Abstract
CRISPR-Cas clinical trials have begun, offering a first glimpse at how DNA and RNA targeting could enable therapies for many genetic and epigenetic human diseases. The speedy progress of CRISPR-Cas from discovery and adoption to clinical use is built on decades of traditional gene therapy research and belies the multiple challenges that could derail the successful translation of these new modalities. Here, we review how CRISPR-Cas therapeutics are translated from technological systems to therapeutic modalities, paying particular attention to the therapeutic cascade from cargo to delivery vector, manufacturing, administration, pipelines, safety, and therapeutic target profiles. We also explore potential solutions to some of the obstacles facing successful CRISPR-Cas translation. We hope to illuminate how CRISPR-Cas is brought from the academic bench toward use in the clinic.
Introduction
Gene therapy holds the promise of treating disease-causing genetic, epigenetic, and transcriptomic aberrations. The first couple of decades of research and development in this field were marked by eager exploration and some devastating setbacks.1 Even so, persistence has begun to pay off with the first generation of approved gene therapies that augment missing or defective genes in conditions such as transfusion-dependent β-thalassemia, spinal muscular atrophy (SMA), and inherited retinal disease.2–4
Fortuitously, the long-awaited emergence of these genetic therapies coincides with the recent advent of genome and transcriptome engineering tools based on the CRISPR-Cas RNA-guided nuclease systems, which have transformed our ability to precisely manipulate nucleic acids. CRISPR-Cas has expanded our genome editing toolkit, enabling precision alteration, replacement, and regulation of genes to therapeutically target disease states and potentially confer disease resistance. Beyond genetic diseases, CRISPR-Cas editing therapies have been used to target acquired diseases such as cancer, cardiovascular disease, infectious disease, and chronic ailments such as Alzheimer's disease (AD) in animal models.5–8 The nexus of these two fields has created extraordinary excitement for the potential of CRISPR-based gene therapies to revolutionize the treatment of many of the most important ailments currently impacting human health.
Although that potential has yet to be fully realized, preclinical studies of CRISPR-based therapies have exploded in recent years. Pioneering CRISPR-Cas gene therapy clinical trials have begun treating some monogenic diseases and severe cancers. Still, the broad application of CRISPR-Cas therapies has a long way to go and hinges on multiple factors. These factors include long-term studies to demonstrate safety without off-target activity; advancement in precise and effective delivery systems; immune system interactions; efficient and cheap manufacturing; and effective regulation to democratize access to these therapies.
In this review, we cover the steps that are necessary to take CRISPR-based gene therapies from concept through drug design and preclinical testing to clinical trials. This process is complex and may be iterative with many decision points and important milestones along the way. We then focus on the most critical challenges facing the development of these therapies, especially in delivery, administration, safety, preclinical study design, drug manufacturing, and economic and regulatory issues. We describe the obstacles they present and how studies to date have approached these issues. We hope to offer both a window into the current state of the field and a map to orient thought and resources toward solving the most significant challenges that may impede the realization of the potential of CRISPR-based gene therapy.
Therapeutic Development Cascade
Development of a gene therapeutic is an enormously complex endeavor,5,9 weighing strategies incorporating unmet medical need, editing approach, needed repair percentages, availability of disease models that faithfully recapitulate human biology, and delivery considerations among a multitude of aspects that are crucial to creating a safe and efficacious medicine. Correspondingly, the path toward enabling a new therapy follows a multifaceted development cascade (Fig. 1).
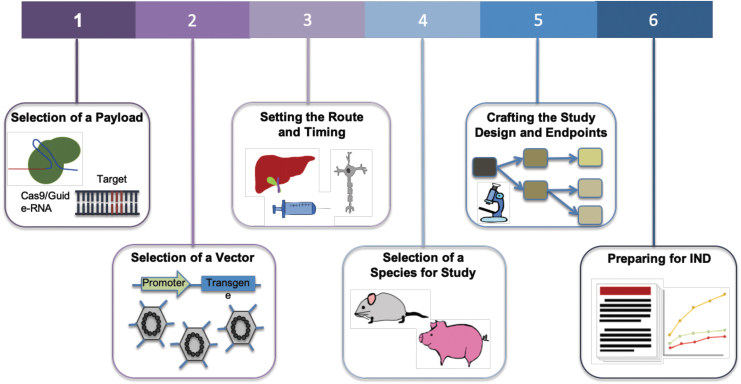
FIG. 1.
Steps toward enabling a gene therapy via CRISPR-Cas. For a disease target, the choice of Cas effector and corresponding gRNA is the first step. Next is to determine CRISPR-Cas delivery options via viral or nonviral methods. Associated with delivery system is also the route of administration. This typically depends on the organ or tissue of treatment and ideally should be limited to that location. Studies are then performed to assess the distribution, persistence, and clearance of the vector and the expression of the therapy in target tissues. Relevant clinical insight is also dependent on the selection of species for study, and a preclinical study design is crafted. Clinical endpoints are set based on safety and toxicology studies. Proof of safety and efficacy in preclinical studies will then lead to an IND application. gRNA, guide-RNA; IND, Investigational New Drug.
(1) Payload: The selection of payload is governed by the nature of the desired edit and considerations of enabling activity at the target locus while ensuring high specificity. The desired therapeutic correction determines the choice of nuclease, editor, or epigenetic modifier. Delivery constraints such as vehicle capacity and interaction with human biology and the immune system, availability of appropriate protospacer-adjacent motifs (PAM) sites, and the dynamic range of therapeutic effect also affect this choice.
(2) Delivery vehicle: Selection of a vehicle for in vivo delivery. Adeno-associated viruses (AAVs) are commonly used as they deliver the payload without insertion into the host genome, maintaining the packaged DNA in an episome. The AAVs work well in postmitotic cells and can be targeted via use of tissue-specific promoters or the selection of appropriate serotypes.10 Efforts have been made to enhance the safety profile of AAVs by understanding how the viruses interact with and evade host immunity11–14 and engineering new capsids to mitigate the immune response.15 Although AAV capacity limits the size of packaged constructs to about 5 kb, recent studies have uncovered Cas proteins that are considerably smaller than Streptococcus pyogenes Cas9, some of which show functional promise.16,17 Besides AAV, CRISPR-Cas can also be delivered via nonviral vehicles, with purified ribonucleoproteins (RNPs) and lipid nanoparticles (LNPs) showing promise for therapeutic translation.
(3) Delivery route and timing: Treatments delivered to the brain, spinal cord, and eye benefit from their immune-privileged status, and thus they are usually more tolerant to foreign antigens used in gene therapies.18 For this reason and also the relative ease of delivery, some of the first clinical trials of CRISPR-Cas9 therapies have been initiated against hematopoietic cells where delivery is administered ex vivo or ocular disorders where delivery is via a subretinal AAV injection (NCT03872479). However, this immune privilege can also be breached when the blood–brain barrier (BBB) is compromised in certain pathological states.
The timing of the administration is also key, as studies have shown that the earlier the treatment is given in the course of disease progression, the higher the potential for efficacy.19 Associated with this are studies to assess the distribution, persistence, and clearance of the delivery vector and expression of the therapy in target tissues.20 A challenge to gene therapies can be the occurrence of off-target tissue effects, where expression occurs in unintended tissues or organs (in contrast to off-target genetic effects where edits occur at unintended genomic locations within the target cells). This can be minimized when therapies are administered to confined spaces, such as the eye or joints,21 and through the incorporation of tissue-specific promoters to drive gene expression.22
(4) Animal model: The fourth consideration is the selection of an appropriate animal model to obtain clinically relevant insights into the efficacy of the therapeutic. There are several challenges associated with testing gene therapies in animal models, including replication of human diseases, response to therapy, and differing immune reactions. However, as a result of advances in genome editing technologies, many animal models with specific alterations that are able to replicate clinical phenotypes have been generated.23 In addition, the use of human immune system mouse models has improved the translatability of animal studies to the human setting.24
(5) Study design: Multiple factors need to be considered when planning a preclinical study, including appropriate controls, number of animals per condition, testing across different dosages, and the times at which samples are taken.25 When utilizing viral vectors for delivery, gene expression can begin after 1–2 weeks, and extend for months, during which samples are assessed.26
Studies must also set endpoints and conduct measurements of clinically relevant phenotypes to assess the treatment's safety and efficacy. Toxicology studies establish potential local and systemic toxicities, safety and feasibility of the delivery system and procedure, immune response against the vector and delivered construct, and the potential for reproductive toxicity. Efficacy is examined via biodistribution studies, whereas clinical signs relevant to the disease are measured.20
(6) Investigational New Drug preparation: An IND application can be prepared once preclinical experiments are analyzed, demonstrating safety and efficacy of the therapy. This application contains information about the animal pharmacology and toxicology, manufacturing and clinical components, and materials and protocols,25 and it is submitted to regulatory agencies as a part of advancing into clinical trials.
Next, we discuss CRISPR-Cas-based gene therapeutics development in the context of this cascade. We highlight the progress made so far, as well as discuss pending challenges and approaches to overcoming them.
Challenges in Developing CRISPR-Cas Therapeutics
Payload
Among the most likely candidates for translating CRISPR-Cas components into functional therapeutics are the RNA-guided DNA nucleases, Cas9 and Cas12a; the RNA-guided RNA nucleases RCas9 and Cas13; and variants of these proteins. Different therapeutic modalities have been built with orthologs and variants from these Cas families, which together unlock a variety of clinical applications. The native nuclease function of Cas9 or Cas12a can be directly applied to genome editing. The resultant double-stranded DNA break is repaired by native cellular machinery, typically through the nonhomologous end-joining pathway27–30 or homologous recombination with an endogenous sequence or an exogenously provided sequence. This can be applied to engineer gene knockouts, edit a regulatory region or transcription factor-binding site, create large deletions, or enable gene repair or replacement. Further therapeutic approaches toward genome editing with Cas9 have been demonstrated with the advent of base editing and prime editing, with the former more efficiently used for single-base transitions and the latter showing considerable potential for a wider range of localized edits, although with lower efficiencies.
Targeting diseases at RNA level is appealing as it prevents any permanent off-target effects, making it less risky for clinical applications. RNA-targeting RCas9 and Cas13 have been used for RNA knockdown with a similar or comparable efficiency to RNA interference (RNAi).31,32 Common RNA-editing strategies rely on naturally occurring human adenosine deaminase acting on RNA (ADAR) enzymes. Modification of ADAR2 and fusing it to a catalytically inactive Cas13, known as RNA editing for Programmable A to I Replacement (REPAIR), recognizes and corrects disease mutations without any PAM sequence constraints.33 There is increasing interest in developing Cas-mediated transcript editing into RNA therapeutics with applications in treating viral infection or alleviating symptoms in neurological disease.34–36
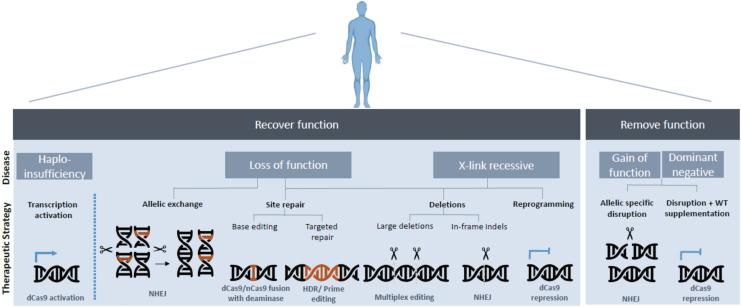
Together, these CRISPR-Cas modalities enable a broad spectrum of strategies toward disease treatment (Fig. 2).
FIG. 2.
CRISPR-Cas therapeutic strategies toward disease-causing mutations.
Delivery
Delivery of the CRISPR-Cas therapeutic cargo represents one of the key challenges in developing a clinically viable CRISPR-Cas therapy.
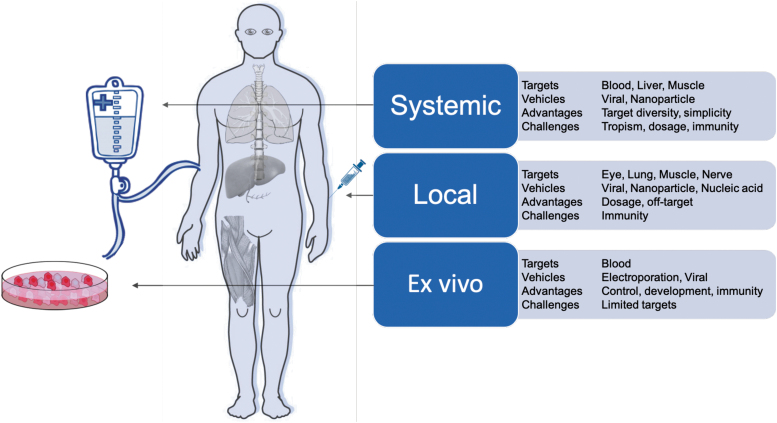
CRISPR-Cas systems can be delivered in three cargo formats: via DNA modalities encoding both the Cas protein and gRNA (such as plasmids, DNA viruses, nanoparticles); RNA modalities encoding the Cas and the gRNA (modified RNA, RNA viruses, in vitro transcribed RNA, nanoparticles); or direct delivery of the purified Cas protein complexed with the gRNA as an RNP or nanoparticle. Each of these cargo formats has been used ex vivo (in cells outside the body) and in vivo (locally within specific tissues or systemically through the body) (Fig. 3). Many of these delivery methods are built on the established framework of traditional gene therapy and, hence, share many of the technological underpinnings and desired properties.
FIG. 3.
Delivery modalities for CRISPR-Cas therapeutics.
The ideal delivery system will likely encompass the following attributes: (1) nonintegrating: limiting unintended integration events in the genome; (2) specific: achieving targeted tissue delivery; (3) low immunogenicity: enabling low toxicity associated with administration and sustained therapeutic effect; (4) option to re-dose: allowing repeated regimens; (5) biodistribution: compatible with target tissue type and tissue accessibility; and (6) scalable: compatible with large-scale manufacturing for both common and orphan diseases. We describe later how these different delivery modalities have been used for CRISPR-Cas and how they might exhibit some of these ideal attributes.
Ex vivo delivery modalities
Ex vivo gene editing allows cell engineering and quality-control regimes that would not be feasible within the human body. It is most compatible with hematological disorders, as blood is much easier to safely remove, treat, and re-infuse than other tissues. A modality successfully used for ex vivo gene editing is Cas proteins complexed with sgRNAs in vitro as RNPs. Unlike DNA- or RNA-encoded CRISPR, RNPs do not rely on cell-driven expression and hence have the quickest onset of activity on administration. In addition, RNPs are not further expressed and degrade over time, offering a transient window of exposure that limits potential off-target effects.37 The most clinically advanced CRISPR-Cas editing strategy relies on ex vivo RNP-mediated editing followed by re-administration of the cells back to the donor.
Advances in the isolation and propagation of hematopoietic stem cells (HSCs) and immune cells have promoted cellular gene therapy as a viable option for the treatment of some monogenic diseases and malignancies. Early successes were complicated by uncontrolled insertional mutagenesis of the vector, resulting in transcriptional activation of nearby proto-oncogenes, chromosomal translocations, and eventual leukemogenesis.38 Unlike traditional gene therapy requiring sustained transgene expression, CRISPR-Cas systems can achieve therapeutic outcomes by transient expression, allowing for less integrative risk and potentially safer methods of delivery. Current ex vivo CRISPR-Cas mediated clinical trials utilize direct delivery of Cas9 RNP via electroporation (NCT03399448, NCT04035434, and NCT03655678).
Curative approaches for β-hemoglobinopathies include allogenic hematopoietic stem cell transplant (HSCT) by using HLA-matched stem cells derived from a donor or autologous transplantation of the patient's own HSCs after receiving the gene therapy ex vivo. Allogenic transplantation is challenging due to dependence on finding an immune-matched donor and mitigating the effects of graft-versus-host response. Autologous transplantation with gene-edited cells avoids much of this immune barrier. Although many indications require varying levels of conditioning treatment to reduce the population of native HSCs to create space for donor cells, autologous ex vivo gene therapy may also allow for a reduced intensity of this process for patients.39 The current approach toward HSC gene therapy uses electroporation of CRISPR-Cas mRNA or RNPs to edit HSC genomes.40 If homology-directed repair is required, the template is simultaneously electroporated or transduced by viral vectors.41 Following this approach of ex vivo editing and re-transplantation into the body, the first two patients with β-thalassemia and sickle cell disease were treated in 2019 by using CRISPR-Cas9-edited HSCs and autologous transplantation.
