phase. J. Med. Chem. 2011, 54, 5013–5030. 10.1021/jm200235u. [DOI] [PubMed] [Google Scholar]
- Perrin D.; van Hille B.; Hill B. T. Differential sensitivities of recombinant human topoisomerase IIalpha and beta to various classes of topoisomerase II-interacting agents. Biochem. Pharmacol. 1998, 56, 503–507. 10.1016/S0006-2952(98)00082-3. [DOI] [PubMed] [Google Scholar]
- Willmore E.; Frank A. J.; Padget K.; Tilby M. J.; Austin C. A. Etoposide targets topoisomerase IIalpha and IIbeta in leukemic cells: isoform-specific cleavable complexes visualized and quantified in situ by a novel immunofluorescence technique. Mol. Pharmacol. 1998, 54, 78–85. 10.1124/mol.54.1.78. [DOI] [PubMed] [Google Scholar]
- Toyoda E.; Kagaya S.; Cowell I. G.; Kurosawa A.; Kamoshita K.; Nishikawa K.; Iiizumi S.; Koyama H.; Austin C. A.; Adachi N. NK314, a topoisomerase II inhibitor that specifically targets the alpha isoform. J. Biol. Chem. 2008, 283, 23711–23720. 10.1074/jbc.M803936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimitsu N.; Adachi N.; Hirai H.; Hossain M. S.; Hamamoto H.; Kobayashi M.; Aratani Y.; Koyama H.; Sekimizu K. Enforced cytokinesis without complete nuclear division in embryonic cells depleting the activity of DNA topoisomerase IIalpha. Genes Cells 2003, 8, 393–402. 10.1046/j.1365-2443.2003.00643.x. [DOI] [PubMed] [Google Scholar]
- Yang X.; Li W.; Prescott E. D.; Burden S. J.; Wang J. C. DNA topoisomerase IIbeta and neural development. Science 2000, 287, 131–134. 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- Sakaguchi A.; Kikuchi A. Functional compatibility between isoform alpha and beta of type II DNA topoisomerase. J. Cell Sci. 2004, 117, 1047–1054. 10.1242/jcs.00977. [DOI] [PubMed] [Google Scholar]
- Kenig S.; Faoro V.; Bourkoula E.; Podergajs N.; Ius T.; Vindigni M.; Skrap M.; Lah T.; Cesselli D.; Storici P.; Vindigni A. Topoisomerase IIbeta mediates the resistance of glioblastoma stem cells to replication stress-inducing drugs. Cancer Cell Int. 2016, 16, 58 10.1186/s12935-016-0339-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerabek-Willemsen M.; Wienken C. J.; Braun D.; Baaske P.; Duhr S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 2011, 9, 342–353. 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel S. A. I.; Dijkman P. M.; Lea W. A.; van den Bogaart G.; Jerabek-Willemsen M.; Lazic A.; Joseph J. S.; Srinivasan P.; Baaske P.; Simeonov A.; Katritch I.; Melo F. A.; Ladbury J. E.; Schreiber G.; Watts A.; Braun D.; Duhr S. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 2013, 59, 301–315. 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienken C. J.; Baaske P.; Rothbauer U.; Braun D.; Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010, 1, 100 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- Perdih A.; Wolber G.; Solmajer T. Molecular dynamics simulation and linear interaction energy study of d-Glu-based inhibitors of the MurD ligase. J. Comput.-Aided Mol. Des. 2013, 27, 723–738. 10.1007/s10822-013-9673-3. [DOI] [PubMed] [Google Scholar]
- Brooks B. R.; Bruccoleri R. E.; Olafson B. D.; States D. J.; Swaminathan S.; Karplus M. Charmm - a Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J. Comput. Chem. 1983, 4, 187–217. 10.1002/jcc.540040211. [DOI] [Google Scholar]
- Albert A.; Goldacre R.; Phillips J. 455. The strength of heterocyclic bases. J. Chem. Soc. 1948, 2240–2249. 10.1039/jr9480002240. [DOI] [Google Scholar]
- Borštnar R.; Repič M.; Kamerlin S. C. L.; Vianello R.; Mavri J. Computational Study of the pKa Values of Potential Catalytic Residues in the Active Site of Monoamine Oxidase B. J. Chem. Theory Comput. 2012, 8, 3864–3870. 10.1021/ct300119u. [DOI] [PubMed] [Google Scholar]
