Abstract
Gastric cancer (GC) is one of the deadliest cancers worldwide, and the progression of gastric carcinogenesis (GCG) covers multiple complicated pathological stages. Molecular mechanisms of GCG are still unclear. Here, we undertook NMR‐based metabolomic analysis of aqueous metabolites extracted from gastric tissues in an established rat model of GCG. We showed that the metabolic profiles were clearly distinguished among 5 histologically classified groups: control, gastritis, low‐grade gastric dysplasia, high‐grade gastric dysplasia (HGD), and GC. Furthermore, we carried out metabolic pathway analysis based on identified significant metabolites and revealed significantly disturbed metabolic pathways closely associated with the 4 pathological stages, including oxidation stress, choline phosphorylation, amino acid metabolism, Krebs cycle, and glycolysis. Three metabolic pathways were continually disturbed during the progression of GCG, including taurine and hypotaurine metabolism, glutamine and glutamate metabolism, alanine, aspartate, and glutamate metabolism. Both the Krebs cycle and glycine, serine, and threonine metabolism were profoundly impaired in both the HGD and GC stages, potentially due to abnormal energy supply for tumor cell proliferation and growth. Furthermore, valine, leucine, and isoleucine biosynthesis and glycolysis were significantly disturbed in the GC stage for higher energy requirement of the rapid growth of tumor cells. Additionally, we identified potential gastric tissue biomarkers for metabolically discriminating the 4 pathological stages, which also showed good discriminant capabilities for their serum counterparts. This work sheds light on the molecular mechanisms of GCG and is of benefit to the exploration of potential biomarkers for clinically diagnosing and monitoring the progression of GCG.
Keywords: gastric carcinogenesis, metabolic pathway, nuclear magnetic resonance, rat model, tissue metabolomics
These results provide new insights into the metabolic mechanisms underlying the progression of gastric carcinogenesis and are beneficial to exploit potential biomarkers for clinically diagnosing and monitoring gastric cancer progression.

1. INTRODUCTION
Gastric cancer (GC) is one of the most prevalent and deadly forms of cancers worldwide, 1 and is especially prevalent in Asian countries. Gastric carcinogenesis (GCG) covers multiple pathological stages. Before progressing to GC, gastric mucosa goes through a series of pathological changes bringing gastritis, atrophy, intestinal metaplasia, and atypical hyperplasia. 2 Until now, molecular mechanisms underlying GC pathogenesis are not yet clear. As one of the most important features of gastric tumors, metabolic disorders are closely connected with the progression of GCG. 3 Gastric tumors differ from their normal counterparts in several biochemical properties, such as increased cell proliferation, cell differentiation, and turnover of nutrients. The unique properties of gastric tumors are closely associated with profoundly impaired metabolism of tumor cells relative to normal cells. 4 , 5 Therefore, a detailed understanding of changing metabolic profiles during the progression of GCG is essential for clarifying the molecular mechanisms of GC pathogenesis. 6 Such investigations would be beneficial to reduce the incidence and mortality of GC.
In our previous work, we established a rat model of GCG, and classified rats into the normal control group (CON) and 4 pathological groups, gastritis (GS), low‐grade gastric dysplasia (LGD), high‐grade gastric dysplasia (HGD), and GC, based on the histological classification of gastric tissues. 7 Using nuclear magnetic resonance (NMR)‐based metabolomic analysis, we identified distinctly changed metabolic profiles and significantly disturbed metabolic pathways associated with the 4 pathological stages of GCG relative to the CON stage. 7 Three metabolic pathways were continually disturbed during GCG, including oxidative stress, choline phosphorylation, and fatty acid degradation. Moreover, amino acid metabolism was profoundly perturbed in gastric dysplasia and GC, and glycine, serine, and threonine metabolism and glycolysis were also dramatically impaired in GC. 7
It is well known that metabolic profiles of tumor tissues are somewhat distinct from those of sera due to different distributions of metabolites in tumor‐located tissues from those in sera. Sera usually reflect global pathological features of diseases. Compared with metabolic profiling of sera, metabolic profiling of tissues is more specifically and closely related to metabolic disorders and disturbed regulatory mechanisms in diseases. 8 , 9 Moreover, significant metabolites identified from tumor tissues could be explored to determine novel potential biomarkers for clinical applications. 10 Furthermore, metabolic differences between tumor cells and their surrounding host cells could provide a relatively accurate understanding of metabolic mechanisms underlying the invasion and metastasis of tumors. 11
In recent years, metabolomics has been extensively used to mechanically understand disease development and progression. By the combination of NMR/LC‐MS/GC‐MS techniques with pattern recognition methods, metabolomic analysis can be applied to address metabolic changes of biological systems in multistep processes of cancer progression. Significantly, NMR spectroscopy is characterized by several unique advantages, such as noninvasive, nonsample destructive, high resolution, and high experimental repeatability. 12 Recently, NMR‐based metabolomics has been applied for undertaking metabolic profiling of GC tissues to both address molecular mechanisms of GC pathogenesis and identify potential biomarkers for the early diagnosis and prognostic prediction of GC. Wang et al carried out metabolic profiling of gastric tissues on a large cohort of GC patients and normal controls. 13 They found that GC patients had distinguished metabolic profiles from normal controls, and also identified 13 differential metabolites between pathological stage‐related tissue samples (stages I‐IV) and normal counterparts. 14 Zhang et al undertook metabolic profiling of GC patients with lymph node metastasis (LNM) and identified branched‐chain amino acids (BCAAs; leucine, isoleucine, and valine), glutathione, and betaine to be potential factors in the diagnosis and prognosis of GC patients with or without LNM. 14 Furthermore, Jung et al compared metabolic profiles of matched tumor and normal stomach tissues, and reported that significantly altered metabolites in GC tissues were associated with amino acid metabolism and lipid metabolism. 15
Gas chromatography‐MS based metabolomics has also been used to undertake metabolic profiling of GC tissues, aiming to mechanistically understand molecular mechanisms of GC pathogenesis and exploit potential biomarkers for GC diagnosis. Wu et al showed the metabolic distinction between malignant and nonmalignant tissues of gastric mucosae in GC patients. 16 They identified 18 significant metabolites in malignant tissues compared with adjacent nonmalignant tissues, and also detected 5 significant metabolites in invasive gastric tumors relative to noninvasive gastric tumors. 16 In addition, Chen et al revealed the metabolic difference between metastatic and nonmetastatic gastric tumors based on animal models of human GC. 17 They identified 20 upregulated and 9 downregulated metabolites in metastatic gastric tumors compared to the nonmetastatic gastric tumors.