The RNP-based approach has also been successful in progressing toward clinical trials. For instance, Cas9 RNPs were electroporated into HSCs to disrupt BCL11A, a silencer of fetal hemoglobin, to generate CTX100 cells. Disruption of BCL11A de-represses fetal hemoglobin expression and rescues sickle-cell defects.42,43 Another therapeutic candidate, CTX110, utilized the multiplexing capability of CRISPR-Cas systems to edit CAR-T cells at multiple loci. This approach results in replacement of the native T cell receptor (TCR) with an anti-CD19 CAR as well as knockdown of MHC I expression via targeting β2M (NCT04035434). Positioned as “off-the-shelf” therapy, using allogenic donor T cells could reduce the cost of single batch production. Another ambitious study used Cas9 RNPs to disrupt PDCD1, TRAC, and TRBC, and it introduced transgenic NY-ESO-1-specific TCR to generate engineered T cells for a clinical trial last year.5 The controlled environment of cell engineering and quality control (QC) relative to in vivo administration is the reason that ex vivo pipelines have been among the first to progress to the clinic, and it will inform the uptake of genome editing for in vivo use.
In vivo delivery modalities
Adeno-associated viruses
Viral vectors are the most common in vivo delivery vehicles for CRISPR-Cas. Viruses have substantial differences in their safety profiles, with AAVs having one of the more favorable profiles. The recently approved gene therapies Luxturna3 and Zolgensma4 deliver their therapeutic transgenes via AAVs. Soon after, the first in vivo CRISPR-Cas therapy entered a clinical trial, in which AAV5-CRISPR-Cas9 was delivered into the eye for safety and efficacy evaluation (NCT03872479). These clinical and commercial successes make AAVs an attractive choice for upcoming therapeutic development pipelines.
AAVs are replication-defective viruses that naturally infect humans and nonhuman primates (NHPs). After infection, the DNA carried by AAVs achieves long-term episomal expression. AAVs are endemically found and have several variants that exhibit different tropism for various tissues, such that a serotype can be selected to best target the tissue and disease indication of interest.44 Significant work has been devoted to AAV capsid protein engineering. For example, rational capsid engineering has been used to increase neuronal transduction by mutating surface-exposed tyrosine residues and phosphorylating threonine residues on the AAV2 capsid.45 Altering vector capsids can also increase tropism toward tissues that are challenging to target, such as microglia.46 Chimeras with new functions can be derived from peptide-domain exchanges among different serotypes, such as AAV2.5, which has improved muscle transduction. Altering the immune profile away from both parental AAV1 and AAV2 serotypes offers a potential strategy for immune bypass and therapeutic re-dosing.47 More complex chimeric capsid libraries can be generated by DNA shuffling of capsid sequences of multiple AAV serotypes to generate greater variation and accelerate the evolutionary process.48 Novel capsids have also been generated by using polymerase chain reaction (PCR)-based mutagenesis and subsequent selection for specific attributes.49 For instance, directed evolution has been an efficient approach to derive tissue tropisms in induced pluripotent stem cells (iPSCs)50 and human ciliated airway epithelium.51
Viral protein engineering has also been used to improve the safety of gene therapy vectors. Even though infection with AAVs is only mildly immunogenic and not associated with disease, the remaining immunogenicity still represents a significant safety and efficacy challenge. The effective use of AAV vectors can be compromised by a pre-existing immune response from natural exposure to AAVs in early life or from prior AAV-mediated therapy.52,53 Adaptive immune response to AAVs leads to production of neutralizing antibodies (NAbs) that block transduction and clear the viruses, whereas adaptive cellular response by CD8+ cytotoxic T lymphocytes (CTL) leads to the eradication of transduced cells and can attenuate treatment efficacy and impose safety risks. One strategy used to overcome these issues involves engineering the AAV capsid epitopes to avoid antibody and T cell recognition. Immune-reactive epitopes have been mapped for some serotypes, and capsid mutagenesis can reduce binding and neutralization effects.54,55 However, it is not always straightforward to engineer epitopes while maintaining desired functionality. Often, such modifications result in loss of function, as exemplified by a study where an AAV9-specific neutralizing epitope was mapped to capsid residues conferring liver tropism; unfortunately, mutations did not provide a successful tradeoff in evasion from polyclonal antibodies.56
In addition to induced or pre-existing adaptive immune responses, AAVs trigger a natural innate immune response through recognition of pathogen-associated molecular patterns inherent to AAV biology.57 Multiple studies have elucidated some of the mechanisms of innate immune activation by AAVs, including binding by TLR9 of unmethylated CpG dinucleotides in the AAV genome, leading to activation of the MyD88/NF-κB pathway,58–60 and recognition by MDA5 of long dsRNAs created by bidirectional transcription from the AAV episome.61 These pro-inflammatory signals result in substantial transcriptional changes, inducing an antiviral state within the target cell, and expression of immune-stimulating cytokines that allow for a robust adaptive immune response.62
In the clinic, patients' cytokine levels are routinely monitored, and sometimes they are modulated with corticosteroids.63 In contrast to other viral vectors, AAVs have not demonstrated a clinically dangerous level of innate immune activation in humans, although certain cell types may be susceptible to damage, thus reducing clinical efficacy.64 Nevertheless, minimizing innate immune induction remains an important goal in AAV vector biology to maximize patient safety and limit adaptive immune induction.
More recently, AAV engineering has expanded beyond protein engineering to include chemical and biophysical methods. Chemical modification introduces synthetic motifs to the AAV surface. An early chemical approach masked exposed arginine residues by a naturally occurring glycation reaction.65 Recently, a site-specific approach was developed by using unnatural amino acids on AAV capsids to attach synthetic ligands through biocompatible small-molecule chemical reactions termed “click chemistry.” This strategy was used to tether ligands on the AAV capsid to achieve tissue-specific targeting and protection against antibodies.66,67 Another approach involves encasing the AAV in a membrane exosome, allowing for greater customization of the external layer of the drug. Exosome-enveloped AAV vectors achieved enhanced transduction and protection against pre-existing neutralization antibodies.68–71 A recent study demonstrated the stabilizing effects of tetraspanin CD9 in increasing exosome production.72 Although it is an attractive solution, manufacturing methods for scalable production of exosomes will be challenging.
Nanoparticles
Nonviral delivery systems are also very promising. Various nanomaterials such as polymers, lipids, proteins, and metals have been explored for in vivo delivery. Following extensive research on LNPs, there are currently more than 10 FDA-approved uses of LNPs for drug delivery.73 Onpattro is a recently approved multi-dose gene therapy utilizing LNP to deliver small interfering RNA for the treatment of polyneuropathies.
LNPs typically comprise cationic lipids that facilitate endocytosis and are released into the cytoplasm and PEGylated lipids that facilitate stabilization and prevent nonspecific interaction.74 PEGylation also reduces immunogenicity and increases circulation time.75 Recently, an LNP-based delivery system incorporating helper lipid and PEG-DMG to encapsulate Cas9 mRNA and modified sgRNA was able to produce durable editing of ∼70% in the liver, benefiting from a multiple dosing regimen.76 Other studies targeting the liver by LNPs showed robust knockdown77 and an HDR-editing rate of 6%.78 Beyond the liver, however, applications might be more limited: Lipid particles are usually taken up and metabolized in the liver during systemic circulation,79 thereby constraining the targetable tissue types for LNPs.
Gold nanoparticles can be used to bind Cas9 RNPs and conjugate with 5′-thio ssDNA for further hybridization with DNA repair templates.80 The nanoparticle is then coated with negatively charged silica for cationic polymer PAsp(DET) encapsulation, after which cytoplasmic glutathione releases the contents from the gold nanoparticle.81 Intramuscular injection of CRISPR Gold Cas9 RNP with template DNA was able to correct 5.4% of dystrophin gene mutations. Intracranial injection of CRISPR Gold Cas9 RNP resulted in localized gene editing of 14.5% of the GRM5 gene and restored 40–50% of the protein's production in the brain and other tissues, rescuing the effects of ASD in a mouse model.82 The gold nanoparticles are well tolerated in the neurons; however, the long-term accumulation and elimination kinetics of these nanoparticles remain to be evaluated.
Ribonucleoproteins
Beyond use as delivery vehicles, established protocols for large-scale production of proteins make direct delivery of the Cas9 RNP complex an attractive clinical proposition. Many polymers are being evaluated for coupling with the Cas9 RNP complex, which has a heterogeneous charge distribution. A mix of cationic and anionic monomers with imidazole can fully encapsulate the RNPs while facilitating endosomal escape. The glutathione cleavable link N,N′-bis(acryloyl)cystamine around the RNP enables cytosolic release in the presence of glutathione, but to boost the efficacy of delivery and therefore editing, a further attachment of tissue-specific ligands to one end of the PEG is necessary.83 Although poly(aspartic) acid (PAsp) based polyplexes are biocompatible with limited toxicity, modified PAsp(DET) bearing 1,2-diaminoethane side chains show higher transfection efficiency.84 Recently, PAsp(DET) assembled nanoparticles were able to effectively deliver Cas9 RNP complexes into muscle and brain tissues.80,82 A screening effort also identified poly(aspartic) acid polymer (PAsp) analogs to efficiently deliver Cas12 RNP in mice muscle fibers.85
Administration
Manufacturing and scale-up challenges
In general, manufacturing of pharmaceutical agents must conform to current Good Manufacturing Practice (cGMP). This often refers to a set of regulatory laws in the U.S. FDA Code of Federal Regulations (CFR) title 21, although other countries and organizations, particularly the WHO and EU, have defined their own similar standards for cGMP. Although the application of GMP may vary depending on a wide range of pharmaceutical agents and production methods, generally it requires the use of dedicated clean production facilities, controlled starting materials and cell lines, and multiple product purification steps with accurate testing at each step. cGMP regulation is often focused on process controls and extensive documentation such that a wide variety of issues, should they arise, can be easily detected and corrected. The components of a cGMP production system will vary depending on the agent being produced and must be evaluated on a case-by-case basis.
Nucleic acid manufacturing
The foundation for generating CRISPR therapeutics is nucleic acid, either as synthetic gRNA, a DNA template for gRNA transcription, plasmid encoding the Cas effector protein, or plasmid encoding delivery vehicle components, for example, viral vectors. The manufacturing specifications required for these nucleic acids may differ depending on the application, but they will generally need to conform at minimum to high quality (HQ) grade manufacturing for vector plasmids and template DNA not used in the final therapeutic, and ideally to full cGMP grade for direct applications.
HQ grade DNA can be produced in dedicated facilities by using strains drawn from a research cell bank, utilizing two to three chromatography steps to eliminate linear, open, and chromosomal DNA and remove LPS and other cellular contaminants. The main differences when upgrading to cGMP production are the necessity of a GMP cell bank starter, a dedicated GMP facility, a validated quality assurance system with in-process control, and full documentation procedures (For a more thorough discussion of these procedures and QC validation requirements see Ref.86).
Despite these stringent requirements, the production of cGMP-compliant, therapeutic-grade nucleic acid is much easier than recombinant protein. Fewer purification steps are required, as the reaction mixtures are much less complex than those involved in bacterial or mammalian cell culture. In addition, similar to protein therapeutics, the production of DNA- and RNA-based therapies has advanced rapidly in the past two decades due to excitement surrounding the potential for nucleic acid vaccines and RNAi drugs. Utilizing many of these same facilities and procedures, several companies offer cGMP preparations of sgRNA for clinical and preclinical CRISPR research, as well as HQ and cGMP viral vector preparations. The cost for these materials is manageable, except for particular cases requiring truly massive quantities of DNA, such as low-yield AAV vectors needed for systemic delivery. Nucleic acid production with cGMP guidelines are a feasible and preferred option for more typical RNP and high-yield AAV serotype delivery modalities.
Manufacturing of nonviral RNPs
The current nonviral formulations for CRISPR-Cas range from simple to difficult for manufacturing, with each carrying its own particular set of challenges for creating therapeutic-quality products. The most clinically advanced nonviral delivery modality is also one of the simplest to produce, with electroporated RNPs consisting of only two purified components, namely a gRNA and a Cas protein. The in vitro transcription or de novo synthesis of gRNAs is well established. Cas proteins such as Cas9, Cas12a, or engineered variants would be recombinantly produced via scalable fermentation in pipelines similar to thousands of other protein therapeutics. After production in fermentation vessels, the resultant product is run through multiple chromatography steps to concentrate the active product and remove impurities.
Much of the initial recombinant protein production for pharmaceuticals was dominated by classic producers such as Escherichia coli and Saccharomyces cerevisiae.87 Since then, most biopharmaceutical production has been deployed by using mammalian cell lines, especially Chinese Hamster Ovary cells or human cells, due to the superior protein-folding intracellular environment and post-translational modifications.88 Still, microbial production retains an important niche owing to its advantages in rapid growth, low nutritional requirements, and ease of genetic manipulation.89 Indeed, Cas effector protein production seems to fit nicely into this niche based on their bacterial origin and independence of complex folding cofactors or post-translational modifications. Process improvements in bacterial production, such as the deployment of an endotoxin-free E. coli strain,90 metabolic engineering efforts,91 and adaptive laboratory evolution,92 will yield additional improvements that are directly applicable to CRISPR-based protein therapeutics. Further, new exploration of alternative manufacturing methods, such as co-production of Cas9 protein and sgRNA in E. coli, may open up new avenues of inexpensive drug production.93
Nevertheless, current manufacturing methodologies are poised to implement recombinant Cas proteins at scale. Commercial recombinant Cas9 preparations, such as Aldevron's SpyFi™ Cas9 nuclease, are available. Importantly, associated procedures should be equally amenable to production of other Cas9 effectors, including dCas9, Cas12a, and variants thereof. Pharmaceutical companies have been highly incentivized to optimize scaled protein production environments,94 including moving to continuous flow rather than batch setups,95 due to lucrative demand for protein therapies, especially monoclonal antibodies. Process advancements brought about by this economic landscape will translate well to CRISPR-based protein drugs.
The RNA component of the CRISPR-Cas RNP can comprise a synthetic crRNA and tracrRNA pair as in the native bacterial system,96 a synthetic combined sgRNA,97 or an in vitro transcribed sgRNA.98 Single gRNA constructs are preferable due to simplicity and elimination of the duplex formation step. Both synthetic and in vitro transcribed gRNAs have associated benefits and drawbacks. Chemically synthesized gRNAs are amenable to modification of one or more of the nucleotide bases, which provides advantages such as increased stability99 and reduced immunogenicity.100 New modifications to further optimize these parameters are currently being explored.101,102 The potential advantages of in vitro transcribed gRNA production include simple and economical scale-up to large volumes, as the generation of templates can be done with PCR. Both in vitro transcribed and chemically synthesized gRNAs are being evaluated in ongoing clinical trials.5,103
AAV manufacturing
AAVs are the prime viral delivery vehicles being explored for in vivo CRISPR therapies given several beneficial properties, including low immunogenicity, nonintegration, different serotypes available for different tissue tropism, and a proven clinical record. Nonetheless, despite years of intense research and development, manufacturing challenges still exist.
Typically, AAVs are produced by using a triple transfection method in which plasmids encoding (1) the recombinant genome to be delivered, (2) the viral packaging genes, and (3) adenoviral helper genes are transfected into cells, most commonly human embryonic kidney (HEK)293 lines by using chemical agents such as polyethylenimine. Many AAV products for clinical trials have been produced by using these methods (NCT02122952, NCT25322757, NCT21031578, NCT27453480). Initially, this was done by using adherent HEK293 cells cultured in 2D on stacks of flasks. These formats can suit typical doses for early trial phases that range from 1011 to 1013 vector genomes (vg) per patient. However, achieving titers high enough for later phases or larger doses would mean hundreds of flask stacks and many labor months.104 This obstacle is most pressing for systemic disease targets such as Duchenne muscular dystrophy (DMD), which require larger doses to achieve good efficacy. Current recommendations for clinical doses are as high as 1015, even 1016 vg per patient. The product volumes required for clinical trials, especially critical phase III trials with large patient cohorts, are simply impractical by using traditional cell culture plasticware.
