Abstract
Exosomal long noncoding RNA (lncRNA) has been found to be associated with the development of cancers. However, the expression characteristics and the biological roles of exosomal lncRNAs in hepatocellular carcinoma (HCC) remain unknown. Here, by RNA sequencing, we found 9440 mRNAs and 8572 lncRNAs were differentially expressed (DE‐) in plasma exosomes between HCC patients and healthy controls. Exosomal DE‐lncRNAs displayed higher expression levels and tissue specificity, lower expression variability and splicing efficiency than DE‐mRNAs. Six candidate DE‐lncRNAs (fold change 6 or more, P ≤ .01) were high in HCC cells and cell exosomes. The knockdown of these candidate DE‐lncRNAs significantly affected the migration, proliferation, and apoptosis in HCC cells. In particular, a novel DE‐lncRNA, RP11‐85G21.1 (lnc85), promoted HCC cellular proliferation and migration by targeted binding and regulating of miR‐324‐5p. More importantly, the level of serum lnc85 was highly expressed in both Alpha‐fetoprotein (AFP)‐positive and AFP‐negative HCC patients and allowed distinguishing AFP‐negative HCC from healthy control and liver cirrhosis (area under the receiver operating characteristic curve, 0.869; sensitivity, 80.0%; specificity, 76.5%) with high accuracy. Our finding offers a new insight into the association between the dysregulation of exosomal lncRNA and HCC, suggesting that lnc85 could be a potential biomarker of HCC.
Keywords: hepatocellular carcinoma, long noncoding RNA, miR‐324‐5p, plasma exosome, RP11‐85G21.1
Long noncoding RNAs (lncRNAs) and mRNAs are aberrantly expressed in plasma exosomes of hepatocellular carcinoma (HCC). Exosomal lncRNAs could have greater potential to serve as the biomarkers of HCC than mRNAs. Exosomal lncRNAs participate in the regulation of cell biology processes. RP11‐85G21.1 promotes proliferation of HCC cell lines by acting as a sponge of microRNA‐324‐5p.
1. INTRODUCTION
Hepatocellular carcinoma (HCC) is the third leading cause of cancer‐related deaths and one of the most aggressive types of human cancers worldwide. 1 , 2 , 3 , 4 In China, the incidence of HCC is relatively high with approximately 360 000 new cases and 350 000 deaths each year. 5 Clinically, due to lack of specific biomarkers and poor clinical symptoms, most HCC patients are already in the advanced stage when diagnosed, and over 80% of diagnosed patients are associated with poor prognosis. 6 In addition, the effects of current therapies are generally unsatisfactory due to recurrence and metastasis, leading to an overall 5‐year survival rate for HCC of only 50%‐70% 7 and the 5‐year survival rate for advanced HCC (less than 15%) is even more dismal. 8 Therefore, it is urgent to investigate the molecular mechanisms involved in HCC initiation and progression in order to develop new diagnosis techniques and therapies. Accumulating evidence indicates that long noncoding RNA (lncRNA) plays a critical role in human disease. 9 , 10 Long noncoding RNA is a member of non‐protein‐coding transcripts composed of more than 200 nucleotides. 11 , 12 Some studies have indicated that lncRNAs are frequently deregulated in various cancers and tightly related to tumorigenesis 13 by their involvement in cell migration, proliferation, and apoptosis. 11 , 14 , 15 Indeed, several lncRNAs have been reported to serve as oncogenes or tumor suppressors for HCC, 16 , 17 , 18 , 19 highlighting the importance of lncRNAs in HCC pathogenesis. Recently, exosomal lncRNA has received increasing attention. Exosomes are RNA‐ and protein‐containing small membrane vesicles (30‐150 nm) that are constantly secreted by cells, 20 and deliver macromolecular messages (including lncRNAs) that enable cell to cell communication. 21 , 22 , 23 However, whether and how exosomal lncRNAs are associated with HCC development have not been comprehensively delineated.
In this study, we used RNA sequencing and quantitative real‐time PCR (qRT‐PCR) to screen and validate differentially expressed (DE‐) lncRNA of plasma exosomes between HCC and healthy controls (HC). We investigated the expressed characteristics of exosomal lncRNAs and mRNAs in HCC, validated the DE‐lncRNAs in HCC cells and cell exosomes, and used loss‐of‐function assays to determine the biological functions of DE‐lncRNAs in HCC cells. Finally, we discovered a novel HCC‐specific lncRNA, RP11‐85G21.1 (lnc85), might play an important role in HCC tumorigenesis as a sponge of microRNA (miR)‐324‐5p (a tumor inhibitor factor) by binding to miR‐324‐5p and inhibiting its function, which has the potential to be a biomarker of HCC.
2. MATERIALS AND METHODS
2.1. Patients and blood sample collection
This study was approved by ethics committee of Guangxi Medical University (protocol no. 20170302‐1) and all methods were undertaken in accordance with the guidelines.
Blood samples from were obtained for RNA sequencing from 4 HCC patients at the First Affiliated Hospital of Guangxi Medical University (FAHGMU) in 2016 (age range, 41‐64 years; male/female = 2/2; Alpha‐fetoprotein [AFP]+/AFP− = 3/1), inclusion criteria were as the follows: (i) diagnosis of HCC by imaging, biochemistry, and pathology; (ii) no chemotherapy, radiotherapy, or other treatments; and (iii) patients were volunteered to participate in this study. Healthy controls (age and sex ratio matched with HCC) were recruited among healthy adults who took routine health examinations at FAHGMU and did not have any type of cancers. Blood samples were taken in the morning using EDTA‐containing tubes and centrifuged at 2890 g at 4°C for 10 minutes, then isolated plasma was stored at −80°C.
Blood samples of 112 HCC patients (age, 40‐60 years; male/female = 103/9; AFP+/AFP− = 30/82) and 43 liver cirrhosis (LC) patients (age, 40‐60 years; male/female = 39/4) were collected for qRT‐PCR from the FAHGMU in 2016. Fifty‐two HC were recruited from individuals who underwent routine health examinations at FAHGMU and did not have any type of cancer (age and sex ratio were matched with HCC patients). The inclusion criteria and sample collection were the same as RNA sequencing. Informed consent was obtained from all subjects.
2.2. Isolation of exosomes
2.2.1. Plasma
Exosomes were isolated from HCC plasma (HCC‐exos) and HC plasma (HC‐exos) by Ribo Exosome Isolation Reagent.