Gastric carcinogenesis is a multistep process connected with several pathological stages. However, few researches have specifically addressed the molecular mechanisms underlying the progression of GCG or exploit potential biomarkers for the early diagnosis of GC by using the gastric tissue. In the present work, we undertook NMR‐based metabolomic analyses of aqueous metabolites extracted from gastric tissues based on the rat model of GCG established previously. 7 We addressed profoundly changed metabolic profiles, identified significant metabolites with markedly altered levels, and significantly disturbed metabolic pathways as well as potential biomarkers, which were associated with the 4 pathological stages of GCG. This work could be beneficial to understanding the molecular mechanisms underlying the progression of GCG.
2. MATERIALS AND METHODS
2.1. Chemicals and animal diets
All used chemicals were the analytical grade. N‐methyl‐N′‐nitro‐N‐nitrosoguanidine (MNNG) was purchased from TCI (Shanghai) Development Co. Sodium azide (NaN3) was obtained from Sangon Biotech (Shanghai) Co. NaH2PO4.2H2O and K2HPO4.3H2O were purchased from Sinopharm Chemical Reagent Co. Sodium 3‐(trimethylsilyl)‐propionate‐2,2,3,3‐d4 (TSP) (99.8% D) was purchased from Cambridge Isotope Laboratories. The custom 8% NaCl chow pellets were obtained from Suzhou ShuangShi Laboratory Animal Feed Science Co. The MNNG was dissolved in water (1 mg/mL) and kept at 4°C. The stock solution of MNNG was diluted to 100 μg/mL by tap water just before use.
2.2. Animal experiments and histopathology
The rat model of GCG was established in accordance with protocols approved by Xiamen University Animal Ethics Committee, and documented in our previous work. 7 A total of 128 Wistar rats (age 3 weeks, male) were randomly divided into the MODEL group (n = 96) and CON group (n = 32). The stomach tissues from MODEL and CON rats were fixed in 10% formalin. After dehydrating, the biopsies embedded in wax were sectioned at 5 μm and stained with H&E for histopathological examination by light microscopy.
2.3. Tissue pretreatments and NMR samples
Pieces of gastric tissues (~100 mg/sample) were disrupted in solvents with an electric homogenizer. Aqueous metabolites were extracted from gastric tissues using the methanol/chloroform/water system. 18 Before NMR experiments, solvents were completely removed using a nitrogen blowing concentrator. Each sample of aqueous extracts was then reconstituted in 450 μL H2O and 50 μL phosphate buffer (1.5 M K2HPO4/NaH2PO4 and 10 mmol/L TSP), mixed uniformly, and centrifuged at 12 000 g for 10 minutes at 4°C to remove any insoluble components. Finally, 500 μL of the resulting supernatant was transferred into a 5 mm NMR tube and analyzed by NMR spectroscopy. 19
2.4. Nuclear magnetic resonance experiments and metabolite assignments
One‐dimensional (1D) 1H‐NMR experiments were carried out on a Bruker AVANCE III 600 MHz spectrometer (Bruker BioSpin) equipped with a BBFO CryoProbe at 298 K. The spectra of aqueous extracts of gastric tissues were recorded on BRUKER NMR Spectrometer by using the pulse sequence NOESYGPPR1D (RD‐90°‐t1‐90°‐τm‐90°‐ACQ) with water suppression during the relaxation delay (RD) and mixing time (τm). 19 The RD was 4 s, t 1 was a short delay (4 μs), and τm was 10 ms. A total of 32 transients were collected into 64K data points using a spectral width of 12 kHz with an acquisition time (ACQ) of 2.73 s.
Furthermore, 2D 1H‐1H total correlation spectroscopy (TOCSY) was acquired for selected NMR samples on a Bruker AVANCE III 850 MHz spectrometer with a TCI CryoProbe at 298K. The experimental parameters were detailed in previous reports. 20 , 21 , 22 Resonance assignments of metabolites were carried out based on the 1D 1H‐spectra using a combination of Chenomx NMR Suite (version 7.1) and the Human Metabolome Data Base (https://hmdb.ca/), referring to relevant published references. The assigned metabolites were confirmed by 2D TOCSY spectrum.
2.5. Data processing
The NMR spectral data processing was undertaken using the MestReNova software (version 9.0; Mestrelab Research). The free induction delay signals were processed by applying an exponential function with a line‐broadening factor of 0.3 Hz prior to Fourier transformation, and then manually phased and baseline‐corrected. The NMR spectra were referenced to the methyl group of TSP (δ 0.00). The spectral regions of δ 9.00‐0.00 were binned by 0.001 ppm. Each 1D 1H‐NMR spectrum was segmented into regions of 0.001 ppm. The region of water resonance δ 5.70‐4.60 was removed from the spectra to eliminate distorted baseline from imperfect water saturation. The icoshift algorithm was executed to align NMR peaks. 23 Both the open source of icoshift and MATLAB scripts can be downloaded from the website (www.models.life.ku.dk ). To reduce concentration differences among samples, we normalized the NMR spectral integrals of metabolites by dividing the total spectral integrals.
2.6. Multivariate statistics
Multivariate statistics were carried out using SIMCA‐P+ software (version 13.0.1; Umetrics). To compensate for differences in tissue sizes, the spectral integrals were normalized by the weight of the tissue sample. Thereafter, pareto scaling was applied to the normalized integrals for increasing the importance of low‐level metabolites without significant amplification of noise.