As a result, both academic and industrial facilities have also adopted more scalable platforms to produce AAVs, particularly in fixed-bed bioreactors with adherent HEK293 cells105 and bioreactors with suspension HEK293 cells.106 Both have provided scalability without a complete process overhaul. Still, yields using these methods have largely been unable to exceed 1014 vg per liter, necessitating thousands of liters for larger clinical trials. In addition, scaling up requires nontrivial quantities of GMP-grade plasmids, which face scalability problems of their own.107 One approach to the transfection scalability issue is to deliver the necessary genetic material by using viral carriers, typically a herpes virus or baculovirus, rather than by chemical transfection.108 This approach has led to a moderate increase (twofold to fivefold) in yield,109 and interestingly, an increase as well in AAV product infectivity in the case of herpes virus production, potentially due to superior genome loading into AAV capsids in the presence of helper genes from the herpes virus.110
This yield challenge has also prompted the use of baculovirus-infected insect Sf9 cells for AAV production, which provides a much higher yield than HEK293 and has been used in multiple clinical trials and production campaigns.111–113 Nonetheless, a recent revelation that Sf9-produced AAVs differ qualitatively from HEK293-produced AAVs creates a potential roadblock that requires further investigation.114
Additional developments, including continuous flow-based production methods and optimization of producer cell lines, may further increase AAV yields. Still, all AAV production methods require major downstream processing, including multiple filtration steps, size exclusion chromatography, ion exchange chromatography, and ultracentrifugation, to achieve the required purity and enrich for properly packaged, infectious vectors.112,115 Even applying numerous recent technical advancements, scalability of AAV vector production remains a huge challenge, especially for relatively common diseases requiring large effective doses such as DMD.
Pre-IND and preclinical development of CRISPR-Cas therapeutics
Successful gene therapies require preclinical validation in animal models. Different preclinical models have been developed to collect data in three main areas: proof of concept for efficacy, determining dose ranges for the chosen route of administration, and evaluating toxicity and safety profiles. For AAV-CRISPR-Cas, four main organs have been targeted most frequently and successfully. This initial success builds on more some three decades of understanding in gene therapy and will itself guide future CRISPR therapeutic development.
Muscle
There is significant morbidity and mortality in hereditary disorders of the muscle, and gene therapy offers promising outcomes for these unmet needs. Many studies have demonstrated the effectiveness of using skeletal muscles as protein factories to express transgenes, contributing to the first gene therapy product (Glybera) being officially approved.116
DMD is one of the most common hereditary muscular diseases in children and is caused by mutations in dystrophin, the largest gene in the human genome. Progress in gene-editing technologies has raised hopes for successful restoration of the dystrophin gene. Conventional gene therapy has been challenging due to the AAV packaging limit; hence, efforts have focused on truncated versions of dystrophin for gene replacement, such as microdystrophin. The therapeutic strategy of AAV-mediated gene therapy in muscles appears safe in preclinical and clinical studies, but so far the therapeutic effects have been limited in DMD individuals.47 Patients with large frameshift deletions also risk T cell-directed destruction against the new epitopes.117,118 The lack of long-term effects could also be due to constant muscle degeneration and regeneration, diluting the expression of the nonintegrated transgene with time. Another strategy is the use of CRISPR-Cas to permanently restore endogenous dystrophin expression. Diverse CRISPR-Cas gene-editing strategies have been employed to treat muscular dystrophy mouse models, including exon skipping, exon deletion, adenine base editors, and HDR (Table 1).119–122
Table 1.
In vivo CRISPR-Cas strategies demonstrating functional rescue in animal models
| Organ | Human disease | Editing strategy | Disease model | Administration | Ref. |
|---|---|---|---|---|---|
| Eye | Retinitis pigmentosa, autosomal recessive, PDE6B mutation | SpCas9/RecA mediated HDR | Pde6b Y347X, nonsense mutation mice | SpCas9, sgRNA, RecA-MS2 Plasmid and ssDNA donor, subretinal and electroporation | 244 |
| Retinitis pigmentosa, autosomal dominant • P23H mutation (class 2) • Class 1 |
Knockdown Rho | Rho-P23H transgenic mice | Cas9-sgRNA plasmids, subretinal and electroporation | 245 | |
| Knockdown mutant Rho | Rho-P23H heterozygous mice | SpCas9-VRQR plasmid and sgRNA plasmid, subretinal and electroporation | 143 | ||
| Rho S334ter rat | Cas9-sgRNA plasmid, subretinal and electroporation | 141 | |||
| Knockdown of transcription factor Neural Leuzine zipper (Nrl) | Rho−/−, Rd10, Rho-P347S mice | AAV8-RK-SpCas9, AAV-sgRNA, subretinal | 246 | ||
| LCA, autosomal recessive • Type 10, IVS26 mutation in CEP20 • Type 2, RPE65 mutation |
NHEJ in-frame indels or HR | Humanized CEP290 IVS26 knock-in mice, NHP | AAV5-sgRNA-GRK1-SaCas9, subretinal | 222 | |
| Rpe65 nonsense mutation rd12 mice | AAV9-EFS-SpCas9, AAV9-sgRNA-Rpe65-donor, subretinal | 247 | |||
| Meesman epithelial corneal dystrophy, dominant negative, KRT12 mutation | Knockdown, allele specific, Deplete mutant KRT12 | KRT12-L132P humanized heterozygous mice | SpCas9 and sgRNA plasmids, Intrastromal ocular | 142 | |
| Neovascular age-related macular degeneration | Knockdown, NHEJ mediated disruption of Vegfa or Hif1a | Laser-induced choroidal neovascularization in mouse eyes | AAV8-CMV-SpCas9, AAV8-sgRNA, subretinal | 146 | |
| AAV9-EFS-LbCpf1-crRNA, intravitreal | 248 | ||||
| Neurological | Huntington's disease (CAG trinucelotide), expansion mutant HTT | Knockdown HTT | HD140Q-knock-in mice, human HTT exon 1 with 140 CAG repeats (homozygous and heterozygous) | AAV-MECP2-Cas9, AAV-gRNA, intracranial | 249 |
| Knockdown mutant HTT | R6/2 mice, transgenic mice, human HTT exon 1 with 115–150 CAG repeats | AAV1-hSyn-SaCas9-sgRNA, intracranial | 196 | ||
| ALS, autosomal dominant, SOD1 mutations | Knockdown human SOD1 | Transgenic human SOD1 G93A mutation ALS neonatal mice | AAV9-CMV-SaCas9-sgRNA, intravenous or intracerebroventricular | 193,194 | |
| BE3 disruption of SOD1 | AAV9-CAG-N-Int-CBE-U6-sgRNA, AAV9-CAG-C-Int-CBE-U6-sgRNA, inthrathecal | 195 | |||
| Fragile X syndrome | Knockdown GRM5 | Fmr1 KO mice, FXS mice model | CRISPR Gold nanoparticle Cas9 RNP and donor DNA, intracranial | 82 | |
| Alzheimer's disease | Knockdown Bace1 | Five familial Alzheimer's disease transgenic mice, APP knock-in Alzheimer's disease mice | Amphiplilic R7L10 peptide nanocomplex Cas9 RNP, intracranial | 7 | |
| Dravet Syndrome, haploinsufficiency, SCN1A loss of function mutation | dCas9-Vp64 mediated Scn1a gene activation (both WT and mutant) | Scn1a heterozygous Dravet Syndrome mice | AAV9-dCas9-VP64, AAV9-sgRNA, intracerebroventricular | 250 | |
| Severe obesity, haploinsufficiency, SIM1 or MC4R | dCas9-Vp64 mediated gene activation of Sim1 | Sim1/Mc4r heterozygous mice | AAVDJ-CMV-dCas9-Vp64, AAVDJ-sgRNA, hypothalamic | 251 | |
| Liver | Hemophilia B, X-linked recessive | HDR, insert hFIX-padua exon 2–8 in mFIX exon 2 | FIX Knockout mice | AAV8-Cas9, AAV8-sgRNA-Donor DNA | 165 |
| HDR, insert into mFIX into murine ROSA26 safe harbor | R333Q Hemophilia mice | Ad5-Cas9-gRNA, Ad5-EF1α-mFIX, intravenous | 252 | ||
| Hemophilia B, Y371D mutation in FIX | HDR | Y381D Hemophilia B mice | Naked DNA, SpCas9, ssODN-HDR Donor, intravenous | 253 | |
| Hemophilia A, X-linked recessive | Insert human B-domain deleted FVIII in intron 13 of liver-specific albumin locus | FVIII Knockout mice | AAV8-SaCas9-sgRNA, AAV8-donor DNA | 166 | |
| OTC deficiency, X-linked recessive | HDR, correct point mutation | spf/ash heterozygous OTC neonate mice | AAV8-TBG-SaCas9, AAV8-sgRNA-donor DNA, intravenous | 168 | |
| HDR, insert codon-optimized human OTC into intron 4 of mouse OTC locus | AAV8-TBG-SaCas9, AAV8-sgRNA-TBG-hOTCco, intravenous injection | 167 | |||
| Hypercholesteolemia | BE3, disruption of mouse W159 Pcsk9 | WT C57BL/6 mice | Ad-BE3-sgRNA, retro-orbital | 242 | |
| BE3, disruption of human W159 PCSK10 or NHEJ-mediated disruption of PCSK10 | Humanized PCSK9 knock-in mice | Ad5-Cas9-sgRNA or Ad5-BE3-sgRNA, Intravenous | 243 | ||
| Atherogenic dyslipidemia, homozygous familial hypercholesterolemia | BE3, disruption of Q135 Angptl3 | Hyperlipidemic Ldlr-knockout mice | Ad-BE3-sgRNA | 254 | |
| Phenylketonuria, autosomal recessive | BE3 | Homozygous point mutation in Pah-F263S, hyperphenylalaninemia mice | AAV8.N-int-BE3 and AAV8.C-int-BE3, intravenous | 255 | |
| Hereditary tyrosinemia type I, autosomal recessive, FAH loss of function mutation | HDR | Fah homozygous mutation in exon 8 mice | SpCas9 mRNA by LNP and AAV8-sgRNA-donor template, intravenous | 78 | |
| HDR with nCas9 | Homozygous 10-bp deletion in exon 2 of Fah in rat HTI model | Ad-nCas9 and Ad-donor template, intravenous | 256 | ||
| Base editing, ABE6.3, tRNA nCas9-adenosine deaminase | Fah homozygous mutation in exon 8 mice | Plasmid by hydrodynamic injection or mRNA/sgRNA by LNP, intravenous | 257 | ||
| Hereditary tyrosinemia type I, autosomal recessive, compound heterozygous mutations in FAH | Allelic exchange by targeting intron 7, NHEJ | compound heterozygous exon 5 insertion/exon 8 mutation in Fah, HTI mouse model | AAV9-SpCas9 and scAAV-sgRNA, intravenous | 258 | |
| Muscle | DMD, exon 50 | Reframing or Exon skipping | ΔEx50 C57/BL6J mice | AAV9-CK8-SpCas9 and AAV9-sgRNA-51, intraperitoneal | 259 |
| ΔEx50 dogs | AAV9-CK8-SpCas9 and AAV9-sgRNA-51, intravenous | 125 | |||
| DMD, exon 44 | Reframing or Exon skipping | ΔEx44 C57BL/6 mice | AAV9-CK8-SpCas9 and AAV9-sgRNA, intraperitoneal | 260 | |
| AAV9-CK8-SpCas9 and scAAV-sgRNA |
126 | ||||
| DMD, exons 45–55 | Excise exon 52 and 53 | mdx4cw mice, nonsense mutation in exon 53 | AAV6-CK8-SpCas9 and AAV6-sgRNA or AAV6-CK8-SaCas9-sgRNA, retro-orbital | 120 | |
| DMD, nonsense mutation in exon 23 | Excise exon 23 | Mdx mice, pt mutation in exon 23 | AAV9-CMV-SaCas9 and AAV9-sgRNA | 119 | |
| AAV8-CMV-SaCas9 and AAV8-sgRNA, Intravenous | 124,261 | ||||
| AAV9-CMV-SaCas9 and AAV9-sgRNA, intravenous | 262 | ||||
| AAV9-miniCMV-spCas9 and AAV9-sgRNA, intraperitoneal | 263 | ||||
| Excise exon 21–23 | Ad-Cas9-sgRNA | 264 | |||
| HDR | CRISPR Gold nanoparticle Cas9 RNP and donor DNA, intramuscular | 80 | |||
| NHEJ, indels to result in reframing | DMD KO C57/BL6J mice, ΔEx23 | AAV9-Spc5-12-CjCas9-sgRNA, intramuscular | 265 | ||
| Wolff-Parkinson-White syndrome, autosomal dominant inherited disease, H530R mutation in PRKAG2 | Disrupt mutant allele | Heart specific H530R transgenic mice, Heterogeneous H530R PRKAG2 knock-in mice | AAV9-Cas9, scAAV9-sgRNA, intravenous or intraventricular | 8 | |
| Ear | Dominant progressive hearing loss (DFN36), TMC1 | Knockdown mutant Tmc1 | Beethoven mice, heterozygous missense mutation in Tmc1 | Cas9-sgRNA RNP, cationic lipid complex, inner ear | 266 |
| Anc80L65-CMV-SaCas9-sgRNA, inner ear | 267 | ||||
| Multiorgan | MPS type I, autosomal recessive | HDR, insert Idua into murine ROSA26 safe harbor | Idua-knock-out MPS I mice | PrecisionX CRISPR/Cas9 SmartNuclease™, Idua donor vector, cationic liposomes, intravenous | 182 |
AAV, adeno-associated virus; ALS, amyotrophic lateral sclerosis; APP, amyloid precursor protein; Bace1, beta-secretase 1; DMD, Duchenne muscular dystrophy; FIX, factor IX; IDUA, α-l-iduronidase; LCA, Leber congenital amaurosis; LNP, lipid nanoparticle; MPS, mucopolysaccharidosis; NHEJ, nonhomologous end-joining; NHP, nonhuman primate; OTC, ornithine transcarbamylase; RNP, ribonucleoprotein; scAAV, self-complementary AAV.
As DMD affects muscle groups throughout the body, the challenge is to develop a single injection that is sufficient for whole-body therapy. Treating DMD systemically via intravascular injection allows targeting of all muscle groups and has been shown to be less immunogenic.123 Durable levels of dystrophin were detected in cardiac and skeletal muscles in systemically treated neonatal mdx mice with AAV-SaCas9.124 Moreover, systemic delivery of AAV9-SpCas9 and AAV9-sgRNA corrected frame-shifted mutant DMD, restoring up to 90% dystrophin expression in a canine model of DMD.125 However, intravascular administration will require high doses of AAV to reach therapeutic levels of gene editing in clinics. The Sarepta Therapeutics clinical trial NCT03375164 efficacy dose is 2 × 1014 vg/kg, where a single treatment of a patient will require 1015 vg of AAV. One approach to reduce AAV dose is the use of self-complementary AAV (scAAV), which in one study has shown up to 20-fold dose reduction.126 However, the large payload requirements with CRISPR-Cas raise limitations on the potential use of this strategy.
Eye
Gene therapy in the eye is attractive due to the anatomical compartmentalization that enables targeting of specific cells with minimal exposure to other organs and the immune system. Specific cell types in the eyes such as retinal pigment epithelium, ganglion, and photoreceptors can be effectively transduced by AAVs, and surgical processes afford access to the various structures in the eye.127 Non-invasive methods have been established for evaluating therapeutic effects, whereas the contralateral eye can be used as a convenient control. Lastly, numerous genetic animal models have been generated for diseases in the retina, paving the way for gene therapy development.128
Leber congenital amaurosis (LCA) is a childhood-onset blindness caused by mutations in at least 1 of 18 different genes. Type II is the most studied form, with mutations in the RPE65 gene. Gene replacement of RPE65 in a blind canine model demonstrated improved vision through simple behavioral tests. More importantly, the therapy demonstrated cortex recovery and response to visual stimulation despite prolonged visual deprivation in the RPE65-mutant dogs.129,130 The results suggested safety of the AAV2 subretinal injection and some level of vision restoration when RPE65 was first introduced in LCA patients.131–133 Re-administration of the therapy in the second eye was also successful in large animal studies despite elevated serum NAb toward AAV2134,135 and proved predictive of the results in human clinical trials.136 AAV2 transfer of RPE65 showed therapeutic efficacy in phase III randomized trials and is marketed as Luxturna.