2.2.2. Cellular supernatant
The cells (purchased from the Chinese Academy of Sciences) were seeded in Petri dishes in DMEM (Gibco) with 10% FBS (Gibco, until the density of cells reached 90%, then the old medium was removed and replaced with FBS‐free medium for 48 hours of culture. Cellular supernatant was collected and exosomes were isolated using ultracentrifugation. Briefly, cells and debris were cleaned from cellular supernatant by 3 rounds of centrifugation (300 g, 10 minutes; 2000 g, 10 minutes; 10 000 ×g, 30 minutes). The prcleared supernatant was centrifuged at 110 000 g for 1.5 hours then discarded the upper liquid. The exosomes were defined as 7702‐exos, HepG2‐exos, and Huh7‐exos.
2.3. Characterization of exosomes
Exosomes were characterized by transmission electron microscopy (TEM), NanoSight, and western blot analysis. For NanoSight, exosomes were resuspended in 30 µL PBS (Gibco). The particle size was measured by the NanoSight NS300 system (Malvern). For western blot analysis, proteins of cell exosomes and plasma exosomes were extracted by using RIPA buffer (Cell Signaling Technology) and quantified by a bicinchoninic acid (BCA) protein quantification kit (Beyotime). Protein was separated by using 10% SDS‐PAGE and electrophoretically transferred to PVDF membranes (Millipore). Membranes were incubated overnight at 4°C with a 1:1000 dilution of primary Abs (Alix, CD9, CD63, and heat shock protein 70 [HSP70]) and GAPDH (Cell Signaling Technology). After incubation with a 1:1000 dilution of anti‐Ig HRP‐linked Ab for 1 hour at room temperature, the immunoreactive bands were visualized by Pierce ECL Western Blotting Substrate (Bio‐Rad). For TEM, exosomes were resuspended with 50 µL PBS then added to the paper, covered with copper mesh, and incubated at room temperature for 2 minutes. Then 1% acetic glaze dye was added for 30 seconds and observed by TEM (H7650; Hitachi).
2.4. RNA sequencing of plasma exosomes
Total RNA was isolated from plasma exosome by TRIzol (Life Technologies). The quantity and quality of RNA were assessed by the NanoDrop ND‐1000 spectrophotometer (Thermo Fisher Scientific). RNA integrity was determined by RNA integrity number with the 2200 TapeStation analyzer (Agilent Technologies), and samples that had an RNA integrity number above 7.0 were considered acceptable. The isolated RNAs were used for preparing cDNA libraries by NEBNext Ultra RNA Library Prep Kit for Illumina (ENB). Products were purified with Agencourt AMPure XP beads (Beckman) and diluted to 10 pM for cluster generation in situ on a HiSeq 3000 paired‐end flow cell. The clustering of the index‐coded samples was carried out using a HiSeq Rapid PE Cluster Kit V2 (Illumina). After cluster generation, the libraries were sequenced (2 × 150 bp) on the HiSeq 3000 platform (RiboBio Sequencing Facility, http://www.ribobio.com).
2.5. Analysis of exosomal sequencing data
The raw read sequences were filtered to remove adapter sequences and low‐quality reads (more than 20% bases with quality less than 20, or reads with more than 10% undefined nucleotides) by using Skewer and the FastX‐Toolkit software. Reads were mapped with human reference rRNA sequences (GenBank and GENCODE v26); reads that did not map with rRNA (clean reads) were subjected to subsequent analysis. Clean reads were aligned to human reference genome assembly (GRCh38) to define mRNA and lncRNA profiles by HISAT2 aligner with the default parameters. Clean reads were also annotated to GENCODE (v26) and the transcript expression level was quantified by the Expectation‐Maximization (RSEM) package. Expression levels were calculated by reads per kilobase of transcript per million mapped reads. Tissue specificity was calculated by comparing the expression level with publicly available RNA sequence data from 34 human cell types (ENCODE https://www.encodeproject.org; Illumina Human Body Map http://www.ebi.ac.uk/gxa/experiments/E‐MTAB‐513). Splicing efficiency was calculated by fragments per kilobase million of the exonic and intronic boundaries of the splice site (discarded exonic less than 0.2) and was defined ranging from 0 (intronic greater than or equal to exonic) to 100 (intronic = 0).
2.6. Quantitative real‐time PCR
Total RNA was isolated by the RNeasy Mini Kit (Qiagen). RNA was reverse‐transcribed to cDNA using a Reverse Transcription Kit (Takara), and qRT‐PCR analysis was carried out by Power SYBR Green (Takara). The PCR reactions were carried out on the StepOne Real‐time PCR System (Applied Biosystems). GAPDH and U6 were used as endogenous controls. The relative fold change in expression was calculated by the 2−ΔΔCt method. Primer sequences are listed in Table S1.
2.7. Western blot analysis
Cellular proteins were extracted with RIPA and quantified by a BCA quantification kit. Protein separation and identification were the same as described under “Characterization of exosomes”. The primary Abs were cyclin D1 and cyclin B1 (1:1000 dilution; Cell Signaling Technology), and GAPDH was the endogenous control.
2.8. Silencing of candidate lncRNAs and biological functions detect in HCC cells
Small interfering RNAs (Table S2) were designed for each candidate DE‐lncRNA. Each siRNA was transfected by Lipofectamine RNAiMAX (Thermo Fisher Scientific). Annexin V‐FITC Apoptosis Detection Kit (Life Technologies), CCK‐8 (Dojindo), and Transwell assay were used to detect the biological functions (cell apoptosis, proliferation, and invasion ability) in HCC cells after siRNA transfection of candidate DE‐lncRNAs.
2.9. Bioinformatics analysis
RegRNA (http://regrna2.mbc.nctu.edu.tw/) was used to predict the target gene of lnc85. 24
2.10. Dual‐luciferase reporter gene assay
The pmirGLO‐lnc85‐WT and pmirGLO‐lnc85‐MUT vectors were constructed and cotransfected into Huh7 and HepG2 cells with miR‐324‐5p mimics (5′‐CGCAUCCCCU AGGGCAUUGGUG‐3′) and miR‐negative control by Lipofectamine 3000 Reagent (Thermo Fisher Scientific), respectively. After 48 hours of transfection, the luciferase activity was detected by dual‐luciferase reporter gene assay kit (Yeasen Biotech).
2.11. Statistical analysis
Data were presented as mean ± SD. Statistical analysis was carried out by SPSS 20.0 (IBM). Student’s t test was used to analyze the difference between 2 groups. One‐way ANOVA was adopted when comparing more than 2 groups. P values less than or equal to .05 were considered to be statistically significant.
3. RESULTS
3.1. Identification of exosomes
The isolated exosomes were analyzed by NanoSight, TEM, and western blot. In general, more than 80% of the particle size was in the range of 30‐150 nm, with the highest peak at 59 nm (Figure 1A). The TEM scan showed that the particles consisted of bilayer membranes (~100 nm; Figure 1B). The biomarkers of exosomes, Alix, HSP70, and CD9, were expressed in cell exosomes, and Alix and CD63 were expressed in plasma exosomes (Figure 1C).