The pareto‐scaled data were analyzed by unsupervised principal component analysis (PCA) to reveal trends, highlight outliers, and show clusters among the tissue samples. 24 Supervised partial least squares to latent structure with discriminant analysis (PLS‐DA) was applied to improve the metabolic classification between the groups of samples, 25 and response permutation testing (RPT) with 600 cycles was utilized to assess the reliability of the sample classification. 26 Furthermore, OPLS‐DA was used to maximize the metabolic separation between the groups of samples, which removed uncorrelated variables within the classes using the orthogonal signal correction filter. Variables that participate in the classification was described on the first predictive principal component in the OPLS‐DA model. 27 , 28 In scores plots of PLS‐DA and OPLS‐DA, we surrounded each class with a 95% confidence ellipse, using the MATLAB function provided by the website (https://stackoverflow.com/questions/3417028/ellipse‐around‐the‐data‐in‐matlab).
2.7. Identification of significant metabolites
The loading plot of the successfully validated OPLS‐DA model was used to identify significant metabolites with 2 criteria. One was the variable importance value (VIP) in the projection, 29 and the other was the correlation coefficients (r) of the variables corresponding to the statistical significance P, relative to the first predictive component (tp1) in the OPLS‐DA model. 30 In the reconstituted OPLS‐DA loading plots, red, orange, and blue colors represent the metabolites with highly significant statistical difference (P < .01 and VIP > 1), significant statistical difference (0.01 ≤ P < .05 and VIP > 1), and no significant statistical difference (P ≥ .05 or VIP < 1), respectively.
2.8. Identification of significantly disturbed metabolic pathways
Based on relative integrals of the significant metabolites, metabolic pathway analysis was carried out to identify significantly disturbed pathways associated with the 4 pathological stages of GCG. The metabolic pathway analysis was carried out by using the Pathway Analysis module of MetaboAnalyst 4.0 (www.metaboanalyst.ca/). 31 Two parameters, statistical P value and pathway impact value (PIV), were used to evaluate the importance of the metabolic pathway. The P values were obtained from the quantitative enrichment analysis, 32 and the PIV values were calculated from the topological analysis with the out‐degree centrality arithmetic. According to the approaches described in our previous work, 7 we identified significantly disturbed metabolic pathways associated with the pathological stages relative to the normal CON stage with P less than 10−5 and PIV greater than 0.3.
2.9. Identification of potential biomarkers based on disturbed metabolic pathways
Metabolomic analysis has been extensively used to exploit potential biomarkers for early diagnosis of tumors. 13 In our work, we randomly selected 66.7% tissue samples to undertake multivariate receiver operating characteristic (ROC) curve analysis 33 for assessing discriminant capabilities of the significant metabolites involved in the significantly disturbed metabolic pathways which were identified from the metabolic pathway analysis. The logistic regression algorithm was applied in the multivariate ROC curve analysis for classification. The area under the ROC curve (AUC) value was used to evaluate the prediction performance of a given biomarker model. The important metabolites with AUC greater than 0.7 were identified to be potential biomarkers for diagnosing a given pathological state. Then we undertook multivariate ROC analyses using the other 33.3% tissue samples to confirm the validities of the identified potential biomarkers. We also evaluated the capabilities of these potential gastric tissue biomarkers for metabolically discriminating the 4 pathological groups of sera derived from the same established rat model of GCG from the CON group using the multivariate ROC curves. The NMR‐based metabonomic analyses of sera derived from the rat model of GCG have been documented in our previous article. 7
3. RESULTS
3.1. Rat model of GCG
In total, 84 rats were used for metabolomic analysis, including 52 MODEL rats and 32 CON rats. Unluckily, the other 44 MODEL rats were lost due to accidental death. None of the CON rats died accidentally. According to histologic examination, the MODEL rats were classified into 4 pathologic groups: GS, LGD, HGD, and GC. The detailed information of the rat model has been described in our previous work. 7
3.2. Metabolic profiles of gastric tissues
Figure S1 shows typical 1D 1H‐spectra of aqueous extracts of gastric tissues derived from the 5 groups of rats. Assigned resonances of metabolites were identified in the spectra and confirmed with 2D 1H‐1H TOCSY spectra of a gastric tissue sample derived from a GC rat (Figure S2).
We used PCA for the NMR data obtained from the 5 groups of rats to obtain a comprehensive comparison of metabolic profiles among the groups. Figure 1 illustrates the PCA score plots with the first 3 principal components (PC1, PC2, and PC3). The scores plot of all rats (Figure 1A) shows that the 5 groups were distinguished from each other. The metabolic profiles of GC and HGD displayed clear separations from those of other 3 groups (CON, GS, and LGD) which were not distinctly distinguishable from one another, with partial overlap (Figure 1A). Moreover, we undertook pairwise PCA for the 5 groups (Figure 1B‐E) to address the changes of metabolic profiles associated with the 4 pathological stages. Overall, the GS group was distinguished from the CON group except for a few samples (Figure 1B). Interestingly, clear separations of metabolic profiles were observed between LGD and GS groups (Figure 1C), HGD and LGD groups (Figure 1D), and GC and HGD groups (Figure 1E). Rats in the GC group showed a metabolic profile remarkably different from HGD rats without any overlap.
FIGURE 1.
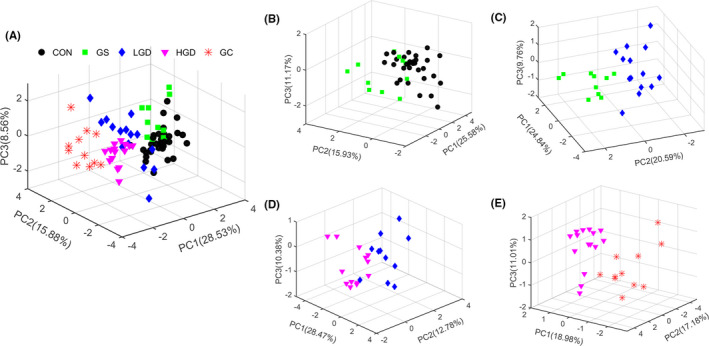
Principal component (PC) analysis scores plots of 1H nuclear magnetic resonance spectral data of aqueous metabolites extracted from gastric tissues. A, All rats. B, Gastritis (GS) rats vs control (CON) rats. C, Low‐grade gastric dysplasia (LGD) rats vs GS rats. D, High‐grade gastric dysplasia (HGD) rats vs LGD rats. E, gastric cancer (GC) rats vs HGD rats
Furthermore, we applied pairwise PLS‐DA to improve metabolic separations between the 5 groups of rats. The scores plots of the PLS‐DA models illustrate that the 5 groups are discriminated from each other (Figure S3). The validation plots of the corresponding RPTs indicate that the 5 PLS‐DA models were reliable (Figure S4).