After the landmark success of Luxturna, the first in vivo CRISPR-Cas gene editing human trial program was developed by Editas Medicine to treat LCA type 10. In this case, subretinal injection of AAV5 delivering SaCas9 is driven by a human rhodopsin kinase (hGRK1) promoter to repair the splicing defect of CEP290. hGRK1 promoter is able to drive robust transgene expression in both cones and rods in NHPs and mice.137–139
A wave of new treatments is being developed for inherited retinopathies. In particular, the CRISPR-Cas editing approach is attractive for retinitis pigmentosa (RP) as 25–30% of the cases are due to autosomal dominant mutations.140 CRISPR-Cas mediates allele-specific editing by recognizing the unique PAM derived from point mutations in dominant disorders such as RP and Meesman epithelial corneal dystrophy.141,142 However, many disease mutations such as RHO P23H do not create a novel PAM. One study improved allele discrimination in heterozygous Rho P23H mice by placing a single-base mutation in the spacer region of the sgRNA coupled with the SpCas9 VRQR variant.143 Interestingly, another study reported that increasing wild-type to mutant Rho expression was able to slow retinal degeneration.144 This suggests that therapeutic benefits can be attained despite incomplete mutant disruption and even with some indiscriminate wild-type downregulation.
Chronic eye diseases such as age-related macular degeneration (AMD), the leading cause of blindness, are another prime target for gene therapy. Current treatment requires regular anti-VEGF intravitreal (IVT) injections but with only modest improvements in patients with advanced neovascular/wet age-related macular degeneration (Wet AMD). Previous clinical trials using AAV2-sFLT01, expressing soluble VEGF receptor in Wet AMD patients, showed variable results,145 so other strategies have been developed to reduce angiogenesis. Clinical trial NCT03748784 is using an IVT injection of AAV7 m8-expressing aflibercept (anti-VEGF protein). In addition, a recent CRISPR-Cas approach targeting VEGF-A using AAV8-SpCas9 suppressed 31% of laser-induced choroidal neovascularization in mice.146 Overall, the pathogenesis of AMD is complex when compared with monogenic disorders and a multimodal approach enabled by CRISPR technologies might be beneficial.
Successful gene therapy treatment is also dependent on effective administration at the target tissue and the resultant immune response. Subretinal injections are efficient for targeting photoreceptors and RPE, whereas IVT injections target cells in the inner retina such as ganglion cells.147 Although IVT administration of AAV provides a broader tissue application in the retina, there is an increased likelihood of generating NAbs against the AAV capsid and transgene as shown in NHPs.148 In addition, T cell responses against AAV2 capsid were also detected during IVT injection in an NHP toxicology study.149
Altogether, studies show that IVT administration increases AAV exposure to the immune system.150 Another consideration is the administration complexity and ensuing complications. The IVT injection is a routine procedure for drug delivery whereas the subretinal injection is more invasive, increasing risk of complications.151 Therefore, multiple studies have tried targeting RPE by using different AAV serotypes via IVT administration. Much research has been done to improve AAV transduction of photoreceptors by AAV capsid engineering or to reduce physical barriers by surgical methods such as vitrectomy or by peeling of the internal limiting membrane.152–154
Liver
The liver is an essential organ that metabolizes and synthesizes intracellular and secreted proteins. Defects in lipid and protein metabolism or detoxification due to liver disease are often destructive to multiple organs, causing complex disorders. Liver-directed gene therapy is an attractive option in the treatment of metabolic diseases, demonstrating safety in acute intermittent porphyria clinical studies as well as monogenic diseases such as hemophilia B.155,156 Hepatocyte-directed gene transfer is further enabled by easy access of the AAV vector through liver sinusoids, as it is small enough to pass through the fenestrae.157
In the treatment of hemophilia and other monogenic diseases, AAV vectors are preferred for liver-directed gene therapy as nonviral and retroviral delivery methods only result in transient expression.158,159 The earliest in vivo AAV vector used was AAV2, which has broad tissue tropism, including hepatocytes. Subsequently, AAV8 was shown to be 10- to 100-fold more efficient in transducing hepatocytes.160–162 Further, humans have a lower prevalence of NAb against AAV5 and AAV8 compared with AAV2,163,164 resulting in a shift favoring AAV8 and AAV5 serotypes for liver-directed gene therapies. Liver gene therapy clinical trials have been tested for a small number of diseases, with hemophilia B studies taking center stage.14
Ultimately, hemophilia patients need to maintain at least 1% plasma circulation for effective hemostasis in their lifetime. AAV-mediated transfer of factor IX (FIX) or factor VIII (FIIIV) in liver, especially in children, will result in dilution of the transgene as the hepatocytes divide. This problem can be mitigated by site-directed genomic integration. A gene-replacement strategy using CRISPR-Cas to integrate the transgene into the albumin locus downstream of the endogenous albumin promoter produced sustained FIX and FIIIV expression in hemophilia mouse models.165,166 AAV8-Cas9 mediated integration of human FIX-padua exon 2–8 in exon 2 of mFIX and a codon-optimized FIIIV into intron 13 of albumin, and it developed persistent FIX or FIIIV levels for 7–8 months with no complications.165,166
A similar strategy proved effective in the treatment of ornithine transcarbamylase (OTC) deficiency, an X-linked urea cycle disorder where a codon-optimized human OTC was inserted into the intron 4 of mouse OTC locus by using AAV8-SaCas9.167 Conversely, Cas9-mediated HDR to correct the mutation had resulted in large deletions and affected endogenous OTC expression in the spf/ash heterozygous OTC mice.168 Cas9 integration of OTC produced both rapid and prolonged gene expression when compared with conventional gene replacement therapy.167
Preclinical studies have shown efficacy in liver-directed gene transfer for lysosomal storage diseases (LSDs) such as Gaucher disease, Fabry disease, and glycogen storage disease type Ib and II.169–172 LSDs are mostly monogenic and include more than 40 different metabolic diseases. The spectrum of disease severity also spans from disorders in the central nervous system (CNS) to systemic multi-organ pathology, as the enzymes are ubiquitously expressed.173 Current treatments such as enzyme replacement therapy (ERT) and HSCT have limited efficacy, especially for those with CNS disease.174,175 Prolonged ERT is associated with poorer quality of life, and intravenous ERT has been insufficient to remedy neurological manifestations,176 whereas HSCT is associated with transplant-related morbidity and mortality rates of 10%.177–179
Interestingly, recent studies in LSDs were able to bring about some neurological benefit by using liver-directed gene editing. The effects in CNS were likely due to enzymes crossing the BBB rather than gene editing in the brain, as editing was driven by a liver-specific promoter. This was demonstrated in AAV-SaCas9-mediated transgene integration into the albumin safe harbor locus in Sandhoff mice.180 Sangamo Therapeutics, in a promising preclinical study, used an AAV2/8 zinc finger nuclease editing strategy to insert α-l-iduronidase (IDUA) into the albumin locus, resulting in cognitive improvements in mice with Hurler syndrome (the most severe form of mucopolysaccharidosis [MPS] I).181 In vivo correction of MPS I has been explored by using CRISPR-Cas to insert the Idua gene in a murine safe harbor locus via HDR.182 IDUA activity was detected in multiple tissues with the use of cationic liposomes, but it could not cross the BBB.182 However, the BBB can be crossed with the use of specific AAV serotypes.183
Central nervous system
Neurological disorders are difficult to treat due to the complexity of the CNS. Although conventional therapy has limited access to the brain, specific AAV serotypes are able to cross the BBB due to their unique tissue tropism.183 A noninvasive mode of delivery by intravenous injection of AAV9 can transduce the spinal cord, including motor neurons and astrocytes in mice, large animals, and NHP.184,185 Intra-cerebrospinal fluid (CSF) delivery is also able to provide widespread distribution in the CNS and reduce transduction of periphery organs.186,187 The third approach is to directly deliver to target regions, limiting therapy to the site of administration; hence, broader delivery will require multiple injections and surgery.188
Zolgensma—the approved CNS targeting gene therapy for SMA—delivers scAAV9-SMN1 systemically to target motor neurons.4 However, the high vector dose has contributed to an exorbitant price for the therapy. Alternatively, the dose can be reduced with intra-CSF administration by intracerebroventricular and intrathecal injections. Gene transfer in a severe SMA mouse model was able to reduce the dose by 10 times with intra-CSF administration to improve survival.189 Safe and robust widespread transduction in the motor neurons of the spinal cord was also demonstrated by an intrathecal injection of AAV9 in pigs.190 As a result, intrathecal administration of scAAV9-SMN1 is currently being evaluated in clinical trials in SMA type 2 patients (NCT03381729).
Amyotrophic lateral sclerosis (ALS) is characterized by motor neuron loss in multiple regions in the CNS.191,192 Dominant SOD1 mutations are most common in familial ALS; CRISPR-Cas editing of mutant Sod1 using AAV9-SaCas9 improved locomotor functions and survival when administered intravenously or via intracerebroventricular injection.193,194 A CSF-administered cytidine base editor was able to disrupt mutant Sod1 in adult ALS transgenic mice and improve survival.195
Localized administration of CRISPR-Cas therapies in the brain is also essential for neurodegenerative diseases, with neuronal loss in specific areas of the brain such as the striatum in HD, limiting potential off-target effects in other tissues. Intracranial delivery of AAV1-SaCas9 targeting the 5′ end of exon 1 reduced mutant HTT expression in the HD mouse model and improved motor function and benefit survival.196
CRISPR-Cas gene-editing therapy was also demonstrated in common progressive neurodegenerative disorders such as AD. Intracranial delivery of Cas9 nanocomplex to disrupt beta-secretase 1 (Bace1) in AD mouse models reduced amyloid plaque accumulation and cognitive deficits.7 Dominant familial AD caused by mutations in amyloid precursor protein (APP), the site of beta-secretase 1 cleavage, results in accumulation of amyloid plaques. AAV9-Cas9 treatment via hippocampus injection specifically disrupted the mutant APP gene, reducing amyloid-β (Aβ) levels.197 However, a possible limitation of the current treatment strategy is the lack of widespread targeting, as neurons in many areas of the brains are progressively affected in AD. Cas expression in the CNS can be driven by neuron-specific promoters while leveraging AAV tropism to increase target cell specificity.197
Immunogenicity of the payload and vehicle
One of the most critical barriers to successful therapeutic translation is immune interaction. Most significantly, there are concerns that the adaptive immune response could inhibit the efficacy of CRISPR-based gene therapies. This could occur through the action of NAbs against either the Cas effector protein or the delivery vehicle, or through cytotoxic T cell responses leading to specific killing of treated cells.
Antibodies toward AAV and CRISPR-Cas9 have been shown to neutralize and negate editing efficacy in mice with just one previous exposure to the therapeutics.198 This is particularly pertinent for clinical application, because multiple studies have now identified pre-existing immunity in the human population toward the commonly used Cas9 orthologs from S. pyogenes and Staphylococcus aureus,199–202 understandably since both are species that frequently colonize the human microbiome and may cause disease. Identification of circulating antibodies reacting to Cas9 could pose a large hurdle toward RNPs, since these purified Cas9 complexes could be neutralized within the bloodstream. Other Cas-encoding DNA and RNA modalities might generally be less inhibited by this obstacle, since the Cas9 protein will be expressed intracellularly and remain less accessible to antibodies from the circulation.
A more substantial risk, however, is the potential for CTL to recognize Cas-expressing cells through the Cas-derived peptides presented at the cell surface via the MHC class I pathway. Given the appropriate costimulatory milieu generated by innate immune signaling in the CTL, the target cell, and the surrounding tissue, this can lead to the killing of target cells expressing the Cas proteins.
Two in vivo mouse studies have highlighted the potential for anti-Cas9 immunity to inhibit the efficacy of CRISPR-based drugs. In one, a single dose of AAV-delivered Cas9 was shown to be sufficient to generate anti-Cas9 T cells in mice and to induce their killing potential.198 Another study showed that prestimulated anti-Cas9 cytotoxic T cells completely negated the therapeutic effect of Cas9 gene editing within 12 weeks.203 Although no human studies have shown an inhibitory effect of the immune system as yet, Cas9-reactive T cells clearly exist in significant fractions of the population.199,201 The interaction of the immune system with CRISPR-based therapies needs further study, but preliminary data suggest that it could become an obstacle to in vivo CRISPR-Cas therapies. Transient immunosuppression or transient transgene expression or Cas engineering might be necessary to maintain patient safety and efficacy.204
Animal models
Several mouse models harboring disease-causing mutations have been generated for the study of CRISPR-Cas mediated gene editing (Table 1). Knockout mouse models have been essential in the study of inherited genetic diseases, including those affecting the CNS.205,206 Similarly, transgenic models bearing sporadic mutations have been generated for AD and Parkinson's disease.207 Mouse models enable testing of the editing strategy, but they often do not fully recapitulate the extent of the disease. Mdx mice, for example, have a much slower disease progression, shortened lifespan relative to DMD patients, exhibiting cardiomyopathy only in older mice.208 Likewise, AD mice exhibit the main pathology of the disease—Aβ aggregation—but overt neurodegeneration is not captured, nor early cognitive and behavioral symptoms.209 In addition, it is harder to extrapolate results of widespread AAV delivery in mice organs such as the brain to humans due to differences in distribution186 and possibly the large disparity in size.
Overall, advanced human diseases are better recapitulated in large animal models. The DMD pig model, for example, embodies severe disease progression such as premature death and cardiomyopathy.210 Large transgenic animal models of HD also display severe phenotypes and early death when compared with rodent models.211 Larger species such as dogs have a more similar brain disease profile to humans, and they develop naturally occurring LSDs.212,213 Large species are also more suitable for surgical manipulations, especially for neurodegenerative diseases.214
Complexity can arise in the choice of animal species when progressing from small to large animal studies. For instance, vector tropism can differ between animal models as shown in the successful transduction of AAV5 in the mouse brain but not in cats.215 In addition, although AAV-PHP.B has an enhanced ability to cross the BBB and increase CNS transduction in mice, this was not replicated in NHPs.216 Extending preclinical studies to larger animals has been essential in better understanding immune responses toward gene therapy. Canine models have been essential in uncovering possible severe immune response against muscle-directed transgene217 and cellular response toward AAV2 and AAV6 capsid.218 Importantly, large animal studies can also inform immunosuppressive regimen for better therapy outcomes.219 Interestingly, naturally AAV-infected rhesus macaques were unable to induce similar T cell responses to AAV capsids on re-exposure during treatment regimen despite the close phylogenetic relationship to humans.220,221
Readout challenges
Designing animal studies to provide an early readout for efficacy and potency is challenging, as shown in the preclinical study of EDIT-101. Editas Medicine used both a disease mouse model and NHP to measure desired physiological response. Well-established tissues in the eye allow for specific isolation, such as the cultivation of the neural retina of adult cadavers into retinal explant cultures for demonstration of editing efficacy in mature human photoreceptors.222 Neural retinas from the knock-in mouse model of LCA10 were isolated to assess dosing to achieve functional rescue for clinical outcome. However, as photoreceptors in mice are predominantly rods, quantitative assessment of the editing rates in foveal cones was validated in a primate model. Surrogate guide RNAs had to be designed for NHPs due to its genetic divergence between humans. Total productive editing of the therapy was subsequently extrapolated from the editing frequency of the tissue sample. This extrapolation, in some instances relying on phenotypic versus genotypic outcomes, becomes particularly critical as obtaining human tissues (e.g., retina) may not always be feasible.
On the other hand, diseases entailing systemic correction such as DMD as opposed to localized editing in the eye will require multi-site sampling to quantify total editing. One of the drawbacks when using surrogate guides would be how representative the editing results will be in determining the correct dose for therapeutic efficacy. Greater confidence can be drawn by comparing with an additional model as shown in this study. For instance, EDIT-101 in the humanized mouse model, when compared with surrogate guides in NHPs, showed similar editing rates in response to increasing doses.
In contrast, the manufacturing of autologous products will vary due to the material received from the donor, but the final product can be well characterized before administration. Stadtmauer et al. quantified unique CRISPR-Cas9 engineered T cells for the frequency of edits by using a PCR assay, assessing for cytotoxic effects against tumor cells with an in vitro potency assay.5 Unlike in vivo targeting, the editing efficiency required to bring about clinical benefit was not established for the targets. Whole blood samples and biopsy of the bone marrow and tumor site in patients provided a direct postinfusion readout of the therapeutic product.
An additional readout challenge is the careful characterization of the off-target genomic and transcriptomic effects of CRISPR-based therapies. In recent years, several sensitive methodologies for assaying off-target effects have been developed.223–229 Even so, the assaying of clinically relevant off-target effects remains difficult because depending on the genomic context and cell type, some off-targets may be completely benign, whereas others could have serious consequences. This is a well-recognized issue in the field and many efforts to manage this are underway, such as engineering the CRISPR payload at both the protein and gRNA level and management of the window of active exposure to the functional RNP complex. For more in-depth reviews on this aspect of CRISPR-based therapies see Refs.230–233
Regulatory concerns and n-of-1 trials
CRISPR-based therapeutics face a host of unique regulatory challenges when compared with other therapeutic methods such as small-molecule drugs or antibodies that typically act at the protein level. There are two layers of information abstraction when moving from DNA to RNA to protein, and often this information may have important implications for therapeutic development.