FIGURE 1.
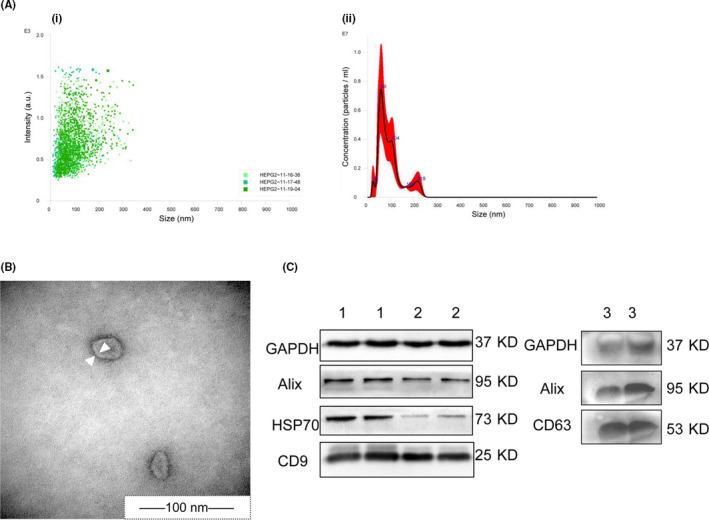
Identification of exosomes. A, NanoSight of cell exosomes. (i) Three fields of view for analysis. (ii) Intensity curve image of exosome size. B, Transmission electron microscopy images of cell exosomes. Arrowheads indicate the bilayer membrane. C, Western blot images of exosome proteins (exos). (1) HepG2‐exos, (2) HL‐7702‐exos. (3) Plasma‐exos. HSP70, heat shock protein 70
3.2. Long noncoding RNAs have higher expression level and tissue specificity and lower variability and splicing efficiency than mRNAs in HCC‐exos
After comparing the expression characteristics between exosomal lncRNAs and exosomal mRNAs, no significant difference in median expression levels between them (1.47 vs. 1.36, respectively; P = .084) was found in HC‐exos. In HCC‐exos, however, the median expression levels of lncRNAs were almost twice that of mRNAs (4.13 vs. 2.21, respectively; P < .005) (Figure 2A). We observed that the rate of specific transcripts of exosomal lncRNAs (15.78%) was significantly higher than exosomal mRNAs (13.28%) in HC‐exos. This phenomenon was stronger in HCC‐exos (25.48% for lncRNAs vs. 18.99% for mRNAs) (Figure 2B). In addition, the splicing efficiency of lncRNAs was lower than mRNAs whether in HCC‐exos (0.38 vs. 0.53, respectively; P < .005) or in HC‐exos (0.50 vs. 0.98, respectively; P < .005) (Figure 2C). Moreover, the expression variability of lncRNAs was significantly lower than mRNAs in HCC‐exos (0.40 vs. 0.68, respectively; P < .005) (Figure 2D).
FIGURE 2.
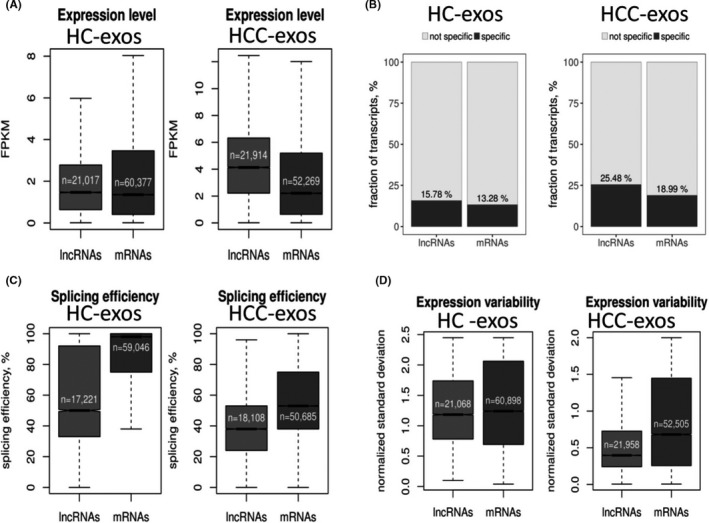
Expression characteristics of exosomal (exos) long noncoding RNAs (lncRNAs) and mRNAs. A, Median expression levels of lncRNA and mRNA transcripts. B, Specific expression lncRNA and mRNA transcript. C, Median splicing efficiency levels of lncRNA and mRNA transcripts. D, Interindividual variability of differentially expressed (DE)‐lncRNA and DE‐mRNA transcripts
3.3. Exosomal lncRNAs and mRNAs are differentially expressed between HC‐exos and HCC‐exos
The expression profiles of exosomal lncRNAs and mRNAs in HCC and HC were examined. According to the criteria of fold change greater than or equal to 2.0 and false discovery rate less than 0.01, we identified a total of 8572 DE‐lncRNAs (fold change 2 or higher, P < .05), in which 8447 were upregulated and 125 were downregulated in HCC‐exos (Figure 3Ai). Likewise, there were 9440 DE‐mRNAs, 8,963 were up‐regulated and 477 were down‐regulated in HCC‐exos (Figure 3Aii). In order to investigate whether these differential exosomal lncRNAs were able to distinguish HCC from HC, volcano plotting (Figure 3A) and hierarchical clustering analyses (Figure 3B) were adopted, and the results consistently showed that these differential exosomal lncRNAs had the ability to distinguish HCC from HC. Similar to exosomal lncRNAs, the expression profiles of exosomal mRNAs in HCC‐exos were also distinct from HC‐exos. The raw expression levels, fold change expression values, and P values of partially differentially expressed genes as determined by RNA Sequencing are summarized in Table S3.
FIGURE 3.