3.3. Significant metabolites in gastric tissues
We undertook pairwise OPLS‐DA for the NMR data of the 5 groups of rats to maximize intergroup metabolic separation, and to identify significant metabolites primarily responsible for the metabolic separation based on the first component (tp1). The OPLS‐DA scores plots show distinct separations between GS and CON groups (Figure S5A), LGD and GS groups (Figure S5B), HGD and LGD groups (Figure S5C), and GC and HGD groups (Figure S5D). We identified significant metabolites based on the loading plots of the OPLS‐DA models with 2 criteria (correlation coefficient r corresponding to P < .05 and VIP > 1) as shown in Figure 2. Detailed information of the identified significant metabolites is shown in Tables S1‐S4. In total, 12, 9, 9, and 12 metabolites were identified from the OPLS‐DA analyses of GS rats vs CON rats (Table S1, 10 increased and 2 decreased metabolites), LGD rats vs GS rats (Table S2, 1 increased and 8 decreased metabolites), HGD rats vs LGD rats (Table S3, 4 increased and 5 decreased metabolites), and GC rats vs HGD rats (Table S4, 10 increased and 2 decreased metabolites).;
FIGURE 2.
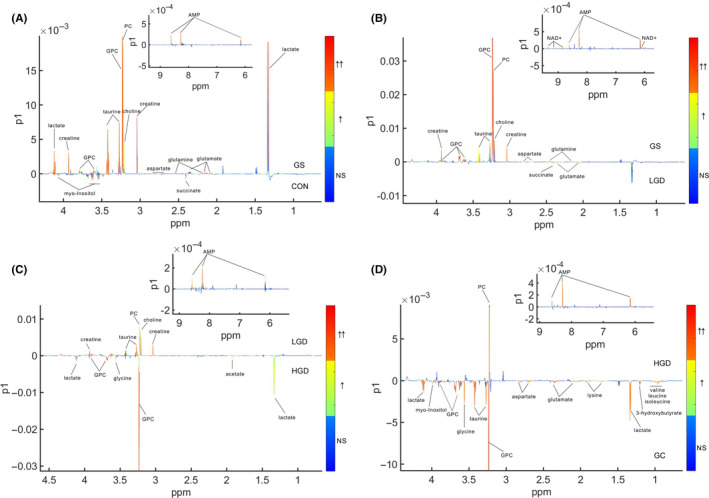
Orthogonal partial least squares discriminant analysis loading plots used to identify significant metabolites primarily responsible for distinguishing gastritis (GS) rats from control (CON) rats (A), low‐grade gastric dysplasia (LGD) rats from GS rats (B), high‐grade gastric dysplasia (HGD) rats from LGD rats (C), and gastric cancer (GC) rats from HGD rats (D). The gradient red color indicates that the variables are very significant (|r|> 0.389 in A, |r| > 0.496 in B, |r| > 0.463 in C, and |r| > 0.496 in D; variable importance value [VIP] > 1). Gradient orange indicates that the variables are significant (0.301 ≤ |r| < 0.389 in A, 0.388 ≤ |r| < 0.496 in B, 0.361 ≤ |r| <0.463 in C, and 0.388 ≤ |r| < 0.496 in D; VIP > 1). Gradient blue indicates that the variables are insignificant (NS). GPC, glycerophosphocholine; PC, phosphocholine
3.4. Relative levels of differential metabolites in gastric tissues
To quantitatively compare metabolite levels among the 5 groups of rats, we calculated the relative integrals of the identified metabolites based on 1D 1H‐NMR spectra of aqueous extracts derived from gastric tissues (Figure S1). The mean and SEM were calculated for each group of rats (Table 1). Then we applied one‐way ANOVA followed by Tukey’s multiple comparisons test, aiming to identify differential metabolites with P less than .05 (Table 1). The differential metabolites identified from one‐way ANOVA were consistent with the significant metabolites identified from the OPLS‐DA loading plots. These remarkably changed metabolites were mostly involved in the following 5 crucial metabolisms: amino acid metabolism, carbohydrate metabolism, fatty acid metabolism, energy metabolism, and quaternary ammonium metabolism.
TABLE 1.