- Dominique S.Dynophores: Novel Dynamic Pharmacophores; Lebenswissenschaftliche Fakultät, Universität zu Berlin, 2015. [Google Scholar]
- Bock A.; Bermudez M.; Krebs F.; Matera C.; Chirinda B.; Sydow D.; Dallanoce C.; Holzgrabe U.; De Amici M.; Lohse M. J.; Wolber G.; Mohr K. Ligand Binding Ensembles Determine Graded Agonist Efficacies at a G Protein-coupled Receptor. J. Biol. Chem. 2016, 291, 16375–16389. 10.1074/jbc.M116.735431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez M.; Rakers C.; Wolber G. Structural characteristics of the allosteric binding site represent a key to subtype selective modulators of muscarinic acetylcholine receptors. Mol. Inf. 2015, 34, 526–530. 10.1002/minf.201500025. [DOI] [PubMed] [Google Scholar]
- Dror R. O.; Dirks R. M.; Grossman J. P.; Xu H.; Shaw D. E. Biomolecular simulation: a computational microscope for molecular biology. Annu. Rev. Biophys. 2012, 41, 429–452. 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- Bergant K.; Janežič M.; Perdih A. Bioassays and in silico methods in the identification of human DNA topoisomerase IIα inhibitors. Curr. Med. Chem. 2018, 25, 3286–3318. 10.2174/0929867325666180306165725. [DOI] [PubMed] [Google Scholar]
- Alpsoy A.; Yasa S.; Gunduz U. Etoposide resistance in MCF-7 breast cancer cell line is marked by multiple mechanisms. Biomed. Pharmacother. 2014, 68, 351–355. 10.1016/j.biopha.2013.09.007. [DOI] [PubMed] [Google Scholar]
- Xie B. S.; Zhao H. C.; Yao S. K.; Zhuo D. X.; Jin B.; Lv D. C.; Wu C. L.; Ma D. L.; Gao C.; Shu X. M.; Ai Z. L. Autophagy inhibition enhances etoposide-induced cell death in human hepatoma G2 cells. Int. J. Mol. Med. 2011, 27, 599–606. 10.3892/ijmm.2011.607. [DOI] [PubMed] [Google Scholar]
- Dasika G. K.; Lin S. C.; Zhao S.; Sung P.; Tomkinson A.; Lee E. Y. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene 1999, 18, 7883–7899. 10.1038/sj.onc.1203283. [DOI] [PubMed] [Google Scholar]
- D’Arcy N.; Gabrielli B. Topoisomerase II Inhibitors and Poisons, and the Influence of Cell Cycle Checkpoints. Curr. Med. Chem. 2017, 24, 1504–1519. 10.2174/0929867323666161205122613. [DOI] [PubMed] [Google Scholar]
- Darpan D.; Joshi G.; Amrutkar S. M.; Baviskar A. T.; Kler H.; Singh S.; Banerjee U. C.; Kumar R. Synthesis and biological evaluation of new 2,5-dimethylthiophene/furan based N-acetyl pyrazolines as selective topoisomerase II inhibitors. RSC Adv. 2016, 6, 14880–14892. 10.1039/C5RA25705K. [DOI] [Google Scholar]
- Islam M. S.; Park S.; Song C.; Kadi A. A.; Kwon Y.; Rahman A. F. Fluorescein hydrazones: A series of novel non-intercalative topoisomerase IIalpha catalytic inhibitors induce G1 arrest and apoptosis in breast and colon cancer cells. Eur. J. Med. Chem. 2017, 125, 49–67. 10.1016/j.ejmech.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Huang H.; Chen Q.; Ku X.; Meng L. H.; Lin L. P.; Wang X.; Zhu C. H.; Wang Y.; Chen Z.; Li M.; Jiang H. L.; Chen K. X.; Ding J.; Liu H. A Series of alpha-Heterocyclic Carboxaldehyde Thiosemicarbazones Inhibit Topoisomerase II alpha Catalytic Activity. J. Med. Chem. 2010, 53, 3048–3064. 10.1021/jm9014394. [DOI] [PubMed] [Google Scholar]
- Henwood J. M.; Brogden R. N. Etoposide. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic potential in combination chemotherapy of cancer. Drugs 1990, 39, 438–490. 10.2165/00003495-199039030-00008. [DOI] [PubMed] [Google Scholar]
- Bruno S.; Darzynkiewicz Z. Cell cycle dependent expression and stability of the nuclear protein detected by Ki-67 antibody in HL-60 cells. Cell Proliferation 1992, 25, 31–40. 10.1111/j.1365-2184.1992.tb01435.x. [DOI] [PubMed] [Google Scholar]