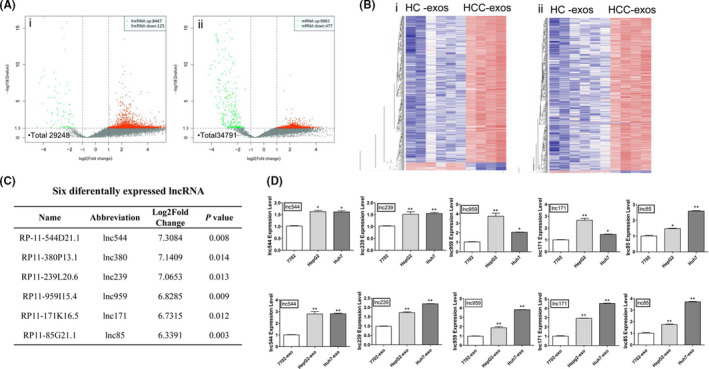
Differentially expressed long noncoding RNAs (DE‐lncRNAs) and DE‐mRNAs between hepatocellular carcinoma (HCC) and healthy controls (HC). A, Volcano plot showing DE‐lncRNAs (i) and DE‐mRNAs (ii) between HCC exosomes (exos) and HC‐exos. Red and green points represent higher and lower expression of transcription (mRNA or lncRNA) with statistical significance (fold change ≥ or ≤2.0, respectively; P ≤ .05) (X axes, fold change values; Y axes, P values). B, Hierarchical clustering of DE‐lncRNA (i) and DE‐mRNA (ii) between HCC‐exos and HC‐exos (red, high expression; blue, low expression). C, Expression levels of 6 DE‐lncRNAs (fold change ≥ 6.0; P ≤ .05) in HCC‐exos. D, Respective expression level of 5 DE‐lncRNAs in HCC cells and cell exosomes by quantitative real‐time PCR. Data are expressed as mean ± SD of 3 independent experiments. *P ≤ .05, **P ≤ .005 vs. control (7702 and 7702‐exo as control, respectively)
3.4. Quantitative real‐time PCR validation of DE‐lncRNAs in HCC cells and cell exosomes
To determine whether the results of RNA sequencing on plasma exosomes could be verified in HCC cells, qRT‐PCR was used to measure the expression levels of 6 DE‐lncRNAs (Figure 3C, upregulated in HCC‐exos measured by RNA sequencing) in HCC cells and their cell exosomes. The results showed that, except for lnc380, the expression levels of 5 DE‐lncRNAs (lnc544, lnc239, lnc959, lnc171, and lnc85) were higher in the 2 HCC cell lines (P < .05) than in 7702 cells. Similar to the results in cells, except for lnc380, the expression levels of the other 5 DE‐lncRNAs were higher in HCC cell exosomes than 7702‐exos (P < .05) (Figure 3D).
3.5. Knockdown of HCC‐related DE‐lncRNAs modified phenotypes of HCC cells
To explore whether 5 candidate DE‐lncRNAs (upregulated in HCC cells and cell exosomes) might play a role in cell proliferation, invasion, and apoptosis, we knocked down these DE‐lncRNAs in HCC cells by using their specific siRNAs. As shown in Figures 4A and S1, 5 candidate DE‐lncRNAs were significantly reduced in cells when transfected with their corresponding siRNA. Inhibiting lnc85 resulted in the impairment of cell proliferation; silencing the other 4 DE‐lncRNAs had little effect (Figures 4B and S2). In Transwell assays, silencing lnc85 resulted in a decreased migration in HCC cells (P < .05) (Figure 4C,D), knockdown of lnc239 led to decreased migration in Huh7 cells (P = .016), and inhibiting lnc959 reduced the migration of HepG2 cells (P = .005). However, silencing lnc544 and lnc171 had no significant effect on cell migration (Figure S3). In the apoptosis analysis, downregulating lnc85 could elevate cell apoptosis in Huh7 cells (P = .014) (Figure 4E,F). Inhibiting lnc171 could significantly increase HepG2 apoptosis (P < .005). Knockdown of lnc544 could slightly increase the apoptosis in Huh7 cells (Figure S4).
FIGURE 4.
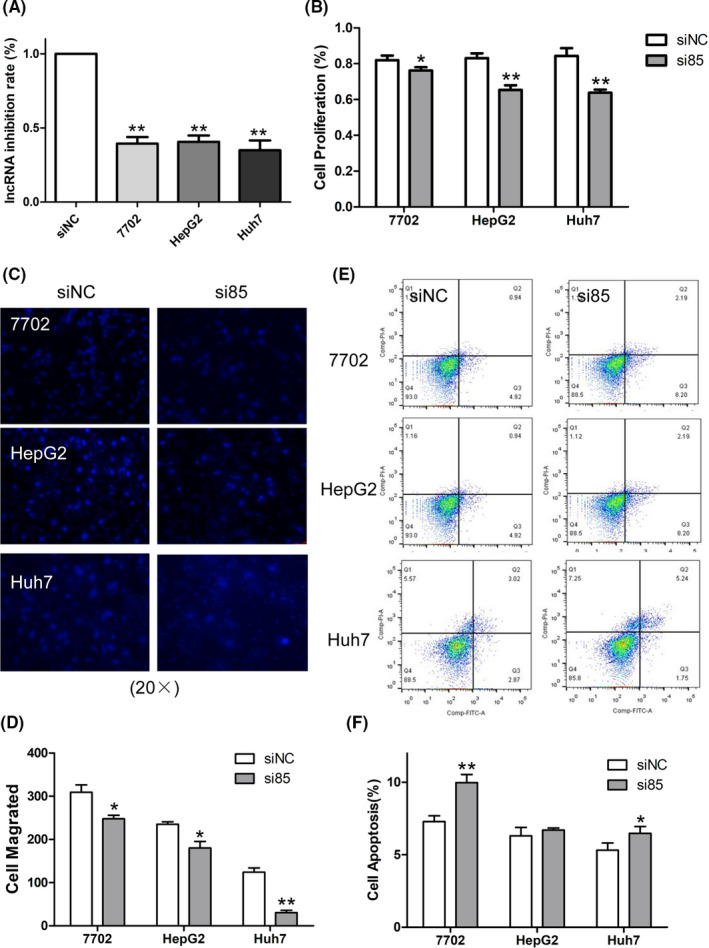
Knockdown of long noncoding RNA 85 (lnc85) significantly regulated cell proliferation, migration, and apoptosis phenotypes in hepatocellular carcinoma cells. A, Inhibition rate of lnc85 after transfection with siRNA. B, CCK‐8 was used to detect cell proliferation. C, D, Transwell assay was used to detect cell migration (20×) (C) and quantification was carried out (D). E, F, Flow cytometry was used to detect cell apoptosis (E) and quantification was carried out (F). Data are expressed as the mean ± SD of 3 independent experiments. *P ≤ .05, **P ≤ .005 vs. siNC (si‐negative control)
3.6. Long noncoding RNA 85 is a sponge of miR‐324‐5p
It is known that lncRNAs might function as competing endogenous RNAs (ceRNA) to modulate the expression and biological function of miRNA and participate in cellular biological processes. 24 To investigate the ceRNA characteristics of DE‐lncRNAs, we focused on lnc85, a novel lncRNA with high upregulation in HCC‐exos (Figure 3C). We found that lnc85 had a higher expression level in both HCC cells and cell exosomes (Figure 3D). By observing the effects of 5 DE‐lncRNAs on cell proliferation, apoptosis, and invasion activity, and it was found that lnc85 showed an obvious promoting effect on the proliferation and invasion ability of HCC cells (Figure 4). Therefore, we decided to study how lnc85 influences the proliferation and invasion of HCC cells.