Comparison of metabolite levels among 4 pathological groups and normal control (CON) group based on relative integrals calculated from 1D 1H‐nuclear magnetic resonance spectra of aqueous extracts of gastric tissues
| Mean and SEM | One‐way ANOVA | Tukey’s multiple comparisons test | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CON | GS | LGD | HGD | GC | P | F | GS vs | LGD vs | HGD vs | GC vs | LGD vs | HGD vs | GC vs | HGD vs | GC vs | GC vs | |
| CON | CON | CON | CON | GS | GS | GS | LGD | LGD | HGD | ||||||||
| Amino acid metabolism | |||||||||||||||||
| Leucine | 0.734 ± 0.064 | 0.802 ± 0.090 | 0.824 ± 0.092 | 0.755 ± 0.101 | 1.005 ± 0.109 | .009 | 3.675 | NS | NS | NS | *** | NS | NS | ** | NS | *** | *** |
| Isoleucine | 0.166 ± 0.013 | 0.176 ± 0.015 | 0.179 ± 0.019 | 0.179 ± 0.022 | 0.243 ± 0.021 | .002 | 4.607 | NS | NS | NS | *** | NS | NS | *** | NS | ** | ** |
| Valine | 0.244 ± 0.018 | 0.220 ± 0.023 | 0.236 ± 0.023 | 0.240 ± 0.0296 | 0.356 ± 0.035 | .000 | 6.201 | NS | NS | NS | *** | NS | * | *** | NS | *** | *** |
| Lysine | 1.313 ± 0.089 | 1.327 ± 0.119 | 1.325 ± 0.131 | 1.312 ± 0.137 | 1.696 ± 0.160 | .000 | 5.736 | NS | NS | NS | *** | NS | NS | *** | NS | *** | *** |
| Aspartate | 0.353 ± 0.032 | 0.466 ± 0.070 | 0.411 ± 0.040 | 0.414 ± 0.061 | 0.539 ± 0.091 | .000 | 9.836 | ** | ** | ** | *** | * | * | *** | NS | *** | *** |
| Glycine | 1.112 ± 0.037 | 1.213 ± 0.088 | 1.205 ± 0.061 | 1.373 ± 0.083 | 1.620 ± 0.093 | .000 | 24.252 | NS | NS | *** | *** | NS | * | *** | * | *** | *** |
| Alanine | 1.846 ± 0.108 | 1.862 ± 0.185 | 1.895 ± 0.130 | 1.771 ± 0.127 | 1.998 ± 0.158 | .561 | 0.750 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| Phenylalanine | 0.192 ± 0.023 | 0.191 ± 0.034 | 0.216 ± 0.035 | 0.207 ± 0.035 | 0.244 ± 0.031 | .024 | 2.989 | NS | NS | NS | * | NS | NS | * | NS | NS | NS |
| Tyrosine | 0.085 ± 0.011 | 0.088 ± 0.017 | 0.094 ± 0.015 | 0.092 ± 0.016 | 0.093 ± 0.019 | .030 | 3.595 | NS | NS | NS | * | NS | NS | NS | NS | * | NS |
| Glutamate | 1.786 ± 0.065 | 2.112 ± 0.173 | 2.553 ± 0.119 | 2.447 ± 0.150 | 2.764 ± 0.184 | .000 | 21.186 | *** | *** | *** | *** | *** | ** | *** | NS | ** | ** |
| Glutamine | 1.382 ± 0.046 | 1.562 ± 0.085 | 1.364 ± 0.060 | 1.353 ± 0.070 | 1.326 ± 0.065 | .008 | 3.738 | *** | NS | NS | NS | *** | *** | *** | NS | NS | NS |
| Glutathione | 1.343 ± 0.051 | 1.345 ± 0.100 | 1.279 ± 0.059 | 1.327 ± 0.077 | 1.265 ± 0.081 | .144 | 1.764 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| Taurine | 7.436 ± 0.204 | 8.517 ± 0.374 | 7.259 ± 0.221 | 6.239 ± 0.305 | 6.607 ± 0.362 | .000 | 9.885 | *** | NS | *** | *** | *** | *** | *** | *** | *** | NS |
| Carbohydrate metabolism | |||||||||||||||||
| Lactate | 7.891 ± 0.423 | 8.852 ± 0.576 | 8.960 ± 0.716 | 9.999 ± 1.302 | 11.274 ± 1.066 | .000 | 15.724 | * | * | *** | *** | NS | ** | *** | * | *** | ** |
| Succinate | 0.669 ± 0.032 | 0.529 ± 0.058 | 0.678 ± 0.033 | 0.687 ± 0.050 | 0.682 ± 0.045 | .000 | 6.307 | *** | NS | NS | NS | *** | *** | *** | NS | NS | NS |
| Pyruvate | 0.123 ± 0.008 | 0.123 ± 0.011 | 0.136 ± 0.010 | 0.127 ± 0.012 | 0.133 ± 0.015 | .134 | 1.814 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| Glucose | 0.094 ± 0.012 | 0.080 ± 0.020 | 0.101 ± 0.021 | 0.081 ± 0.005 | 0.074 ± 0.017 | .040 | 3.015 | NS | NS | NS | * | NS | NS | NS | NS | * | NS |
| Formate | 0.046 ± 0.006 | 0.045 ± 0.007 | 0.037 ± 0.007 | 0.040 ± 0.0088 | 0.037 ± 0.004 | .353 | 1.120 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| Fumarate | 0.006 ± 0.001 | 0.004 ± 0.002 | 0.004 ± 0.001 | 0.004 ± 0.002 | 0.004 ± 0.001 | .360 | 1.105 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| Acetate | 0.621 ± 0.062 | 0.692 ± 0.137 | 0.658 ± 0.101 | 0.699 ± 0.126 | 0.699 ± 0.174 | .042 | 2.194 | NS | NS | * | * | NS | NS | NS | * | * | NS |
| Creatine | 2.878 ± 0.088 | 3.352 ± 0.162 | 2.829 ± 0.118 | 2.231 ± 0.134 | 2.254 ± 0.163 | .000 | 15.128 | *** | NS | *** | *** | *** | *** | *** | *** | *** | NS |
| Fatty acid metabolism | |||||||||||||||||
| Glycerol | 2.040 ± 0.078 | 2.191 ± 0.138 | 2.160 ± 0.129 | 2.09 ± 0.131 | 2.055 ± 0.148 | .332 | 1.167 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| Myo‐inositol | 1.511 ± 0.079 | 1.210 ± 0.147 | 1.337 ± 0.080 | 1.261 ± 0.104 | 1.598 ± 0.129 | .000 | 7.961 | *** | * | *** | NS | NS | NS | *** | NS | *** | *** |
| 3‐Hydroxybutyrate | 0.367 ± 0.036 | 0.387 ± 0.068 | 0.374 ± 0.047 | 0.403 ± 0.040 | 0.706 ± 0.090 | .007 | 3.764 | NS | NS | NS | *** | NS | NS | *** | NS | *** | *** |
| Energy metabolism | |||||||||||||||||
| AMP | 0.240 ± 0.019 | 0.333 ± 0.051 | 0.250 ± 0.028 | 0.166 ± 0.022 | 0.083 ± 0.007 | .000 | 22.730 | *** | NS | *** | *** | *** | *** | *** | *** | *** | *** |
| NAD+ | 0.094 ± 0.008 | 0.105 ± 0.018 | 0.086 ± 0.014 | 0.081 ± 0.013 | 0.073 ± 0.010 | .000 | 6.287 | NS | NS | NS | *** | ** | ** | *** | NS | NS | NS |
| ATP | 0.105 ± 0.019 | 0.096 ± 0.040 | 0.123 ± 0.031 | 0.099 ± 0.012 | 0.097 ± 0.020 | .102 | 2.005 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| Nicotinurate | 0.017 ± 0.003 | 0.018 ± 0.007 | 0.018 ± 0.005 | 0.019 ± 0.005 | 0.018 ± 0.004 | .628 | 0.650 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| Quaternary ammonium group metabolism | |||||||||||||||||