One such example is the fact that even a single genetic disease caused by knockout of a single gene may have multiple underlying mutations in different patients. Often a single mutation, such as the deltaF508 in cystic fibrosis, the maternal allele deletion of chromosome 15q11-13 in Angelman syndrome, and the 1278insTATC mutation in the HEXA gene in Tay-Sachs disease, accounts for a majority of cases, but in each of these diseases, tens to hundreds of other mutations affecting the same genes also give rise to the disease. Correction of these mutations by CRISPR-Cas genome editing would likely require different gRNA constructs, or even different Cas effector modalities, even with the same delivery mechanism. These different gRNAs would naturally have different safety and efficacy profiles as a result of the varying functional activities and off-target effects of distinct gRNAs. Under standard regulatory frameworks, this would seem to require many individual therapies based on the same platform to treat the same disease to undergo their own arduous and expensive approval studies.
Oligonucleotide-based drugs, particularly antisense oligos (ASOs), although ripe with therapeutic potential, face a similar regulatory problem. To address this, companies such as Ionis Therapeutics have pioneered n-of-1 trials with personalized ASOs for patient-specific mutations, causing a variety of diseases including ALS, Batten disease, and cystic fibrosis.234–236 Despite the success of several of these trials, it is an arduous process for which the FDA is not streamlined. These trials to date have relied on right-to-try clauses, expanded access INDs, and perhaps most critically, heroic fundraising efforts on behalf of the patients. There remains no clear regulatory path for these kinds of hyper-personalized medicines to be tested at scale. However, due to the trailblazing of ASO therapies and the promise of CRISPR-based drugs, the FDA and other global regulatory agencies are acutely aware of this problem and may well deploy new guidelines to mitigate it.237,238
Clearly, safety and efficacy must be demonstrated, even for n-of-1 trials and personalized therapies.239 However, the process by which that can be accomplished must be simplified and streamlined for drugs that have an established platform. Many aspects of safety testing can be ported between similar drugs that may use the same delivery and mechanism, but with different underlying nucleic acid targets. In addition, inexpensive, fast, and robust efficacy testing platforms must be developed, which fortunately is an area of significant research interest. Although the landscape of current drug development models is not designed for the dynamic, personalized potential of the future of therapeutic breakthroughs, including CRISPR-based drugs, deft regulatory work and new guidelines can address this problem and pave the way to unlock that potential.
Conclusions
We are currently witnessing the birth of a new therapeutic paradigm based on CRISPR-Cas to precisely target genetic and epigenetic disease in a powerful and previously inaccessible manner. Given the rapid development and adoption of the CRISPR-Cas system, the explosion of preclinical studies, and the progress in clinical trials in the past few years, the near future will likely see the approval and deployment of several CRISPR-based therapies targeting diverse pathologies, some previously untreatable, and likely encompassing multiple therapeutic and delivery modalities. Despite this exciting prognosis, significant challenges remain, especially precise and high-quantity delivery to target tissues, demonstrated safety at the level of both off-target effects and immune interactions, and scalable, affordable deployment of the drugs. The substantial research efforts mounted toward all of these issues have yielded rapid advancements on every front, but much more remains to be done.
Although first-generation gene therapy Zolgensma garnered the epithet of “most expensive drug ever” at more than $2.1 million, we do not believe this must be a lasting attribute of this class of drugs. Although aspects such as AAV manufacturing challenges, R&D costs, regulatory burdens, and pharmaceutical market economics are important drivers of high price tags, the pace of technical progress can rapidly alter what once seemed set in stone. The precision and deftness with which we can manipulate genomes using CRISPR systems was virtually unthinkable just 17 years ago when the Human Genome Project was completed, just one year after the acronym CRISPR was first proposed.240 Now, we have a thousand dollar genome and a million dollar gene therapy. Given the explosion of research and development into CRISPR-based gene therapy and the expansion of its application from rare genetic disease to large-market ubiquitous conditions such as chronic pain,241 cardiovascular disease,242,243 and Alzheimer's,7 it is not entirely implausible nor unprecedented in the decades to come to hope for a few thousand dollar gene therapy.
Optimizing the translation of new technologies to rapidly enable disease treatment is of critical importance but is far from easy. Many therapeutic modalities offering huge benefits have been stymied by unforeseen or unappreciated setbacks on the path to approval, often taking decades to address. We have attempted here to outline the current state and translational path forward of CRISPR therapeutics, highlighting areas where additional research is required, or potential challenges may occur to help orient thinking and optimize resources to solve these challenges. Expediting the development of these therapies will require vigilance, insight, and cooperation among scientists of multiple backgrounds, pharmaceutical companies, and regulatory stakeholders to ensure that the potential of CRISPR-based drugs may be realized as efficiently and safely as possible.
Author Disclosure Statement
P.M. is a scientific co-founder of Navega Therapeutics, Boundless Biosciences, Seven Therapeutics, Engine Biosciences, and Shape Therapeutics. The terms of these arrangements have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. W.L.C. is a scientific co-founder of Seven Therapeutics.
Funding Information
This work was generously supported by NIH grants (R01HG009285, RO1CA222826, RO1GM123313) and by the Agency for Science, Technology, and Research under its Industrial Alignment Fund (Pre-Positioning) (H17/01/a0/012).
References
- 1. Dunbar CE, High KA, Joung JK, et al. . Gene therapy comes of age. Science. 2018;359:eaan4672 DOI: 10.1126/science.aan4672 [DOI] [PubMed] [Google Scholar]
- 2. Schuessler-Lenz M, Enzmann H, Vamvakas S. Regulators' advice can make a difference: European Medicines Agency approval of Zynteglo for beta thalassemia. Clin Pharmacol Ther. 2020;107:492–494. DOI: 10.1002/cpt.1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Russell S, Bennett J, Wellman JA, et al. . Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 2017;390:849–860. DOI: 10.1016/S0140-6736(17)31868-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mendell JR, Al-Zaidy S, Shell R, et al. . Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med. 2017;377:1713–1722. DOI: 10.1056/NEJMoa1706198 [DOI] [PubMed] [Google Scholar]
- 5. Stadtmauer EA, Fraietta JA, Davis MM, et al. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020:eaba7365. DOI: 10.1126/science.aba7365 [DOI] [PMC free article] [PubMed]
- 6. Strich JR, Chertow DS. CRISPR-Cas biology and its application to infectious diseases. J Clin Microbiol. 2019;57:e01307–e01318. DOI: 10.1128/JCM.01307-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park H, Oh J, Shim G, et al. . In vivo neuronal gene editing via CRISPR–Cas9 amphiphilic nanocomplexes alleviates deficits in mouse models of Alzheimer's disease. Nat Neurosci. 2019;22:524–528. DOI: 10.1038/s41593-019-0352-0 [DOI] [PubMed] [Google Scholar]
- 8. Xie C, Zhang Y-P, Song L, et al. . Genome editing with CRISPR/Cas9 in postnatal mice corrects PRKAG2 cardiac syndrome. Cell Res. 2016;26:1099–1111. DOI: 10.1038/cr.2016.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu Y, Xue J, Deng T, et al. . Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat Med. 2020;26:732–740. DOI: 10.1038/s41591-020-0840-5 [DOI] [PubMed] [Google Scholar]
- 10. Lino CA, Harper JC, Carney JP, et al. . Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25:1234–1257. DOI: 10.1080/10717544.2018.1474964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tseng Y-S, Agbandje-McKenna M. Mapping the AAV capsid host antibody response toward the development of second generation gene delivery vectors. Front Immunol. 2014;5:9 DOI: 10.3389/fimmu.2014.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mays LE, Wilson JM. The complex and evolving story of T cell activation to AAV vector-encoded transgene products. Mol Ther. 2011;19:16–27. DOI: 10.1038/mt.2010.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ronzitti G, Gross D-A, Mingozzi F. Human immune responses to adeno-associated virus (AAV) vectors. Front Immunol. 2020;11:670 DOI: 10.3389/fimmu.2020.00670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Verdera HC, Kuranda K, Mingozzi F. AAV vector immunogenicity in humans: a long journey to successful gene transfer. Mol Ther. 2020;28:723–746. DOI: 10.1016/j.ymthe.2019.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barnes C, Scheideler O, Schaffer D. Engineering the AAV capsid to evade immune responses. Curr Opin Biotechnol. 2019;60:99–103. DOI: 10.1016/j.copbio.2019.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu J-J, Orlova N, Oakes BL, et al. . CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature. 2019;566:218–223. DOI: 10.1038/s41586-019-0908-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pausch P, Al-Shayeb B, Bisom-Rapp E, et al. . CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science. 2020;369:333–337. DOI: 10.1126/science.abb1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sack BK, Herzog RW. Evading the immune response upon in vivo gene therapy with viral vectors. Curr Opin Mol Ther. 2009;11:493–503 [PMC free article] [PubMed] [Google Scholar]
- 19. Fu H, Cataldi MP, Ware TA, et al. . Functional correction of neurological and somatic disorders at later stages of disease in MPS IIIA mice by systemic scAAV9-hSGSH gene delivery. Mol Ther Methods Clin Dev. 2016;3:16036–16036. DOI: 10.1038/mtm.2016.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Silva Lima B, Videira MA. Toxicology and biodistribution: the clinical value of animal biodistribution studies. Mol Ther Methods Clin Dev. 2018;8:183–197. DOI: 10.1016/j.omtm.2018.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Petit L, Khanna H, Punzo C. Advances in gene therapy for diseases of the eye. Hum Gene Ther. 2016;27:563–579. DOI: 10.1089/hum.2016.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng C, Baum BJ. Evaluation of promoters for use in tissue-specific gene delivery. Methods Mol Biol. 2008;434:205–219. DOI: 10.1007/978-1-60327-248-3_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Q, Qin Z, Wang Q, et al. . Applications of genome editing technology in animal disease modeling and gene therapy. Comput Struct Biotechnol J. 2019;17:689–698. DOI: 10.1016/j.csbj.2019.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gonzalez L, Strbo N, Podack ER. Humanized mice: novel model for studying mechanisms of human immune-based therapies. Immunol Res. 2013;57:326–334. DOI: 10.1007/s12026-013-8471-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Husain SR, Han J, Au P, et al. . Gene therapy for cancer: regulatory considerations for approval. Cancer Gene Ther. 2015;22:554–563. DOI: 10.1038/cgt.2015.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zincarelli C, Soltys S, Rengo G, et al. . Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. DOI: 10.1038/mt.2008.76 [DOI] [PubMed] [Google Scholar]
- 27. Mali P, Yang L, Esvelt KM, et al. . RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. DOI: 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jinek M, East A, Cheng A, et al. . RNA-programmed genome editing in human cells. Elife. 2013;2:e00471 DOI: 10.7554/eLife.00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cong L, Ran FA, Cox D, et al. . Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. DOI: 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen X, Xu F, Zhu C, et al. . Dual sgRNA-directed gene knockout using CRISPR/Cas9 technology in Caenorhabditis elegans. Sci Rep. 2014;4:7581 DOI: 10.1038/srep07581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O'Connell MR, Oakes BL, Sternberg SH, et al. . Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266. DOI: 10.1038/nature13769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Konermann S, Lotfy P, Brideau NJ, et al. . Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173:665–676.e614. DOI: 10.1016/j.cell.2018.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cox DBT, Gootenberg JS, Abudayyeh OO, et al. . RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. DOI: 10.1126/science.aaq0180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Price AA, Sampson TR, Ratner HK, et al. . Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc Natl Acad Sci U S A. 2015;112:6164–6169. DOI: 10.1073/pnas.1422340112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Freije CA, Myhrvold C, Boehm CK, et al. . Programmable inhibition and detection of RNA viruses using Cas13. Mol Cell. 2019;76:826–837.e811. DOI: 10.1016/j.molcel.2019.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou H, Su J, Hu X, et al. . Glia-to-neuron conversion by CRISPR-CasRx alleviates symptoms of neurological disease in mice. Cell. 2020;181:590–603.e516. DOI: 10.1016/j.cell.2020.03.024 [DOI] [PubMed] [Google Scholar]
- 37. DeWitt MA, Corn JE, Carroll D. Genome editing via delivery of Cas9 ribonucleoprotein. Methods. 2017;121–122:9–15. DOI: 10.1016/j.ymeth.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. . Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. DOI: 10.1172/JCI35700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Naldini L. Genetic engineering of hematopoiesis: Current stage of clinical translation and future perspectives. EMBO Mol Med. 2019;11:e9958 DOI: 10.15252/emmm.201809958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. De Ravin SS, Li L, Wu X, et al. . CRISPR-Cas9 gene repair of hematopoietic stem cells from patients with X-linked chronic granulomatous disease. Sci Transl Med. 2017;9:eaah3480 DOI: 10.1126/scitranslmed.aah3480 [DOI] [PubMed] [Google Scholar]
- 41. Dever DP, Bak RO, Reinisch A, et al. . CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. DOI: 10.1038/nature20134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu J, Peng C, Sankaran VG, et al. . Correction of Sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science. 2011;334:993–996. DOI: 10.1126/science.1211053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sankaran VG, Menne TF, Xu J, et al. . Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. DOI: 10.1126/science.1165409 [DOI] [PubMed] [Google Scholar]
- 44. Srivastava A. In vivo tissue-tropism of adeno-associated viral vectors. Curr Opin Virol. 2016;21:75–80. DOI: 10.1016/j.coviro.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kanaan NM, Sellnow RC, Boye SL, et al. . Rationally engineered AAV capsids improve transduction and volumetric spread in the CNS. Mol Ther Nucleic Acids. 2017;8:184–197. DOI: 10.1016/j.omtn.2017.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rosario AM, Cruz PE, Ceballos-Diaz C, et al. . Microglia-specific targeting by novel capsid-modified AAV6 vectors. Mol Ther Methods Clin Dev. 2016;3:16026 DOI: 10.1038/mtm.2016.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bowles DE, McPhee SWJ, Li C, et al. . Phase 1 gene therapy for Duchenne muscular dystrophy using a translational optimized AAV vector. Mol Ther. 2012;20:443–455. DOI: 10.1038/mt.2011.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li W, Asokan A, Wu Z, et al. . Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles. Mol Ther. 2008;16:1252–1260. DOI: 10.1038/mt.2008.100 [DOI] [PubMed] [Google Scholar]
- 49. Maheshri N, Koerber JT, Kaspar BK, et al. . Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat Biotechnol. 2006;24:198–204. DOI: 10.1038/nbt1182 [DOI] [PubMed] [Google Scholar]