The potential targets of lnc85 were predicted by RegRNA, which showed that the 3′‐UTR ends of lnc85 contained a predicted binding site for miR‐324‐5p. Therefore, we used a dual‐luciferase reporter gene assay to evaluate whether miR‐324‐5p could be targeted by lnc85 in HCC cells (Figure 5A). The results showed that miR‐324‐5p mimic significantly inhibited the luciferase activity of lnc85 WT 3′UTR in HepG2 and Huh7 cells (Figure 5B), whereas miR‐324‐5p mimic had no significant effect on the luciferase activity of mutant 3′UTR. In addition, qRT‐PCR analysis confirmed that the expression of miR‐324‐5p was negatively correlated with lnc85 in HCC cells (Figure 5C). Transfection of si85 suppressed the expression levels of lnc85, but it also upregulated miR‐324‐5p expression levels in HCC cells (Figure 5D), indicating that miR‐324‐5p might serve as an inhibitory target of lnc85.
FIGURE 5.
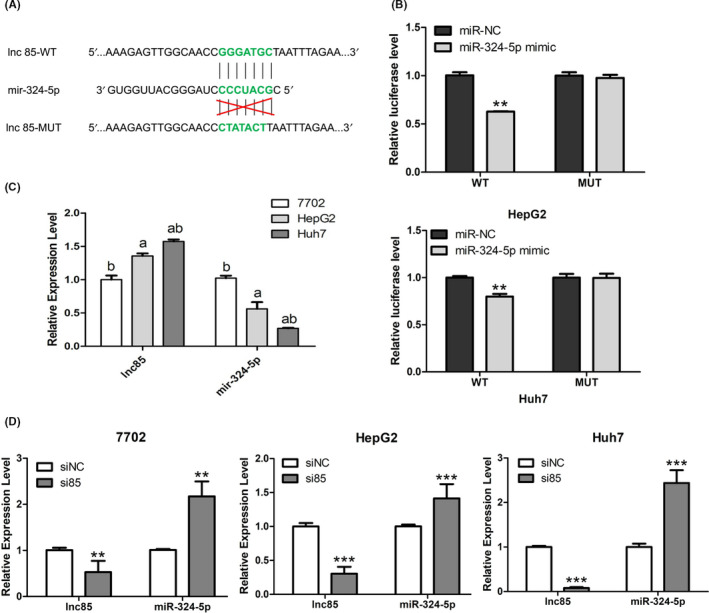
Targeting relationship between long noncoding RNA 85 (lnc85) and microRNA (miR)‐324‐5p. A, miR‐324‐5p putative binding site on lnc85. B, Relative luciferase activity was detected in HepG2 and Huh7 cells cotransfected with pmirGLO‐lnc85‐WT or pmirGLO‐lnc85‐MUT vectors and mir‐324‐5p mimic or miR‐negative control (miR‐NC). C, D, Expression level of lnc85 and miR‐324‐5p before (C) and after (D) silencing of lnc85 in HL‐7702, HepG2, and Huh7 cells. Data are expressed as the mean ± SD of 3 independent experiments. *P ≤ .05, **P ≤ .005, ***P ≤ .0005 vs. control
3.7. Long noncoding RNA 85 promotes proliferation, metastasis, and apoptosis of HCC cells by regulating the expression of miR‐324‐5p‐associated target genes
MicroRNA‐324‐5p, which is generated from a hairpin RNA structure, 25 serves as a tumor inhibitor in many cancers. 26 , 27 , 28 , 29 , 30 , 31 Overexpression of miR‐324‐5p significantly reduced the growth and invasive ability of cancer cells. As we had confirmed that miR‐324‐5p was targeted by lnc85, the effect on the expression level of miR‐324‐5p downstream target genes after silencing lnc85 was investigated. The results showed that silencing lnc85 significantly inhibited the mRNA expression level of several proliferation proteins (cyclin D1, cyclin B1, and c‐myc), which were downstream of miR‐324‐5p 31 , 32 , 33 , 34 , 35 in HCC cells. Similar results were also observed for Bcl‐2, one of the antiapoptotic proteins. 33 , 34 Moreover, inhibiting lnc85 led to the downregulation of miR‐324‐5p targets (MMP2 and MMP9) that are involved in pro‐invasion/metastasis 29 (Figure 6A‐C). Based on the results above, we could see that silencing lnc85 affected the mRNA expression level of proliferation, antiapoptotic, and metastasis genes, which were downstream of mir‐324‐5p, especially the proliferation and invasion genes. The mechanism of mir‐324‐5p inhibiting the invasion and migration of HCC cells by regulating the activities of MMP2 and MMP9 has been reported in detail. 29 Here, we studied the protein expression levels of proliferation‐related proteins (cyclin D1 and cyclin B1) after silencing of lnc85 by western blot. As shown in Figure 6D,E, compared to the siNC group, decreased levels of cyclin D1 and cyclin B1 were found in the siRNA group of Huh7 and HepG2 cells. Taken together, these results show that lnc85 might modify the proliferation, migration, and apoptosis of these cells by regulating the expression of miR‐324‐5p target genes (Figure 6F).
FIGURE 6.