| Choline | 1.946 ± 0.237 | 2.507 ± 0.640 | 1.756 ± 0.270 | 1.044 ± 0.129 | 1.078 ± 0.054 | .000 | 23.725 | * | NS | *** | *** | ** | *** | *** | *** | *** | NS |
| Phosphocholine | 3.968 ± 0.224 | 4.854 ± 0.362 | 3.628 ± 0.221 | 2.874 ± 0.260 | 2.125 ± 0.203 | .000 | 32.205 | *** | NS | *** | *** | *** | *** | *** | *** | *** | *** |
| Glycerophosphocholine | 6.443 ± 0.482 | 10.044 ± 1.153 | 7.000 ± 0.609 | 9.427 ± 1.533 | 12.973 ± 1.389 | .000 | 50.923 | *** | NS | ** | *** | *** | NS | *** | *** | *** | *** |
| Betaine | 0.622 ± 0.027 | 0.611 ± 0.063 | 0.590 ± 0.028 | 0.570 ± 0.036 | 0.610 ± 0.047 | .181 | 1.605 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| Dimethylamine | 0.126 ± 0.022 | 0.099 ± 0.024 | 0.123 ± 0.025 | 0.099 ± 0.022 | 0.118 ± 0.036 | .530 | 0.798 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| TMAO | 0.490 ± 0.015 | 0.480 ± 0.032 | 0.501 ± 0.020 | 0.491 ± 0.023 | 0.494 ± 0.025 | .912 | 0.244 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS |
Red and blue colors denote that the difference is positive (ie leucine was increased in gastric cancer [GC] relative to CON) and negative, respectively.
Abbreviations: GS, gastritis; HGD, high‐grade gastric dysplasia; LGD, low‐grade gastric dysplasia; TMAO, trimethylamine N‐oxide.
Differences between 2 groups: ***P < .001, highly significant; **P < .01, very significant; *P < .05, significant; NS, P > .05, insignificant.
3.5. Amino acid metabolism
Branched‐chain amino acids (leucine, isoleucine, and valine) and lysine were only changed in GC rats with significantly upregulated levels. Aspartate was profoundly enhanced in MODEL rats relative to CON rats; GS rats showed an aspartate level a little higher than LGD and HGD rats, but remarkably lower than GC rats. Glycine showed the highest level in GC rats, and a slightly higher level compared to LGD, GS, and CON rats. Glutamate was gradually increased during GCG and showed almost the same levels in LGD and HGD rats. Glutamine was only changed in GS rats with a distinctly increased level. Taurine showed the highest level in GS rats, and lower levels in HGD and GC rats relative to CON and LGD rats. Phenylalanine and tyrosine were only changed in GC rats with slightly enhanced levels. Alanine and glutathione kept relative stable levels during GCG.
3.6. Carbohydrate metabolism
Lactate was gradually increased during GCG and showed slightly enhanced levels in HGD and GC rats. Succinate was only altered in GS rats with a markedly decreased level. Glucose was only changed with a slightly declined level in GC rats. Acetate was not significantly changed during GCG with gently increased levels in HGD and GC rats. Creatine showed the highest level in GS rats and slightly decreased levels in HD and GC rats relative to CON and LGD rats. Pyruvate, formate, and fumarate were not obviously changed during GCG.
3.7. Fatty acid metabolism
3‐Hydroxybutyrate was only altered in GC rats with a greatly enhanced level. Myo‐inositol was decreased in GS, LGD, and HGD rats relative to CON rats; GC rats showed an increase in myo‐inositol level roughly identical to CON rats. Glycerol did not show distinctly changed levels in the 5 pathological stages of GCG.
3.8. Energy metabolism
Adenosine 5′‐monophosphate was markedly increased in GS rats, thereafter gradually decreased in LGD, HGD, and GC rats; GC rats showed the lowest AMP level. Regarding NAD+, GS rats did not show a significantly changed level compared with CON rats, but showed a significantly higher level than LGD, HGD, and GC rats, which had almost identical levels. Notably, NAD+ in GC rats was profoundly decreased relative to CON rats. Both ATP and nicotinurate kept relatively stable levels during GCG.
3.9. Quaternary ammonium metabolism
Choline and phosphocholine (PC) were dramatically increased in GS rats, and then gradually decreased remarkably in LGD, HGD, and GC rats. Rats with GC showed almost the same PC level as HGD rats. Glycerophosphocholine (GPC) was profoundly increased in GS rats, and significantly decreased in LGD rats, and then increased again in HGD and GC rats. Rats with GC showed the highest GPC level. Betaine, dimethylamine, and trimethylamine N‐oxide were not distinctly changed in MODEL rats relative to CON rats.
3.10. Significantly disturbed metabolic pathways in gastric tissues
Based on the significant metabolites, we identified significantly disturbed metabolic pathways associated with the 4 pathological stages of GCG relative to the CON stage (Figure 3). Both the GS and LGD stages showed 3 significantly disturbed metabolic pathways, including taurine and hypotaurine metabolism, glutamine and glutamate metabolism, and alanine, aspartate, and glutamate metabolism (Figure 3A,B). The HGD stage displayed 5 significantly disturbed metabolic pathways, including the 3 pathways identified in the LGD stage and 2 extra pathways (glycine, serine, and threonine metabolism; Krebs cycle) (Figure 3C). The GC stage showed 7 significantly disturbed metabolic pathways, including the 5 pathways identified in the HGD stage and 2 extra pathways (glycolysis; valine, leucine, and isoleucine biosynthesis) (Figure 3D).