- 50. Asuri P, Bartel MA, Vazin T, et al. . Directed evolution of adeno-associated virus for enhanced gene delivery and gene targeting in human pluripotent stem cells. Mol Ther. 2012;20:329–338. DOI: 10.1038/mt.2011.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li W, Zhang L, Johnson JS, et al. . Generation of novel AAV variants by directed evolution for improved CFTR delivery to human ciliated airway epithelium. Mol Ther. 2009;17:2067–2077. DOI: 10.1038/mt.2009.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang L, Calcedo R, Bell P, et al. . Impact of pre-existing immunity on gene transfer to nonhuman primate liver with adeno-associated virus 8 vectors. Hum Gene Ther. 2011;22:1389–1401. DOI: 10.1089/hum.2011.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rivière C, Danos O, Douar AM. Long-term expression and repeated administration of AAV type 1, 2 and 5 vectors in skeletal muscle of immunocompetent adult mice. Gene Ther. 2006;13:1300–1308. DOI: 10.1038/sj.gt.3302766 [DOI] [PubMed] [Google Scholar]
- 54. Tse LV, Klinc KA, Madigan VJ, et al. . Structure-guided evolution of antigenically distinct adeno-associated virus variants for immune evasion. Proc Natl Acad Sci U S A. 2017;114:E4812–E4821. DOI: 10.1073/pnas.1704766114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lochrie MA, Tatsuno GP, Christie B, et al. . Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J Virol. 2006;80:821–834. DOI: 10.1128/jvi.80.2.821-834.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Giles AR, Govindasamy L, Somanathan S, et al. . Mapping an adeno-associated virus 9-specific neutralizing epitope to develop next-generation gene delivery vectors. J Virol. 2018;92:e01011–e01018. DOI: 10.1128/jvi.01011-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dauletbekov DL, Pfromm JK, Fritz AK, et al. . Innate immune response following AAV administration. Adv Exp Med Biol. 2019;1185:165–168. DOI: 10.1007/978-3-030-27378-1_27 [DOI] [PubMed] [Google Scholar]
- 58. Butterfield JSS, Biswas M, Shirley JL, et al. . TLR9-activating CpG-B ODN but not TLR7 agonists triggers antibody formation to factor IX in muscle gene transfer. Hum Gene Ther Methods. 2019;30:81–92. DOI: 10.1089/hgtb.2019.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ashley SN, Somanathan S, Giles AR, et al. . TLR9 signaling mediates adaptive immunity following systemic AAV gene therapy. Cell Immunol. 2019;346:103997 DOI: 10.1016/j.cellimm.2019.103997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Faust SM, Bell P, Cutler BJ, et al. . CpG-depleted adeno-associated virus vectors evade immune detection. J Clin Invest. 2013;123:2994–3001. DOI: 10.1172/JCI68205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shao W, Earley LF, Chai Z, et al. . Double-stranded RNA innate immune response activation from long-term adeno-associated virus vector transduction. JCI Insight. 2018;3:e120474 DOI: 10.1172/jci.insight.120474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rabinowitz J, Chan YK, Samulski RJ. Adeno-associated Virus (AAV) versus Immune Response. Viruses. 2019;11:102 DOI: 10.3390/v11020102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Calcedo R, Chichester JA, Wilson JM. Assessment of humoral, innate, and T-cell immune responses to adeno-associated virus vectors. Hum Gene Ther Methods. 2018;29:86–95. DOI: 10.1089/hgtb.2018.038 [DOI] [PubMed] [Google Scholar]
- 64. Xiong W, Wu DM, Xue Y, et al. . AAV cis-regulatory sequences are correlated with ocular toxicity. Proc Natl Acad Sci U S A. 2019;116:5785–5794. DOI: 10.1073/pnas.1821000116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Horowitz ED, Weinberg MS, Asokan A. Glycated AAV vectors: Chemical redirection of viral tissue tropism. Bioconjug Chem. 2011;22:529–532. DOI: 10.1021/bc100477g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Katrekar D, Moreno AM, Chen G, et al. . Oligonucleotide conjugated multi-functional adeno-associated viruses. Sci Rep. 2018;8:3589 DOI: 10.1038/s41598-018-21742-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kelemen RE, Mukherjee R, Cao X, et al. . A precise chemical strategy to alter the receptor specificity of the adeno-associated virus. Angew Chem Int Ed Engl. 2016;55:10645–10649. DOI: 10.1002/anie.201604067 [DOI] [PubMed] [Google Scholar]
- 68. Hudry E, Martin C, Gandhi S, et al. . Exosome-associated AAV vector as a robust and convenient neuroscience tool. Gene Ther. 2016;23:380–392. DOI: 10.1038/gt.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Meliani A, Boisgerault F, Fitzpatrick Z, et al. . Enhanced liver gene transfer and evasion of preexisting humoral immunity with exosome-enveloped AAV vectors. Blood Adv. 2017;1:2019–2031. DOI: 10.1182/bloodadvances.2017010181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. György B, Sage C, Indzhykulian AA, et al. . Rescue of hearing by gene delivery to inner-ear hair cells using exosome-associated AAV. Mol Ther. 2017;25:379–391. DOI: 10.1016/j.ymthe.2016.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wassmer SJ, Carvalho LS, György B, et al. . Exosome-associated AAV2 vector mediates robust gene delivery into the murine retina upon intravitreal injection. Sci Rep. 2017;7:45329 DOI: 10.1038/srep45329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schiller LT, Lemus-Diaz N, Rinaldi Ferreira R, et al. . Enhanced production of exosome-associated AAV by overexpression of the tetraspanin CD9. Mol Ther Methods Clin Dev. 2018;9:278–287. DOI: 10.1016/j.omtm.2018.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Akinc A, Maier MA, Manoharan M, et al. . The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat Nanotechnol. 2019;14:1084–1087. DOI: 10.1038/s41565-019-0591-y [DOI] [PubMed] [Google Scholar]
- 74. Semple SC, Akinc A, Chen J, et al. . Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. DOI: 10.1038/nbt.1602 [DOI] [PubMed] [Google Scholar]
- 75. Mishra S, Webster P, Davis ME. PEGylation significantly affects cellular uptake and intracellular trafficking of non-viral gene delivery particles. Eur J Cell Biol. 2004;83:97–111. DOI: 10.1078/0171-9335-00363 [DOI] [PubMed] [Google Scholar]
- 76. Finn JD, Smith AR, Patel MC, et al. . A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018;22:2227–2235. DOI: 10.1016/j.celrep.2018.02.014 [DOI] [PubMed] [Google Scholar]
- 77. Jiang C, Mei M, Li B, et al. . A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 2017;27:440–443. DOI: 10.1038/cr.2017.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yin H, Song C-Q, Dorkin JR, et al. . Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34:328–333. DOI: 10.1038/nbt.3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Rensen PCN, de Vrueh RLA, Kuiper J, et al. . Recombinant lipoproteins: lipoprotein-like lipid particles for drug targeting. Adv Drug Deliv Rev. 2001;47:251–276. DOI: 10.1016/S0169-409X(01)00109-0 [DOI] [PubMed] [Google Scholar]
- 80. Lee K, Conboy M, Park HM, et al. . Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat Biomed Eng. 2017;1:889–901. DOI: 10.1038/s41551-017-0137-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ding Y, Jiang Z, Saha K, et al. . Gold nanoparticles for nucleic acid delivery. Mol Ther. 2014;22:1075–1083. DOI: 10.1038/mt.2014.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lee B, Lee K, Panda S, et al. . Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat Biomed Eng. 2018;2:497–507. DOI: 10.1038/s41551-018-0252-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chen G, Abdeen AA, Wang Y, et al. . A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat Nanotechnol. 2019;14:974–980. DOI: 10.1038/s41565-019-0539-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Miyata K, Oba M, Nakanishi M, et al. . Polyplexes from poly(aspartamide) bearing 1,2-diaminoethane side chains induce pH-selective, endosomal membrane destabilization with amplified transfection and negligible cytotoxicity. J Am Chem Soc. 2008;130:16287–16294. DOI: 10.1021/ja804561g [DOI] [PubMed] [Google Scholar]
- 85. Park HM, Liu H, Wu J, et al. . Extension of the crRNA enhances Cpf1 gene editing in vitro and in vivo. Nat Commun. 2018;9:3313 DOI: 10.1038/s41467-018-05641-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Schmeer M, Schleef M. Pharmaceutical grade large-scale plasmid DNA manufacturing process. Methods Mol Biol. 2014;1143:219–240. DOI: 10.1007/978-1-4939-0410-5_14 [DOI] [PubMed] [Google Scholar]
- 87. Ferrer-Miralles N, Domingo-Espín J, Corchero JL, et al. . Microbial factories for recombinant pharmaceuticals. Microb Cell Fact. 2009;8:17 DOI: 10.1186/1475-2859-8-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhu J. Mammalian cell protein expression for biopharmaceutical production. Biotechnol Adv. 2012;30:1158–1170. DOI: 10.1016/j.biotechadv.2011.08.022 [DOI] [PubMed] [Google Scholar]
- 89. Sanchez-Garcia L, Martín L, Mangues R, et al. . Recombinant pharmaceuticals from microbial cells: a 2015 update. Microb Cell Fact. 2016;15:33 DOI: 10.1186/s12934-016-0437-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mamat U, Wilke K, Bramhill D, et al. . Detoxifying Escherichia coli for endotoxin-free production of recombinant proteins. Microb Cell Fact. 2015;14:57 DOI: 10.1186/s12934-015-0241-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Nieß A, Siemann-Herzberg M, Takors R. Protein production in Escherichia coli is guided by the trade-off between intracellular substrate availability and energy cost. Microb Cell Fact. 2019;18:8 DOI: 10.1186/s12934-019-1057-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sandberg TE, Salazar MJ, Weng LL, et al. . The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab Eng. 2019;56:1–16. DOI: 10.1016/j.ymben.2019.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Qiao J, Li W, Lin S, et al. . Co-expression of Cas9 and single-guided RNAs in Escherichia coli streamlines production of Cas9 ribonucleoproteins. Commun Biol. 2019;2:161 DOI: 10.1038/s42003-019-0402-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tripathi NK, Shrivastava A. Recent Developments in bioprocessing of recombinant proteins: expression hosts and process development. Front Bioeng Biotechnol. 2019;7:420 DOI: 10.3389/fbioe.2019.00420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Karst DJ, Steinebach F, Morbidelli M. Continuous integrated manufacturing of therapeutic proteins. Curr Opin Biotechnol. 2018;53:76–84. DOI: 10.1016/j.copbio.2017.12.015 [DOI] [PubMed] [Google Scholar]
- 96. Seki A, Rutz S. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J Exp Med. 2018;215:985–997. DOI: 10.1084/jem.20171626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hendel A, Bak RO, Clark JT, et al. . Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. 2015;33:985–989. DOI: 10.1038/nbt.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Schumann K, Lin S, Boyer E, et al. . Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci U S A. 2015;112:10437–10442. DOI: 10.1073/pnas.1512503112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kelley ML, Strezoska Ž, He K, et al. . Versatility of chemically synthesized guide RNAs for CRISPR-Cas9 genome editing. J Biotechnol. 2016;233:74–83. DOI: 10.1016/j.jbiotec.2016.06.011 [DOI] [PubMed] [Google Scholar]
- 100. Schubert MS, Cedrone E, Neun B, et al. . Chemical modification of CRISPR gRNAs eliminate type i interferon responses in human peripheral blood mononuclear cells. J Cytokine Biol. 2018;3:121 DOI: 10.4172/2576-3881.1000121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Basila M, Kelley ML, Smith AvB. Minimal 2'-O-methyl phosphorothioate linkage modification pattern of synthetic guide RNAs for increased stability and efficient CRISPR-Cas9 gene editing avoiding cellular toxicity. PLoS One. 2017;12:e0188593 DOI: 10.1371/journal.pone.0188593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Scott T, Soemardy C, Morris KV. Development of a facile approach for generating chemically modified CRISPR/Cas9 RNA. Mol Ther Nucleic Acids. 2020;19:1176–1185. DOI: 10.1016/j.omtn.2020.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Xu L, Wang J, Liu Y, et al. . CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia. N Engl J Med. 2019;381:1240–1247. DOI: 10.1056/NEJMoa1817426 [DOI] [PubMed] [Google Scholar]
- 104. Allay JA, Sleep S, Long S, et al. . Good manufacturing practice production of self-complementary serotype 8 adeno-associated viral vector for a hemophilia B clinical trial. Hum Gene Ther. 2011;22:595–604. DOI: 10.1089/hum.2010.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Powers AD, Piras BA, Clark RK, et al. . Development and optimization of AAV hFIX particles by transient transfection in an iCELLis® Fixed-Bed Bioreactor. Hum Gene Ther Methods. 2016;27:112–121. DOI: 10.1089/hgtb.2016.021 [DOI] [PubMed] [Google Scholar]
- 106. Grieger JC, Soltys SM, Samulski RJ. Production of recombinant adeno-associated virus vectors using suspension HEK293 cells and continuous harvest of vector from the culture media for GMP FIX and FLT1 clinical vector. Mol Ther. 2016;24:287–297. DOI: 10.1038/mt.2015.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Schmeer M, Buchholz T, Schleef M. Plasmid DNA manufacturing for indirect and direct clinical applications. Hum Gene Ther. 2017;28:856–861. DOI: 10.1089/hum.2017.159 [DOI] [PubMed] [Google Scholar]
- 108. Flotte TR, Trapnell BC, Humphries M, et al. . Phase 2 clinical trial of a recombinant adeno-associated viral vector expressing α1-antitrypsin: interim results. Hum Gene Ther. 2011;22:1239–1247. DOI: 10.1089/hum.2011.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Thomas DL, Wang L, Niamke J, et al. . Scalable recombinant adeno-associated virus production using recombinant herpes simplex virus type 1 coinfection of suspension-adapted mammalian cells. Hum Gene Ther. 2009;20:861–870. DOI: 10.1089/hum.2009.004 [DOI] [PubMed] [Google Scholar]
- 110. Adamson-Small L, Potter M, Falk DJ, et al. . A scalable method for the production of high-titer and high-quality adeno-associated type 9 vectors using the HSV platform. Mol Ther Methods Clin Dev. 2016;3:16031 DOI: 10.1038/mtm.2016.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Urabe M, Ding C, Kotin RM. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum Gene Ther. 2002;13:1935–1943. DOI: 10.1089/10430340260355347 [DOI] [PubMed] [Google Scholar]
- 112. Clément N, Grieger JC. Manufacturing of recombinant adeno-associated viral vectors for clinical trials. Mol Ther Methods Clin Dev. 2016;3:16002 DOI: 10.1038/mtm.2016.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Smith RH, Levy JR, Kotin RM. A simplified baculovirus-AAV expression vector system coupled with one-step affinity purification yields high-titer rAAV stocks from insect cells. Mol Ther. 2009;17:1888–1896. DOI: 10.1038/mt.2009.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Rumachik NG, Malaker SA, Poweleit N, et al. Methods matter—standard production platforms for recombinant AAV can produce chemically and functionally distinct vectors. bioRxiv. 2019:640169. DOI: 10.1101/640169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wright JF, Zelenaia O. Vector characterization methods for quality control testing of recombinant adeno-associated viruses. Methods Mol Biol. 2011;737:247–278. DOI: 10.1007/978-1-61779-095-9_11 [DOI] [PubMed] [Google Scholar]
- 116. Ylä-Herttuala S. Endgame: glybera finally recommended for approval as the first gene therapy drug in the European union. Mol Ther. 2012;20:1831–1832. DOI: 10.1038/mt.2012.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Flanigan KM, Campbell K, Viollet L, et al. . Anti-dystrophin T cell responses in Duchenne muscular dystrophy: prevalence and a glucocorticoid treatment effect. Hum Gene Ther. 2013;24:797–806. DOI: 10.1089/hum.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Mendell JR, Campbell K, Rodino-Klapac L, et al. . Dystrophin immunity in Duchenne's muscular dystrophy. N Engl J Med. 2010;363:1429–1437. DOI: 10.1056/NEJMoa1000228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Tabebordbar M, Zhu K, Cheng JKW, et al. . In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. DOI: 10.1126/science.aad5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Bengtsson NE, Hall JK, Odom GL, et al. . Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun. 2017;8:14454 DOI: 10.1038/ncomms14454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zhang Y, Long C, Li H, et al. . CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci Adv. 2017;3:e1602814 DOI: 10.1126/sciadv.1602814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ryu S-M, Koo T, Kim K, et al. . Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol. 2018;36:536–539. DOI: 10.1038/nbt.4148 [DOI] [PubMed] [Google Scholar]