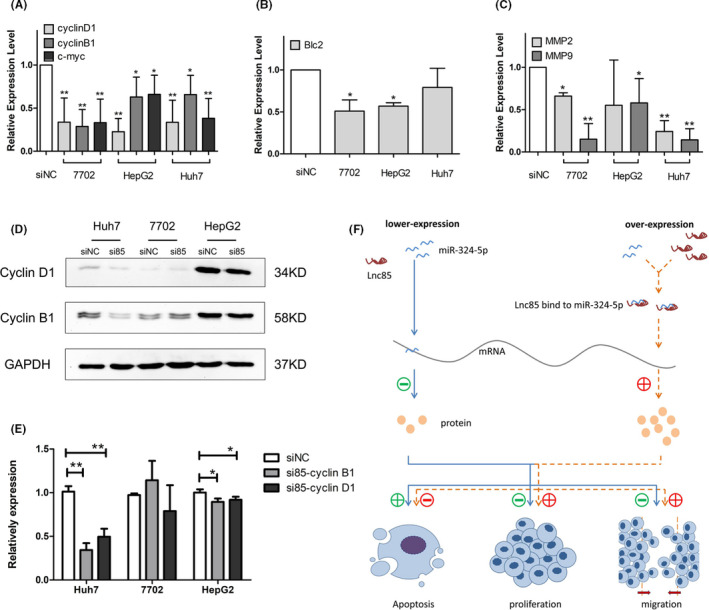
Scarcity of long noncoding RNA 85 (lnc85) enhances the suppression ability of microRNA (miR)‐324‐5p to its downstream gene. A‐C, mRNA expression of proliferation genes (A), antiapoptosis gene (B), and premigration genes (C) is decreased when lnc85 is silenced. D, E, Western blot images (D) and quantification (E) show downregulation of proliferation proteins after silencing of lnc85. F, Working model of lnc85 in hepatocellular carcinoma. Data are expressed as mean ± SD of 3 independent experiments. *P ≤ .05, **P ≤ .005 vs. siNC (si‐negative control)
3.8. Long noncoding RNA 85 is a biomarker for HCC
Some of the DE‐lncRNAs have been confirmed to be biomarkers for human disease. 36 In RNA sequencing, lnc85 was one of the DE‐lncRNAs that was highly expressed in both AFP+ and AFP− HCC‐exos (Figure 7A) and showed potential to be a biomarker of HCC. To explore whether lnc85 could serve as a biomarker of HCC, qRT‐PCR was used to detect the expression level of lnc85 in sera of 112 HCC, 43 LC, and 52 HC samples. As showed in Figure 7B, lnc85 was highly expressed in HCC compared to HC and LC (P < .005). More importantly, the expression level of lnc85 was not only significantly high in AFP+ HCC, but also highly expressed in AFP− HCC (P < .005). The results of receiver operating characteristic curve analyses were used to analyze the diagnostic value of lnc85 (Figure 7C,D). When HC and LC were combined into 1 group (HC + LC), the area under the curve (AUC) of lnc85 was 0.873 (P < .005; 95% confidence interval [CI], 0.828‐0.918) for HCC, and the sensitivity and specificity were 80.0% and 74.5% (cut‐off = 1.563). To distinguish between LC and HCC, the AUC of lnc85 was 0.888 (P < .005; 95% CI, 0.800‐0.938), and the sensitivity and specificity of lnc85 were 80.0% and 74.4% (cut‐off = 1.563). As mentioned above, lnc85 was highly expressed in serum of AFP− HCC. This outcome suggested that lnc85 might partially compensate for the diagnosis of AFP− HCC. Therefore, the diagnostic value of lnc85 in AFP+ and AFP− HCC was analyzed. Our results showed that when HC and LC were combined into 1 group (HC + LC), the AUC of lnc85 was 0.883 (P < .005; 95% CI, 0.835‐0.931) for AFP+ HCC, and 0.869 (P < .005; 95% CI, 0.816‐0.937) for AFP− HCC. The sensitivity and specificity of lnc85 in diagnosing AFP+ HCC were 80.5% and 76.5%, respectively (cut‐off = 1.650); in AFP− HCC the sensitivity and specificity were 80.0% and 76.5%, respectively (cut‐off = 1.645). To distinguish between LC and AFP+ or AFP− HCC, the AUC of lnc85 was 0.897 (P < .005; 95% CI, 0.846‐0.949) for AFP+ HCC, and 0.883 (P < .005; 95% CI, 0.809‐0.957) for AFP− HCC. The sensitivity and specificity of lnc85 in diagnosing AFP+ HCC were 80.5% and 76.7%, respectively (cut‐off = 1.650); in AFP− HCC, sensitivity and specificity were 80.0% and 76.7%, respectively (cut‐off = 1.645). This meant that lnc85 showed a compelling diagnostic potential in both AFP+ and AFP− HCC. As a result, lnc85 could be used as an independent biomarker for HCC diagnosis or in combination with other biomarkers to improve the diagnostic of AFP− HCC.
FIGURE 7.
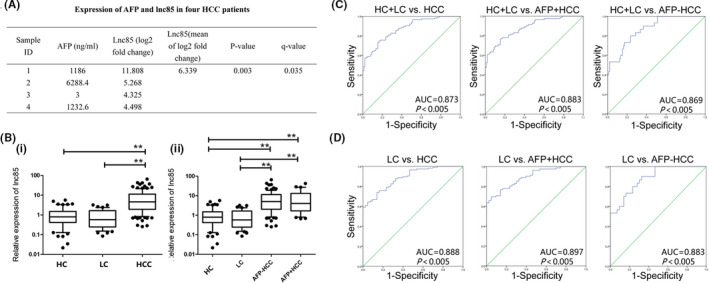
Long noncoding RNA 85 (lnc85) as a biomarker of hepatocellular carcinoma (HCC). A, Expression levels of α‐fetoprotein (AFP) and lnc85 in HCC exosomes. B, Lnc85 expression is higher in HCC patients’ serum (i), whether it is AFP‐negative (−) or AFP‐positive (+) (ii), than in healthy controls (HC) and patients with liver cirrhosis (LC). C, Receiver operating characteristic (ROC) curves of lnc85 diagnosis of HC, LC, and HCC patients. D, ROC curves of lnc85 diagnosis of LC and HCC patients. *P ≤ .05, **P ≤ .005 vs. HC
4. DISCUSSION
Recently, several reports have shown that a number of lncRNAs are aberrantly expressed in tumor tissues of HCC and play critical roles in tumor biological processes. 36 For example, lncRNA‐NNT‐AS1 promotes the cellular proliferation and migration of HCC by downregulating miR‐363. 37 Linc‐USP16 suppresses HCC cellular migration by increasing the expression of phosphatase and PTEN. 38 Cell exosomes are known to carry lncRNAs and transfer RNA cargo to recipient cells, but the potential roles of exosomal lncRNAs in HCC have not been systematically examined. In the present study, we revealed a significant difference in the expression characteristics between lncRNAs and mRNAs in HCC‐exos. To the best of our knowledge, this is the first study that systematically quantifies the expression features of exosomal lncRNAs and mRNAs by using RNA sequencing data in HCC‐exos. We observed that exosomal lncRNAs had higher expression levels and tissue specificity, and lower variability and splicing efficiency than exosomal mRNAs. The features suggested that exosomal lncRNAs could have greater potential to serve as biomarkers of HCC compared to exosomal mRNAs.