FIGURE 3.
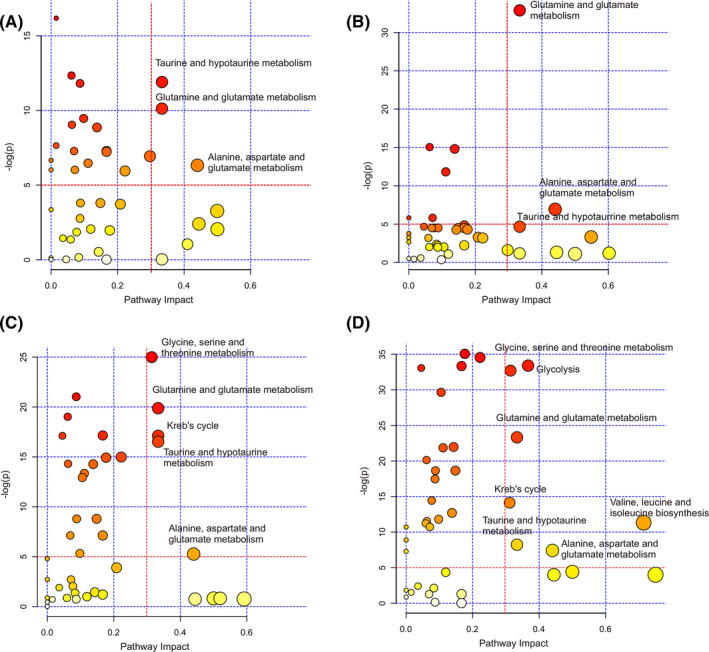
Significantly disturbed metabolic pathways associated with the 4 pathological stages of gastric carcinogenesis relative to the normal control (CON) stage. A, Gastritis vs CON. B, Low‐grade gastric dysplasia vs CON. C, High‐grade gastric dysplasia vs CON. D, Gastric cancer vs CON. Based on the significant metabolites, significantly disturbed metabolic pathways were identified with pathway impact values > 0.3 and P values < 10−5, using the Pathway Analysis module provided by MetaboAnalyst 4.0
3.11. Potential biomarkers in gastric tissues
Based on the discriminant capabilities of the significant metabolites predicted from the multivariate ROC analysis (Figures 4, 5, 6, 7), we identified potential gastric tissue biomarkers that could metabolically discriminate the 4 pathological stages from the CON stage. In the GS stage, 3 significant pathways displayed good discriminant capabilities with larger AUC values of 0.9405 for taurine and hypotaurine metabolism, 0.8839 for glutamine and glutamate metabolism, and 0.9077 for alanine, aspartate, and glutamate metabolism (Figure 4). More significantly, taurine and glutamine had the top 2 AUC values (0.8839 and 0.8423) in these pathways. We then applied the multivariate ROC analysis based on the 2 metabolites, obtaining high AUC values: all, 0.9700; taurine, 0.9300; and glutamine, 0.8500 (Figure S6A). This result suggests that taurine and glutamine could be exploited to be potential gastric tissue biomarkers for GS diagnosis. Significantly, the combination of the 2 metabolites also showed a good capability for metabolically discriminating the GS sera from the CON sera with higher AUC values: all, 0.8655; taurine, 0.8082; and glutamine, 0.7933 (Figure S7A).
FIGURE 4.
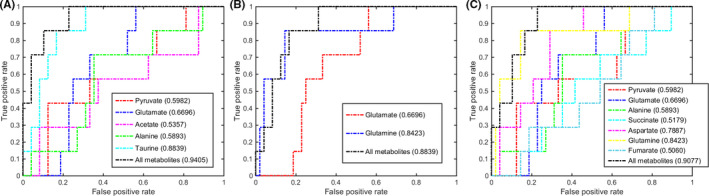
Multivariate receiver operating characteristic (ROC) curves assessing capabilities of significantly disturbed metabolic pathways in gastric tissues for metabolically discriminating the gastritis stage from the control stage. The area under the ROC curve values, shown in brackets, were used to evaluate prediction performances of the biomarker models. A, Taurine and hypotaurine metabolism. B, Glutamine and glutamate metabolism. C, Alanine, aspartate, and glutamate metabolism
FIGURE 5.
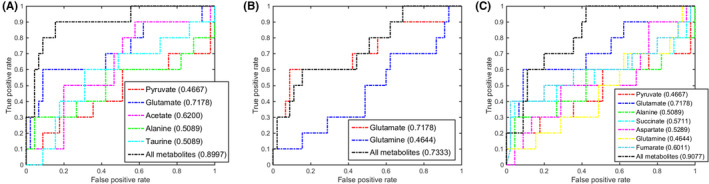
Multivariate receiver operating characteristic (ROC) curves assessing capabilities of significantly disturbed metabolic pathways in gastric tissues for metabolically discriminating the low‐grade gastric dysplasia stage from the control stage. The area under the ROC curve values, shown in brackets, were used to evaluate prediction performances of the biomarker models. A, Taurine and hypotaurine metabolism. B, Glutamine and glutamate metabolism. C, Alanine, aspartate and glutamate metabolism
FIGURE 6.