- 123. Le Guiner C, Servais L, Montus M, et al. . Long-term microdystrophin gene therapy is effective in a canine model of Duchenne muscular dystrophy. Nat Commun. 2017;8:16105 DOI: 10.1038/ncomms16105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Nelson CE, Wu Y, Gemberling MP, et al. . Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat Med. 2019;25:427–432. DOI: 10.1038/s41591-019-0344-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Amoasii L, Hildyard JCW, Li H, et al. . Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362:86–91. DOI: 10.1126/science.aau1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Zhang Y, Li H, Min Y-L, et al. . Enhanced CRISPR-Cas9 correction of Duchenne muscular dystrophy in mice by a self-complementary AAV delivery system. Sci Adv. 2020;6:eaay6812 DOI: 10.1126/sciadv.aay6812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Lee SH, Kim YS, Nah SK, et al. . Transduction patterns of adeno-associated viral vectors in a laser-induced choroidal neovascularization mouse model. Mol Ther Methods Clin Dev. 2018;9:90–98. DOI: 10.1016/j.omtm.2018.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Fauser S, Luberichs J, Schüttauf F. Genetic animal models for retinal degeneration. Surv Ophthalmol. 2002;47:357–367. DOI: 10.1016/S0039-S6257(02)00314-4 [DOI] [PubMed] [Google Scholar]
- 129. Acland GM, Aguirre GD, Ray J, et al. . Gene therapy restores vision in a canine model of childhood blindness. Nat Genet. 2001;28:92–95. DOI: 10.1038/ng0501-92 [DOI] [PubMed] [Google Scholar]
- 130. Aguirre GK, Komáromy AM, Cideciyan AV, et al. . Canine and human visual cortex intact and responsive despite early retinal blindness from RPE65 mutation. PLoS Med. 2007;4:e230 DOI: 10.1371/journal.pmed.0040230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Maguire AM, Simonelli F, Pierce EA, et al. . Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358:2240–2248. DOI: 10.1056/NEJMoa0802315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Hauswirth WW, Aleman TS, Kaushal S, et al. . Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. Hum Gene Ther. 2008;19:979–990. DOI: 10.1089/hum.2008.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Bainbridge JWB, Smith AJ, Barker SS, et al. . Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med. 2008;358:2231–2239. DOI: 10.1056/NEJMoa0802268 [DOI] [PubMed] [Google Scholar]
- 134. Amado D, Mingozzi F, Hui D, et al. . Safety and efficacy of subretinal readministration of a viral vector in large animals to treat congenital blindness. Sci Transl Med. 2010;2:21ra16 DOI: 10.1126/scitranslmed.3000659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Annear MJ, Bartoe JT, Barker SE, et al. . Gene therapy in the second eye of RPE65-deficient dogs improves retinal function. Gene Ther. 2011;18:53–61. DOI: 10.1038/gt.2010.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Bennett J, Ashtari M, Wellman J, et al. . AAV2 gene therapy readministration in three adults with congenital blindness. Sci Transl Med. 2012;4:120ra115 DOI: 10.1126/scitranslmed.3002865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Boye SE, Alexander JJ, Boye SL, et al. . The human rhodopsin kinase promoter in an AAV5 vector confers rod- and cone-specific expression in the primate retina. Hum Gene Ther. 2012;23:1101–1115. DOI: 10.1089/hum.2012.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Sun X, Pawlyk B, Xu X, et al. . Gene therapy with a promoter targeting both rods and cones rescues retinal degeneration caused by AIPL1 mutations. Gene Ther. 2010;17:117–131. DOI: 10.1038/gt.2009.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Beltran WA, Cideciyan AV, Lewin AS, et al. . Gene therapy rescues photoreceptor blindness in dogs and paves the way for treating human X-linked retinitis pigmentosa. Proc Natl Acad Sci U S A. 2012;109:2132–2137. DOI: 10.1073/pnas.1118847109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Daiger SP, Bowne SJ, Sullivan LS. Genes and mutations causing autosomal dominant retinitis pigmentosa. Cold Spring Harb Perspect Med. 2014;5:a017129 DOI: 10.1101/cshperspect.a017129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Bakondi B, Lv W, Lu B, et al. . In vivo CRISPR/Cas9 gene editing corrects retinal dystrophy in the S334ter-3 rat model of autosomal dominant retinitis pigmentosa. Mol Ther. 2016;24:556–563. DOI: 10.1038/mt.2015.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Courtney DG, Moore JE, Atkinson SD, et al. . CRISPR/Cas9 DNA cleavage at SNP-derived PAM enables both in vitro and in vivo KRT12 mutation-specific targeting. Gene Ther. 2016;23:108–112. DOI: 10.1038/gt.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Li P, Kleinstiver BP, Leon MY, et al. . Allele-specific CRISPR-Cas9 genome editing of the single-base P23H mutation for rhodopsin-associated dominant retinitis pigmentosa. CRISPR J. 2018;1:55–64. DOI: 10.1089/crispr.2017.0009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Mao H, James T Jr., Schwein A, et al. . AAV delivery of wild-type rhodopsin preserves retinal function in a mouse model of autosomal dominant retinitis pigmentosa. Hum Gene Ther. 2011;22:567–575. DOI: 10.1089/hum.2010.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Heier JS, Kherani S, Desai S, et al. . Intravitreous injection of AAV2-sFLT01 in patients with advanced neovascular age-related macular degeneration: a phase 1, open-label trial. Lancet. 2017;390:50–61. DOI: 10.1016/S0140-6736(17)30979-0 [DOI] [PubMed] [Google Scholar]
- 146. Chung SH, Mollhoff IN, Nguyen U, et al. . Factors impacting efficacy of AAV-mediated CRISPR-based genome editing for treatment of choroidal neovascularization. Mol Ther Methods Clin Dev. 2020;17:409–417. DOI: 10.1016/j.omtm.2020.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Chadderton N, Palfi A, Millington-Ward S, et al. . Intravitreal delivery of AAV-NDI1 provides functional benefit in a murine model of Leber hereditary optic neuropathy. Eur J Hum Genet. 2013;21:62–68. DOI: 10.1038/ejhg.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kotterman MA, Yin L, Strazzeri JM, et al. . Antibody neutralization poses a barrier to intravitreal adeno-associated viral vector gene delivery to non-human primates. Gene Ther. 2015;22:116–126. DOI: 10.1038/gt.2014.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. MacLachlan TK, Lukason M, Collins M, et al. . Preclinical safety evaluation of AAV2-sFLT01—a gene therapy for age-related macular degeneration. Mol Ther. 2011;19:326–334. DOI: 10.1038/mt.2010.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Seitz IP, Michalakis S, Wilhelm B, et al. . Superior retinal gene transfer and biodistribution profile of subretinal versus intravitreal delivery of AAV8 in nonhuman primates. Invest Ophthal Vis Sci. 2017;58:5792–5801. DOI: 10.1167/iovs.17-22473 [DOI] [PubMed] [Google Scholar]
- 151. Ochakovski GA, Bartz-Schmidt KU, Fischer MD. Retinal gene therapy: surgical vector delivery in the translation to clinical trials. Front Neurosci. 2017;11:174 DOI: 10.3389/fnins.2017.00174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Takahashi K, Igarashi T, Miyake K, et al. . Improved intravitreal AAV-mediated inner retinal gene transduction after surgical internal limiting membrane peeling in cynomolgus monkeys. Mol Ther. 2017;25:296–302. DOI: 10.1016/j.ymthe.2016.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Kay CN, Ryals RC, Aslanidi GV, et al. . Targeting photoreceptors via intravitreal delivery using novel, capsid-mutated AAV vectors. PLoS One. 2013;8:e62097 DOI: 10.1371/journal.pone.0062097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Dalkara D, Byrne LC, Klimczak RR, et al. . In vivo–directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci Transl Med. 2013;5:189ra176 DOI: 10.1126/scitranslmed.3005708 [DOI] [PubMed] [Google Scholar]
- 155. D'Avola D, López-Franco E, Sangro B, et al. . Phase I open label liver-directed gene therapy clinical trial for acute intermittent porphyria. J Hepatol. 2016;65:776–783. DOI: 10.1016/j.jhep.2016.05.012 [DOI] [PubMed] [Google Scholar]
- 156. Nathwani AC, Reiss UM, Tuddenham EGD, et al. . Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med. 2014;371:1994–2004. DOI: 10.1056/NEJMoa1407309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Jacobs F, Gordts SC, Muthuramu I, et al. . The liver as a target organ for gene therapy: State of the art, challenges, and future perspectives. Pharmaceuticals (Basel). 2012;5:1372–1392. DOI: 10.3390/ph5121372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Glover DJ, Lipps HJ, Jans DA. Towards safe, non-viral therapeutic gene expression in humans. Nat Rev Genet. 2005;6:299–310. DOI: 10.1038/nrg1577 [DOI] [PubMed] [Google Scholar]
- 159. Izembart A, Aguado E, Gauthier O, et al. . In vivo retrovirus-mediated gene transfer to the liver of dogs results in transient expression and induction of a cytotoxic immune response. Hum Gene Ther. 1999;10:2917–2925. DOI: 10.1089/10430349950016339 [DOI] [PubMed] [Google Scholar]
- 160. Gao G-P, Alvira MR, Wang L, et al. . Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci U S A. 2002;99:11854–11859. DOI: 10.1073/pnas.182412299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Cooper M, Nayak S, Hoffman BE, et al. . Improved induction of immune tolerance to factor IX by hepatic AAV-8 gene transfer. Hum Gene Ther. 2009;20:767–776. DOI: 10.1089/hum.2008.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Davidoff AM, Gray JT, Ng CYC, et al. . Comparison of the ability of adeno-associated viral vectors pseudotyped with serotype 2, 5, and 8 capsid proteins to mediate efficient transduction of the liver in murine and nonhuman primate models. Mol Ther. 2005;11:875–888. DOI: 10.1016/j.ymthe.2004.12.022 [DOI] [PubMed] [Google Scholar]
- 163. Calcedo R, Vandenberghe LH, Gao G, et al. . Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J Infect Dis. 2009;199:381–390. DOI: 10.1086/595830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Boutin S, Monteilhet V, Veron P, et al. . Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther. 2010;21:704–712. DOI: 10.1089/hum.2009.182 [DOI] [PubMed] [Google Scholar]
- 165. Wang L, Yang Y, Breton CA, et al. . CRISPR/Cas9-mediated in vivo gene targeting corrects hemostasis in newborn and adult factor IX–knockout mice. Blood. 2019;133:2745–2752. DOI: 10.1182/blood.2019000790 [DOI] [PubMed] [Google Scholar]
- 166. Chen H, Shi M, Gilam A, et al. . Hemophilia A ameliorated in mice by CRISPR-based in vivo genome editing of human Factor VIII. Sci Rep. 2019;9:16838 DOI: 10.1038/s41598-019-53198-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Wang L, Yang Y, Breton C, et al. . A mutation-independent CRISPR-Cas9-mediated gene targeting approach to treat a murine model of ornithine transcarbamylase deficiency. Sci Adv. 2020;6:eaax5701 DOI: 10.1126/sciadv.aax5701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Yang Y, Wang L, Bell P, et al. . A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34:334–338. DOI: 10.1038/nbt.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Kwon JH, Lee YM, Cho J-H, et al. . Liver-directed gene therapy for murine glycogen storage disease type Ib. Hum Mol Genet. 2017;26:4395–4405. DOI: 10.1093/hmg/ddx325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Jeyakumar J, Kia A, McIntosh J, et al. . Liver-directed gene therapy corrects Fabry disease in mice. Mol Genet Metab. 2019;126:S80 DOI: 10.1016/j.ymgme.2018.12.196 [DOI] [Google Scholar]
- 171. Miranda CJ, Canavese M, Chisari E, et al. . Liver-directed AAV gene therapy for Gaucher disease. Blood. 2019;134:3354 DOI: 10.1182/blood-2019-124280 [DOI] [Google Scholar]
- 172. Franco LM, Sun B, Yang X, et al. . Evasion of immune responses to introduced human acid α-glucosidase by liver-restricted expression in glycogen storage disease type II. Mol Ther. 2005;12:876–884. DOI: 10.1016/j.ymthe.2005.04.024 [DOI] [PubMed] [Google Scholar]
- 173. Sands MS, Davidson BL. Gene therapy for lysosomal storage diseases. Mol Ther. 2006;13:839–849. DOI: 10.1016/j.ymthe.2006.01.006 [DOI] [PubMed] [Google Scholar]
- 174. Wraith JE, Scarpa M, Beck M, et al. . Mucopolysaccharidosis type II (Hunter syndrome): a clinical review and recommendations for treatment in the era of enzyme replacement therapy. Eur J Pediatr. 2008;167:267–277. DOI: 10.1007/s00431-007-0635-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Guffon N, Bertrand Y, Forest I, et al. . Bone marrow transplantation in children with Hunter syndrome: outcome after 7 to 17 years. J Pediatr. 2009;154:733–737. DOI: 10.1016/j.jpeds.2008.11.041 [DOI] [PubMed] [Google Scholar]
- 176. Wyatt K, Henley W, Anderson L, et al. . The effectiveness and cost of enzyme replacement and substrate reduction therapies: A longitudinal cohort study of people with lysosomal storage disorders. Health Technol Assess. 2012;16:1–543. DOI: 10.3310/hta16390 [DOI] [PubMed] [Google Scholar]
- 177. Boelens JJ, Orchard PJ, Wynn RF. Transplantation in inborn errors of metabolism: Current considerations and future perspectives. Br J Haematol. 2014;167:293–303. DOI: 10.1111/bjh.13059 [DOI] [PubMed] [Google Scholar]
- 178. Boelens JJ, Wynn RF, O'Meara A, et al. . Outcomes of hematopoietic stem cell transplantation for Hurler's syndrome in Europe: A risk factor analysis for graft failure. Bone Marrow Transplant. 2007;40:225–233. DOI: 10.1038/sj.bmt.1705718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Aldenhoven M, Boelens J, de Koning TJ. The clinical outcome of Hurler syndrome after stem cell transplantation. Biol Blood Marrow Transplant. 2008;14:485–498. DOI: 10.1016/j.bbmt.2008.01.009 [DOI] [PubMed] [Google Scholar]
- 180. Ou L, Przybilla MJ, Tăbăran A-F, et al. . A novel gene editing system to treat both Tay-Sachs and Sandhoff diseases. Gene Ther. 2020;27:226–236. DOI: 10.1038/s41434-019-0120-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Ou L, DeKelver RC, Rohde M, et al. . ZFN-mediated in vivo genome editing corrects murine Hurler syndrome. Mol Ther. 2019;27:178–187. DOI: 10.1016/j.ymthe.2018.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Schuh RS, Poletto É, Pasqualim G, et al. . In vivo genome editing of mucopolysaccharidosis I mice using the CRISPR/Cas9 system. J Control Release. 2018;288:23–33. DOI: 10.1016/j.jconrel.2018.08.031 [DOI] [PubMed] [Google Scholar]
- 183. Zhang H, Yang B, Mu X, et al. . Several rAAV vectors efficiently cross the blood-brain barrier and transduce neurons and astrocytes in the neonatal mouse central nervous system. Mol Ther. 2011;19:1440–1448. DOI: 10.1038/mt.2011.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Duque S, Joussemet B, Riviere C, et al. . Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol Ther. 2009;17:1187–1196. DOI: 10.1038/mt.2009.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Samaranch L, Salegio EA, San Sebastian W, et al. . Adeno-associated virus serotype 9 transduction in the central nervous system of nonhuman primates. Hum Gene Ther. 2012;23:382–389. DOI: 10.1089/hum.2011.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Passini MA, Wolfe JH. Widespread gene delivery and structure-specific patterns of expression in the brain after intraventricular injections of neonatal mice with an adeno-associated virus vector. J Virol. 2001;75:12382–12392. DOI: 10.1128/jvi.75.24.12382-12392.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Haurigot V, Marcó S, Ribera A, et al. . Whole body correction of mucopolysaccharidosis IIIA by intracerebrospinal fluid gene therapy. J Clin Invest. 2013;123:3254–3271. DOI: 10.1172/JCI66778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Souweidane MM, Fraser JF, Arkin LM, et al. . Gene therapy for late infantile neuronal ceroid lipofuscinosis: neurosurgical considerations. J Neurosurg Pediatr. 2010;6:115–122. DOI: 10.3171/2010.4.PEDS09507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Meyer K, Ferraiuolo L, Schmelzer L, et al. . Improving single injection CSF delivery of AAV9-mediated gene therapy for SMA: a dose–response study in mice and nonhuman primates. Mol Ther. 2015;23:477–487. DOI: 10.1038/mt.2014.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Federici T, Taub JS, Baum GR, et al. . Robust spinal motor neuron transduction following intrathecal delivery of AAV9 in pigs. Gene Ther. 2012;19:852–859. DOI: 10.1038/gt.2011.130 [DOI] [PubMed] [Google Scholar]
- 191. Ragagnin AMG, Shadfar S, Vidal M, et al. . Motor neuron susceptibility in ALS/FTD. Front Neurosci. 2019;13:532 DOI: 10.3389/fnins.2019.00532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Rosen DR, Siddique T, Patterson D, et al. . Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. DOI: 10.1038/362059a0 [DOI] [PubMed] [Google Scholar]
- 193. Gaj T, Ojala DS, Ekman FK, et al. . In vivo genome editing improves motor function and extends survival in a mouse model of ALS. Sci Adv. 2017;3:eaar3952 DOI: 10.1126/sciadv.aar3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Duan W, Guo M, Yi L, et al. . The deletion of mutant SOD1 via CRISPR/Cas9/sgRNA prolongs survival in an amyotrophic lateral sclerosis mouse model. Gene Ther. 2020;27:157–169. DOI: 10.1038/s41434-019-0116-1 [DOI] [PubMed] [Google Scholar]