We found 125 downregulated and 8447 upregulated exosomal DE‐lncRNAs in HCC‐exos compared to HC‐exos. Although the biological functions of these lncRNAs were mostly unclear, several lines of evidence suggested that our findings were biologically plausible. First, several DE‐lncRNAs detected in this study had been biologically characterized in other studies. For instance, LINC00640 was found as a new nonsyndromic cleft lip with palate risk loci. 39 ZNF571‐AS1 was involved in acute myeloid leukemia through the STAT signaling pathway by regulating KIT and STAT5, and could be served as prognostic biomarkers. 40 CASC9 was upregulated in esophageal squamous cell carcinoma (ESCC) and considered as a valuable biomarker for ESCC diagnosis. 41 The DE‐lncRNAs that showed the largest difference in expression levels (fold change of 6 or higher) in HCC were also found to be upregulated in both HCC cells and cell exosomes. Second, loss‐of‐function experiments found that some of these DE‐lncRNAs contributed to HCC cell proliferation, migration, and apoptosis, suggesting DE‐lncRNAs might play important roles in HCC pathogenesis. Finally, the regulatory effects of dysregulated lncRNAs on particular miRNAs and their downstream target genes indicated that lncRNAs might contribute to carcinogenesis, acting as oncogenes or tumor suppressor genes. 42 , 43 Further studies are needed to uncover the details of molecular mechanisms on how these lncRNAs, individually or cooperatively, contribute to the carcinogenesis of HCC.
Upregulated in HCC cells and cell exosomes, as well as in HCC‐exos, lnc85 is a novel lncRNA with unidentified biological functions. In this study, we found that silencing lnc85 led to the inhibition of proliferation and migration in HCC cells. Long noncoding RNA 85 targeted miR‐324‐5p and regulated its expression through a ceRNA mechanism, 44 and subsequently modulated the expression of mRNAs that were critical for cell proliferation, apoptosis, and invasion. 45 , 46 , 47 MicroRNA‐324‐5p was derived through a maturation process in which pre‐miR‐324 was divided by RNase III endonuclease DICER, a key enzyme for miRNA biogenesis. 25 Until now, only a few studies have investigated the biological functions of miR‐324 in human cancer progression. Gu et al reported that miR‐324‐5p was capable of inhibiting proliferation and invasion activities of colorectal cancer cells. 27 Sun et al found that downregulating miR‐324‐5p caused cytoskeleton remodeling and tumorigenesis of colorectal epithelium. 28 Xu et al indicated that miR‐324‐5p inhibited the proliferation of glioma cells through GLI1 silencing. 31 More importantly, miR‐324‐5p overexpression could lead to the reduction of migration and invasion by modulating MMP2, MMP9, ETS1, and SP1 gene expression in HCC. 29 We discovered that when lnc85 was inhibited, the expression of miR‐324‐5p was upregulated, leading to the downregulation of miR‐324‐5p‐targeted mRNAs. In summary, our data suggested that lnc85 could promote the cell proliferation of HCC by functioning as a sponge of miR‐324‐5p, resulting in the dysregulation of miR‐324‐5p‐targeted genes.
Finally, we discussed the possibility that lnc85 could be used as a biomarker for HCC. Clinically, AFP is the most widely used serum biomarker for HCC diagnosis. However, the diagnostic sensitivity of AFP is partially impacted by AFP− HCC. 48 Our study found that lnc85 was significantly increased in both AFP+ and AFP− HCC, and it distinguished AFP− HCC from HC and LC. All of these findings suggested that lnc85 is able to serve as a potential biomarker of HCC, and helps to improve the sensitivity of diagnosis in AFP− HCC.
In conclusion, our study systematically characterized the expression features of exosomal lncRNAs in HCC‐exos and, at the same time, identified specific DE‐lncRNAs that play important roles in the regulation of cell proliferation, migration, and apoptosis. Long noncoding RNA 85 could function as an oncogenic lncRNA for HCC tumorigenesis by regulating miR‐324‐5p expression. Finally, lnc85 was found to be elevated in HCC serum and showed a compelling potential in HCC diagnosis. Our findings support that exosomal lncRNA plays an important role in HCC and facilitates the development of exosomal lncRNA‐related biomarkers or therapeutics targets.
CONFLICT OF INTEREST
The authors have no conflict of interest.
Supporting information
Figs S1‐S4
Table S1
Table S2
Table S3
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81760612, 81572908), Program of the Key Laboratory of High‐Incidence Tumor Prevention and Treatment (Guangxi Medical University), Ministry of Education, China (GKE2019‐03), the Science and Technology Department of Guangxi (2018GXNSFAA281165, ZY1949025), the Peacock Initiative Program of Shenzhen Municipality (827‐000157), the Science and Innovation Committee of Shenzhen Municipality (KQCX2015032416521307, JCYJ20160427105140594), and the Innovation Project of Guangxi Graduate Education (30/02407217014C).
Huang X, Sun L, Wen S, et al. RNA sequencing of plasma exosomes revealed novel functional long noncoding RNAs in hepatocellular carcinoma. Cancer Sci. 2020;111:3338–3349. 10.1111/cas.14516
Xuejing Huang, Liyuan Sun, and Sha Wen contributed equally to this work.
Contributor Information
Jianning Jiang, Email: jjianning@163.com.
Rihong Zhai, Email: rzhai@szu.edu.cn.
Min He, Email: hemin@gxmu.edu.cn.