- Smart D. J.; Halicka H. D.; Schmuck G.; Traganos F.; Darzynkiewicz Z.; Williams G. M. Assessment of DNA double-strand breaks and gammaH2AX induced by the topoisomerase II poisons etoposide and mitoxantrone. Mutat. Res., Fundam. Mol. Mech. Mutagen. 2008, 641, 43–47. 10.1016/j.mrfmmm.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelnikova O. A.; Rogakou E. P.; Panyutin I. G.; Bonner W. M. Quantitative detection of (125)IdU-induced DNA double-strand breaks with gamma-H2AX antibody. Radiat. Res. 2002, 158, 486–492. 10.1667/0033-7587(2002)158[0486:QDOIID]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Rogakou E. P.; Boon C.; Redon C.; Bonner W. M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999, 146, 905–916. 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G.; Willett P.; Glen R. C.; Leach A. R.; Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- Kirchmair J.; Markt P.; Distinto S.; Wolber G.; Langer T. Evaluation of the performance of 3D virtual screening protocols: RMSD comparisons, enrichment assessments, and decoy selection - What can we learn from earlier mistakes?. J. Comput.-Aided Mol. Des. 2008, 22, 213–228. 10.1007/s10822-007-9163-6. [DOI] [PubMed] [Google Scholar]
- Wolber G.; Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 2005, 45, 160–169. 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- Jo S.; Kim T.; Iyer V. G.; Im W. Software news and updates - CHARNIM-GUI: A web-based grraphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- MacKerell A. D.; Bashford D.; Bellott M.; Dunbrack R. L.; Evanseck J. D.; Field M. J.; Fischer S.; Gao J.; Guo H.; Ha S.; Joseph-McCarthy D.; Kuchnir L.; Kuczera K.; Lau F. T. K.; Mattos C.; Michnick S.; Ngo T.; Nguyen D. T.; Prodhom B.; Reiher W. E.; Roux B.; Schlenkrich M.; Smith J. C.; Stote R.; Straub J.; Watanabe M.; Wiorkiewicz-Kuczera J.; Yin D.; Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Mackerell A. D.; Feig M.; Brooks C. L. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004, 25, 1400–1415. 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- Vanommeslaeghe K.; Hatcher E.; Acharya C.; Kundu S.; Zhong S.; Shim J.; Darian E.; Guvench O.; Lopes P.; Vorobyov I.; MacKerell A. D. CHARMM general force field: A force field for drug-like molecules compatible with the charmm all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: Visual molecular dynamics. J. Mol. Graphics 1996, 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Loboda K. B.; Valjavec K.; Stampar M.; Wolber G.; Zegura B.; Filipic M.; Dolenc M. S.; Perdih A. Design and synthesis of 3,5-substituted 1,2,4-oxadiazoles as catalytic inhibitors of human DNA topoisomerase IIalpha. Bioorg. Chem. 2020, 99, 103828 10.1016/j.bioorg.2020.103828. [DOI] [PubMed] [Google Scholar]
- GraphPad Software, L. J. C. U. www.graphpad.com (accessed Jan 5, 2020).
- Osheroff N.; Mary-Ann B.. DNA Topoisomerase Protocols, II: Enzymology and Drugs; Humana Press Inc.: New Jersey, 2001. [Google Scholar]
- Hammonds T. R.; Maxwell A. The DNA dependence of the ATPase activity of human DNA topoisomerase IIalpha. J. Biol. Chem. 1997, 272, 32696–32703. 10.1074/jbc.272.51.32696. [DOI] [PubMed] [Google Scholar]
- Campbell S.; Maxwell A. The ATP-operated clamp of human DNA topoisomerase IIalpha: hyperstimulation of ATPase by “piggy-back” binding. J. Mol. Biol. 2002, 320, 171–188. 10.1016/S0022-2836(02)00461-8. [DOI] [PubMed] [Google Scholar]
- R Core Team . A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2012. [Google Scholar]
- Wickham H. Reshaping Data with the reshape Package. J. Stat. Software 2007, 21, 1–20. 10.18637/jss.v021.i12. [DOI] [Google Scholar]
- Wickham H.ggplot2: Elegant Graphics for Data Analysis; Springer Publishing Company, 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.