- 195. Lim CKW, Gapinske M, Brooks AK, et al. . Treatment of a mouse model of ALS by in vivo base editing. Mol Ther. 2020;28:1177–1189. DOI: 10.1016/j.ymthe.2020.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Ekman FK, Ojala DS, Adil MM, et al. . CRISPR-Cas9-mediated genome editing increases lifespan and improves motor deficits in a Huntington's disease mouse model. Mol Ther Nucleic Acids. 2019;17:829–839. DOI: 10.1016/j.omtn.2019.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. György B, Lööv C, Zaborowski MP, et al. . CRISPR/Cas9 mediated disruption of the Swedish APP allele as a therapeutic approach for early-onset Alzheimer's disease. Mol Ther Nucleic Acids. 2018;11:429–440. DOI: 10.1016/j.omtn.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Moreno AM, Palmer N, Alemán F, et al. . Immune-orthogonal orthologues of AAV capsids and of Cas9 circumvent the immune response to the administration of gene therapy. Nat Biomed Eng. 2019;3:806–816. DOI: 10.1038/s41551-019-0431-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Wagner DL, Amini L, Wendering DJ, et al. . High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat Med. 2019;25:242–248. DOI: 10.1038/s41591-018-0204-6 [DOI] [PubMed] [Google Scholar]
- 200. Simhadri VL, McGill J, McMahon S, et al. . Prevalence of pre-existing antibodies to CRISPR-associated nuclease Cas9 in the USA population. Mol Ther Methods Clin Dev. 2018;10:105–112. DOI: 10.1016/j.omtm.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Charlesworth CT, Deshpande PS, Dever DP, et al. . Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med. 2019;25:249–254. DOI: 10.1038/s41591-018-0326-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Crudele JM, Chamberlain JS. Cas9 immunity creates challenges for CRISPR gene editing therapies. Nat Commun. 2018;9:3497 DOI: 10.1038/s41467-018-05843-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Li A, Tanner MR, Lee CM, et al. . AAV-CRISPR gene editing is negated by pre-existing immunity to Cas9. Mol Ther. 2020;28:1432–1441. DOI: 10.1016/j.ymthe.2020.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Ferdosi SR, Ewaisha R, Moghadam F, et al. . Multifunctional CRISPR-Cas9 with engineered immunosilenced human T cell epitopes. Nat Commun. 2019;10:1842 DOI: 10.1038/s41467-019-09693-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Roca C, Motas S, Marcó S, et al. . Disease correction by AAV-mediated gene therapy in a new mouse model of mucopolysaccharidosis type IIID. Hum Mol Genet. 2017;26:1535–1551. DOI: 10.1093/hmg/ddx058 [DOI] [PubMed] [Google Scholar]
- 206. Passini MA, Macauley SL, Huff MR, et al. . AAV vector-mediated correction of brain pathology in a mouse model of Niemann–Pick a disease. Mol Ther. 2005;11:754–762. DOI: 10.1016/j.ymthe.2005.01.011 [DOI] [PubMed] [Google Scholar]
- 207. Ohno M, Cole SL, Yasvoina M, et al. . BACE1 gene deletion prevents neuron loss and memory deficits in 5XFAD APP/PS1 transgenic mice. Neurobiol Dis. 2007;26:134–145. DOI: 10.1016/j.nbd.2006.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. McGreevy JW, Hakim CH, McIntosh MA, et al. . Animal models of Duchenne muscular dystrophy: From basic mechanisms to gene therapy. Dis Model Mech. 2015;8:195–213. DOI: 10.1242/dmm.018424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Tu Z, Yang W, Yan S, et al. . CRISPR/Cas9: A powerful genetic engineering tool for establishing large animal models of neurodegenerative diseases. Mol Neurodegener. 2015;10:35 DOI: 10.1186/s13024-015-0031-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Moretti A, Fonteyne L, Giesert F, et al. . Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat Med. 2020;26:207–214. DOI: 10.1038/s41591-019-0738-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Chang R, Liu X, Li S, et al. . Transgenic animal models for study of the pathogenesis of Huntington's disease and therapy. Drug Des Devel Ther. 2015;9:2179–2188. DOI: 10.2147/DDDT.S58470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Menon KP, Tieu PT, Neufeld EF. Architecture of the canine IDUA gene and mutation underlying canine mucopolysaccharidosis I. Genomics. 1992;14:763–768. DOI: 10.1016/S0888-7543(05)80182-X [DOI] [PubMed] [Google Scholar]
- 213. Ellinwood NM, Vite CH, Haskins ME. Gene therapy for lysosomal storage diseases: The lessons and promise of animal models. J Gene Med. 2004;6:481–506. DOI: 10.1002/jgm.581 [DOI] [PubMed] [Google Scholar]
- 214. Vite CH, McGowan JC, Niogi SN, et al. . Effective gene therapy for an inherited CNS disease in a large animal model. Ann Neurol. 2005;57:355–364. DOI: 10.1002/ana.20392 [DOI] [PubMed] [Google Scholar]
- 215. Vite CH, Passini MA, Haskins ME, et al. . Adeno-associated virus vector-mediated transduction in the cat brain. Gene Ther. 2003;10:1874–1881. DOI: 10.1038/sj.gt.3302087 [DOI] [PubMed] [Google Scholar]
- 216. Matsuzaki Y, Konno A, Mochizuki R, et al. . Intravenous administration of the adeno-associated virus-PHP.B capsid fails to upregulate transduction efficiency in the marmoset brain. Neurosci Lett. 2018;665:182–188. DOI: 10.1016/j.neulet.2017.11.049 [DOI] [PubMed] [Google Scholar]
- 217. Yuasa K, Yoshimura M, Urasawa N, et al. . Injection of a recombinant AAV serotype 2 into canine skeletal muscles evokes strong immune responses against transgene products. Gene Ther. 2007;14:1249–1260. DOI: 10.1038/sj.gt.3302984 [DOI] [PubMed] [Google Scholar]
- 218. Wang Z, Allen JM, Riddell SR, et al. . Immunity to adeno-associated virus-mediated gene transfer in a random-bred canine model of Duchenne muscular dystrophy. Hum Gene Ther. 2007;18:18–26. DOI: 10.1089/hum.2006.093 [DOI] [PubMed] [Google Scholar]
- 219. Wang Z, Kuhr CS, Allen JM, et al. . Sustained AAV-mediated Dystrophin expression in a canine model of Duchenne muscular dystrophy with a brief course of immunosuppression. Mol Ther. 2007;15:1160–1166. DOI: 10.1038/sj.mt.6300161 [DOI] [PubMed] [Google Scholar]
- 220. Nathwani AC, Gray JT, McIntosh J, et al. . Safe and efficient transduction of the liver after peripheral vein infusion of self-complementary AAV vector results in stable therapeutic expression of human FIX in nonhuman primates. Blood. 2007;109:1414–1421. DOI: 10.1182/blood-2006-03-010181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Li H, Lasaro MO, Jia B, et al. . Capsid-specific T-cell responses to natural infections with adeno-associated viruses in humans differ from those of nonhuman primates. Mol Ther. 2011;19:2021–2030. DOI: 10.1038/mt.2011.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Maeder ML, Stefanidakis M, Wilson CJ, et al. . Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat Med. 2019;25:229–233. DOI: 10.1038/s41591-018-0327-9 [DOI] [PubMed] [Google Scholar]
- 223. Wienert B, Wyman SK, Richardson CD, et al. . Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science. 2019;364:286–289. DOI: 10.1126/science.aav9023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Akcakaya P, Bobbin ML, Guo JA, et al. . In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature. 2018;561:416–419. DOI: 10.1038/s41586-018-0500-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Cameron P, Fuller CK, Donohoue PD, et al. . Mapping the genomic landscape of CRISPR–Cas9 cleavage. Nat Methods. 2017;14:600–606. DOI: 10.1038/nmeth.4284 [DOI] [PubMed] [Google Scholar]
- 226. Kim D, Bae S, Park J, et al. . Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. DOI: 10.1038/nmeth.3284 [DOI] [PubMed] [Google Scholar]
- 227. Tsai SQ, Nguyen NT, Malagon-Lopez J, et al. . CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR–Cas9 nuclease off-targets. Nat Methods. 2017;14:607–614. DOI: 10.1038/nmeth.4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Yan WX, Mirzazadeh R, Garnerone S, et al. . BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat Commun. 2017;8:15058 DOI: 10.1038/ncomms15058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Zuo E, Sun Y, Wei W, et al. . Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364:289–292. DOI: 10.1126/science.aav9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Manghwar H, Li B, Ding X, et al. . CRISPR/Cas systems in genome editing: methodologies and tools for sgRNA design, off-target evaluation, and strategies to mitigate off-target effects. Adv Sci (Weinh). 2020;7:1902312 DOI: 10.1002/advs.201902312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Han HA, Pang JKS, Soh B-S. Mitigating off-target effects in CRISPR/Cas9-mediated in vivo gene editing. J Mol Med (Berl). 2020;98:615–632. DOI: 10.1007/s00109-020-01893-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Wang D, Zhang F, Gao G. CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors. Cell. 2020;181:136–150. DOI: 10.1016/j.cell.2020.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Dai W-J, Zhu L-Y, Yan Z-Y, et al. . CRISPR-Cas9 for in vivo gene therapy: promise and hurdles. Mol Ther Nucleic Acids. 2016;5:e349 DOI: 10.1038/mtna.2016.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. McGarry ME, Illek B, Ly NP, et al. . In vivo and in vitro ivacaftor response in cystic fibrosis patients with residual CFTR function: N-of-1 studies. Pediatr Pulmonol. 2017;52:472–479. DOI: 10.1002/ppul.23659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Peabody Lever JE, Mutyam V, Hathorne HY, et al. . Ataluren/ivacaftor combination therapy: two N-of-1 trials in cystic fibrosis patients with nonsense mutations. Pediatr Pulmonol. 2020;55:1838–1842. DOI: 10.1002/ppul.24764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Kim J, Hu C, Moufawad El Achkar C, et al. . Patient-customized oligonucleotide therapy for a rare genetic disease. N Engl J Med. 2019;381:1644–1652. DOI: 10.1056/NEJMoa1813279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Marks P, Witten C. Toward a new framework for the development of individualized therapies. Gene Ther. 2020. [Epub ahead of print]; DOI: 10.1038/s41434-020-0143-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Stunnenberg BC, Deinum J, Nijenhuis T, et al. . N-of-1 trials: evidence-based clinical care or medical research that requires IRB approval? A practical flowchart based on an ethical framework. Healthcare (Basel). 2020;8:49 DOI: 10.3390/healthcare8010049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Blackston JW, Chapple AG, McGree JM, et al. . Comparison of aggregated n-of-1 trials with parallel and crossover randomized controlled trials using simulation studies. Healthcare (Basel). 2019;7:137 DOI: 10.3390/healthcare7040137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Jansen R, Embden JDAv, Gaastra W, et al. . Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. DOI: 10.1046/j.1365-2958.2002.02839.x [DOI] [PubMed] [Google Scholar]
- 241. Moreno AM, Catroli GF, Alemán F, et al. . Long-lasting analgesia via targeted in vivo epigenetic repression of Nav1.7. bioRxiv. 2019;711812 DOI: 10.1101/711812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Chadwick AC, Wang X, Musunuru K. In vivo base editing of PCSK9 (proprotein convertase subtilisin/kexin type 9) as a therapeutic alternative to genome editing. Arterioscler Thromb Vasc Biol. 2017;37:1741–1747. DOI: 10.1161/ATVBAHA.117.309881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Carreras A, Pane LS, Nitsch R, et al. . In vivo genome and base editing of a human PCSK9 knock-in hypercholesterolemic mouse model. BMC Biol. 2019;17:4 DOI: 10.1186/s12915-018-0624-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Cai Y, Cheng T, Yao Y, et al. . In vivo genome editing rescues photoreceptor degeneration via a Cas9/RecA-mediated homology-directed repair pathway. Sci Adv. 2019;5:eaav3335 DOI: 10.1126/sciadv.aav3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Latella MC, Di Salvo MT, Cocchiarella F, et al. . In vivo editing of the human mutant Rhodopsin gene by electroporation of plasmid-based CRISPR/Cas9 in the mouse retina. Mol Ther Nucleic Acids. 2016;5:e389 DOI: 10.1038/mtna.2016.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Yu W, Mookherjee S, Chaitankar V, et al. . Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat Commun. 2017;8:14716 DOI: 10.1038/ncomms14716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Jo DH, Song DW, Cho CS, et al. . CRISPR-Cas9-mediated therapeutic editing of Rpe65 ameliorates the disease phenotypes in a mouse model of Leber congenital amaurosis. Sci Adv. 2019;5:eaax1210 DOI: 10.1126/sciadv.aax1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Koo T, Park SW, Jo DH, et al. . CRISPR-LbCpf1 prevents choroidal neovascularization in a mouse model of age-related macular degeneration. Nat Commun. 2018;9:1855 DOI: 10.1038/s41467-018-04175-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Yang S, Chang R, Yang H, et al. . CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington's disease. J Clin Invest. 2017;127:2719–2724. DOI: 10.1172/JCI92087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Colasante G, Lignani G, Brusco S, et al. . dCas9-based Scn1a gene activation restores inhibitory interneuron excitability and attenuates seizures in Dravet syndrome mice. Mol Ther. 2020;28:235–253. DOI: 10.1016/j.ymthe.2019.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Matharu N, Rattanasopha S, Tamura S, et al. . CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science. 2019;363:eaau0629 DOI: 10.1126/science.aau0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Stephens CJ, Lauron EJ, Kashentseva E, et al. . Long-term correction of hemophilia B using adenoviral delivery of CRISPR/Cas9. J Control Release. 2019;298:128–141. DOI: 10.1016/j.jconrel.2019.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Guan Y, Ma Y, Li Q, et al. . CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. EMBO Mol Med. 2016;8:477–488. DOI: 10.15252/emmm.201506039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Chadwick AC, Evitt NH, Lv W, et al. . Reduced blood lipid levels with in vivo CRISPR-Cas9 base editing of ANGPTL3. Circulation. 2018;137:975–977. DOI: 10.1161/CIRCULATIONAHA.117.031335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Villiger L, Grisch-Chan HM, Lindsay H, et al. . Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med. 2018;24:1519–1525. DOI: 10.1038/s41591-018-0209-1 [DOI] [PubMed] [Google Scholar]
- 256. Shao Y, Wang L, Guo N, et al. . Cas9-nickase-mediated genome editing corrects hereditary tyrosinemia in rats. J Biol Chem. 2018;293:6883–6892. DOI: 10.1074/jbc.RA117.000347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Song C-Q, Jiang T, Richter M, et al. . Adenine base editing in an adult mouse model of tyrosinaemia. Nat Biomed Eng. 2020;4:125–130. DOI: 10.1038/s41551-019-0357-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Wang D, Li J, Song C-Q, et al. . Cas9-mediated allelic exchange repairs compound heterozygous recessive mutations in mice. Nat Biotechnol. 2018;36:839–842. DOI: 10.1038/nbt.4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Amoasii L, Long C, Li H, et al. . Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci Transl Med. 2017;9:eaan8081 DOI: 10.1126/scitranslmed.aan8081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Min Y-L, Li H, Rodriguez-Caycedo C, et al. . CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci Adv. 2019;5:eaav4324 DOI: 10.1126/sciadv.aav4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Nelson CE, Hakim CH, Ousterout DG, et al. . In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. DOI: 10.1126/science.aad5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Hakim CH, Wasala NB, Nelson CE, et al. . AAV CRISPR editing rescues cardiac and muscle function for 18 months in dystrophic mice. JCI Insight. 2018;3:e124297 DOI: 10.1172/jci.insight.124297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Long C, Amoasii L, Mireault AA, et al. . Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. DOI: 10.1126/science.aad5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Xu L, Park KH, Zhao L, et al. . CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol Ther. 2016;24:564–569. DOI: 10.1038/mt.2015.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Koo T, Lu-Nguyen NB, Malerba A, et al. . Functional rescue of dystrophin deficiency in mice caused by frameshift mutations using Campylobacter jejuni Cas9. Mol Ther. 2018;26:1529–1538. DOI: 10.1016/j.ymthe.2018.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Gao X, Tao Y, Lamas V, et al. . Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature. 2018;553:217–221. DOI: 10.1038/nature25164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. György B, Nist-Lund C, Pan B, et al. . Allele-specific gene editing prevents deafness in a model of dominant progressive hearing loss. Nat Med. 2019;25:1123–1130. DOI: 10.1038/s41591-019-0500-9 [DOI] [PMC free article] [PubMed] [Google Scholar]