REFERENCES
- 1. Venook AP, Papandreou C, Furuse J, de Guevara LL. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist. 2010;15(Suppl 4):5‐13. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69‐90. [DOI] [PubMed] [Google Scholar]
- 3. Chiang JK, Koo M, Kuo TB, Fu CH. Association between cardiovascular autonomic functions and time to death in patients with terminal hepatocellular carcinoma. J Pain Symptom Manage. 2010;39:673‐679. [DOI] [PubMed] [Google Scholar]
- 4. Yang X, Xie X, Xiao YF, et al. The emergence of long non‐coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett. 2015;360:119‐124. [DOI] [PubMed] [Google Scholar]
- 5. Chen JG, Zhang SW. Liver cancer epidemic in China: past, present and future. Semin Cancer Biol. 2011;21:59‐69. [DOI] [PubMed] [Google Scholar]
- 6. Dai M, Chen S, Wei X, et al. Diagnosis, prognosis and bioinformatics analysis of lncRNAs in hepatocellular carcinoma. Oncotarget. 2017;8:95799‐95809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang Y, Sun A, Zhao Y, et al. Proteomics identifies new therapeutic targets of early‐stage hepatocellular carcinoma. Nature. 2019;567(7747):257‐261. [DOI] [PubMed] [Google Scholar]
- 8. DiStefano JK, Davis B. Diagnostic and prognostic potential of AKR1B10 in human hepatocellular carcinoma. Cancers. 2019;11(4):486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tano K, Akimitsu N. Long non‐coding RNAs in cancer progression. Front Genet. 2012;3:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354‐361.21550244 [Google Scholar]
- 11. Fatica A, Bozzoni I. Long non‐coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7‐21. [DOI] [PubMed] [Google Scholar]
- 12. Cui M, Zheng M, Sun B, Wang Y, Ye L, Zhang X. A long noncoding RNA perturbs the circadian rhythm of hepatoma cells to facilitate hepatocarcinogenesis. Neoplasia. 2015;17:79‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang F, Zhang L, Zhang C. Long noncoding RNAs and tumorigenesis: genetic associations, molecular mechanisms, and therapeutic strategies. Tumour Biol. 2016;37:163‐175. [DOI] [PubMed] [Google Scholar]
- 14. Li J, Xuan Z, Liu C. Long non‐coding RNAs and complex human diseases. Int J Mol Sci. 2013;14:18790‐18808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904‐914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lai MC, Yang Z, Zhou L, et al. Long non‐coding RNA MALAT‐1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810‐1816. [DOI] [PubMed] [Google Scholar]
- 17. Shibata C, Otsuka M, Kishikawa T, et al. Diagnostic and therapeutic application of noncoding RNAs for hepatocellular carcinoma. World. J Hepatol. 2015;7:1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ding C, Yang Z, Lv Z, et al. Long non‐coding RNAPVT1 is associated with tumor progression and predicts recurrence in hepatocellular carcinoma patients. Oncol Lett. 2015;9:955‐963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let‐7 sponge. J Exp Clin Cancer Res. 2015;34:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li M, Zeringer E, Barta T, Schageman J, Cheng A, Vlassov AV. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos Trans R Soc Lond B Biol Sci. 2014;369(1652):20130502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Skog J, Würdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumor growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470‐1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taylor DD, Gercel‐Taylor C. The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Front Genet. 2013;4:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu D, Tahara H. The role of exosomes and microRNAs in senescence and aging. Adv Drug Deliv Rev. 2013;65:368‐375. [DOI] [PubMed] [Google Scholar]
- 24. Bonora M, Wieckowsk MR, Chinopoulos C, et al. Molecular mechanisms of cell death: central implication of ATP synthase in mitochondrial permeability transition. Oncogene. 2015;34:1608. [DOI] [PubMed] [Google Scholar]
- 25. Kuo WT, Yu SY, Li SC, et al. MicroRNA‐324 in human cancer: miR‐324‐5p and miR‐324‐3p have distinct biological functions in human cancer. Anticancer Res. 2016;36:5189‐5196. [DOI] [PubMed] [Google Scholar]
- 26. Jiang H, Huang G, Zhao N, et al. Long non‐coding RNA TPT1‐AS1 promotes cell growth and metastasis in cervical cancer via acting AS a sponge for miR‐324‐5p. J Exp Clin Cancer Res. 2018;37:169. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27. Gu C, Zhang M, Sun W, Dong C. Up‐regulation of miR‐324‐5p inhibits proliferation and invasion of colorectal cancer cells by targeting ELAVL1. Oncol Res. 2019;27(5):515‐524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun LN, Xing C, Zhi Z, et al. Dicer suppresses cytoskeleton remodeling and tumorigenesis of colorectal epithelium by miR‐324‐5p mediated suppression of HMGXB3 and WASF‐2. Oncotarget. 2017;8:55776‐55789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cao L, Xie B, Yang X, et al. MiR‐324‐5p suppresses hepatocellular carcinoma cell invasion by counteracting ECM degradation through post‐transcriptionally downregulating ETS1 and SP1. PLoS One. 2015;10:e0133074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chiam K, Wang T, Watson DI, et al. Circulating serum exosomal miRNAs as potential biomarkers for esophageal adenocarcinoma. J Gastrointest Surg. 2015;19:1208‐1215. [DOI] [PubMed] [Google Scholar]
- 31. Xu HS, Zong HL, Shang M, et al. MiR‐324‐5p inhibits proliferation of glioma by target regulation of GLI1. Eur Rev Med Pharmacol Sci. 2014;18:828‐832. [PubMed] [Google Scholar]
- 32. Wang YZ, Han JJ, Fan SQ, et al. miR‐132 weakens proliferation and invasion of glioma cells via the inhibition of Gli1. Eur Rev Med Pharmacol Sci. 2018;22:1971‐1978. [DOI] [PubMed] [Google Scholar]
- 33. Tang CT, Lin XL, Wu S, et al. NOX4‐driven ROS formation regulates proliferation and apoptosis of gastric cancer cells through the GLI1 pathway. Cell Signal. 2018;46:52‐63. [DOI] [PubMed] [Google Scholar]
- 34. Lin Z, Sheng H, You C, et al. Inhibition of the cyclin D1 promoter in response to sonic hedgehog signaling pathway transduction is mediated by Gli1. Exp Ther Med. 2017;13:307‐314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun Y, Guo W, Ren T, et al. Gli1 inhibition suppressed cell growth and cell cycle progression and induced apoptosis as well as autophagy depending on ERK1/2 activity in human chondrosarcoma cells. Cell Death Dis. 2014;5:e979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hu J, Song C, Duan B, et al. LncRNA‐SVUGP2 suppresses progression of hepatocellular carcinoma. Oncotarget. 2017;8:97835‐97850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu YB, Jiang Q, Yang MY, Zhou JX, Zhang Q. Long noncoding RNA NNT‐AS1 promotes hepatocellular carcinoma progression and metastasis through miR‐363/CDK6 axis. Oncotarget. 2017;8:88804‐88814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sui J, Yang X, Qi W, et al. Long non‐coding RNA linc‐USP16 functions as a tumour suppressor in hepatocellular carcinoma by regulating PTEN expression. Cell Physiol Biochem. 2017;44:1188‐1198. [DOI] [PubMed] [Google Scholar]
- 39. Yu Y, Zuo X, He M, et al. Genome‐wide analyses of non‐syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat Commun. 2017;8:14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pan JQ, Zhang YQ, Wang JH, Xu P, Wang W. lncRNA co‐expression network model for the prognostic analysis of acute myeloid leukemia. Int J Mol Med. 2017;39:663‐671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu Y, Hu L, Liang Y, et al. Up‐regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer. 2017;16:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Braconi C, Kogure T, Valeri N, et al. microRNA‐29 can regulate expression of the long non‐coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750‐4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non‐coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39:2119‐2128. [DOI] [PubMed] [Google Scholar]
- 44. Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. 2013;425:3723‐3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR‐331‐3p in gastric cancer. Mol Cancer. 2014;13:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cao C, Zhang T, Zhang D, et al. The long non‐coding RNA, SNHG6‐003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene. 2017;36:1112‐1122. [DOI] [PubMed] [Google Scholar]
- 47. Zhou X, Zhang W, Jin M, Chen J, Xu W, Kong X. lncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR‐22‐3p in diabetic cardiomyopathy. Cell Death Dis. 2017;8:e2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trevisani F, Garuti F, Neri A. Alpha‐fetoprotein for diagnosis, prognosis, and transplant selection. Semin Liver Dis. 2019;39(2):163‐177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs S1‐S4
Table S1
Table S2
Table S3