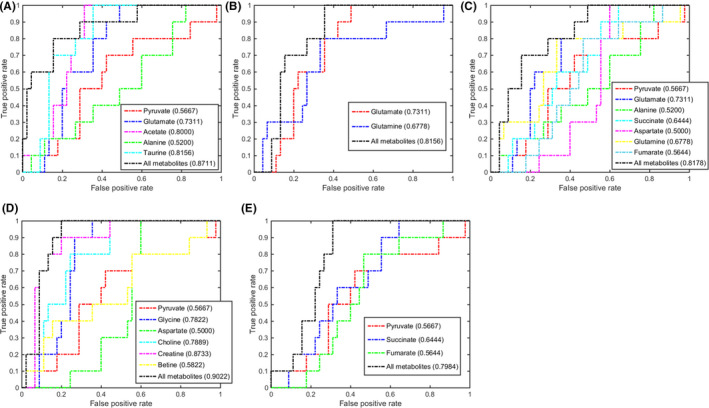
Multivariate receiver operating characteristic (ROC) curves assessing capabilities of significantly disturbed metabolic pathways in gastric tissues for metabolically discriminating the high‐grade gastric dysplasia stage from the control stage. The area under the ROC curve values, shown in brackets, were used to evaluate prediction performances of the biomarker models. A, Taurine and hypotaurine metabolism. B, Glutamine and glutamate metabolism. C, Alanine, aspartate, and glutamate metabolism. D, Glycine, serine, and threonine metabolism. E, Krebs cycle
FIGURE 7.
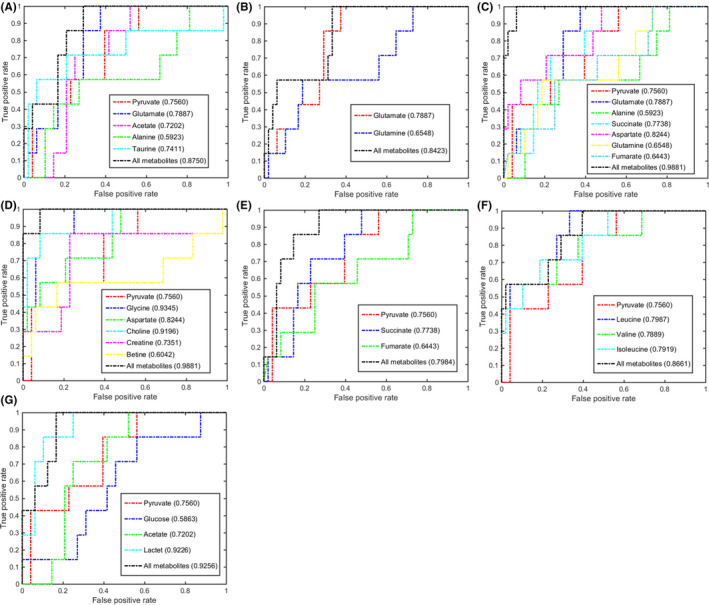
Multivariate receiver operating characteristic (ROC) curves assessing capabilities of significantly disturbed metabolic pathways in gastric tissues for metabolically discriminating the gastric cancer stage from the control stage. The area under the ROC curve values, shown in brackets, were used to evaluate prediction performances of the biomarker models. A, Taurine and hypotaurine metabolism. B, Glutamine and glutamate metabolism. C, Alanine, aspartate, and glutamate metabolism. D, Glycine, serine, and threonine metabolism. E, Krebs cycle. F, Valine, leucine, and isoleucine biosynthesis. G, Glycolysis
In the LGD stage, the 3 significant pathways showed also good discriminant capabilities with larger AUC values of 0.8997 for taurine and hypotaurine metabolism, 0.7333 for glutamine and glutamate metabolism, and 0.9077 for alanine, aspartate, and glutamate metabolism (Figure 5). Notably, glutamate showed the largest AUC value of 0.7178 in these pathways. The high AUC value of 0.8833 calculated from the multivariate ROC analysis was only based on glutamate, suggesting that glutamate could be also exploited to be a potential gastric tissue biomarker for LGD diagnosis (Figure S6B). However, glutamate did not show a high capability for metabolically discriminating the LGD sera from the CON sera with an AUC value of 0.6599 (Figure S7B).
In the HGD stage, the 5 significant pathways displayed good discriminant capabilities with larger AUC values of 0.8711 for taurine and hypotaurine metabolism, 0.8156 for glutamine and glutamate metabolism, 0.8178 for alanine, aspartate, and glutamate metabolism, 0.9022 for glycine, serine, and threonine metabolism, and 0.7984 for the Krebs cycle (Figure 6). The glycine, serine, and threonine metabolism and taurine and hypotaurine metabolism ranked the top 2 AUC values. Three metabolites had the top 3 AUC values: creatine, 0.8733; taurine, 0.8156; and acetate, 0.8000 (Figure 6). Moreover, the multivariate ROC analysis based on the 3 metabolites showed higher AUC values: all, 0.9283; creatine, 0.8833; taurine, 0.8167; and acetate, 0.7250. These results suggest that creatine, taurine, and acetate could be exploited to be potential gastric tissue biomarkers for HGD diagnosis (Figure S6C). We also calculated the corresponding AUC values for the HGD sera by undertaking multivariate ROC analysis on the 3 metabolites, obtaining AUC values: all, 0.9066; creatine, 0.8010; taurine, 0.7411; and acetate, 0.6947 (Figure S7C). These results suggest that the combination of the 3 metabolites had an excellent capability for metabolically discriminating the HGC sera from the CON sera (Figure S7C).
In the GC stage, the 7 significant pathways showed good discriminant capabilities with larger AUC values of 0.8750 for taurine and hypotaurine metabolism, 0.8423 for glutamine and glutamate metabolism, 0.9881 for alanine, aspartate, and glutamate metabolism, 0.9881 for glycine, serine, and threonine metabolism, 0.7984 for the Krebs cycle, 0.8661 for valine, leucine, and isoleucine biosynthesis, and 0.9256 for glycolysis (Figure 7). Three metabolic pathways (alanine, aspartate, and glutamate metabolism, glycine, serine, and threonine metabolism, and glycolysis) had excellent discriminant capabilities with the top 3 AUC values (0.9881, 0.9881, and 0.9256). Three metabolites had the top 3 AUC values: glycine, 0.9345; lactate, 0.9226; and choline, 0.9191. The multivariate ROC analysis based on the 3 metabolites showed high AUC values: all, 1.000; glycine, 0.9800; lactate, 0.9200; and choline, 0.8900 (Figure S6D). These results suggest that glycine, lactate, and choline could be exploited to be potential gastric tissue biomarkers for GC diagnosis. More significantly, the combination of the 3 metabolites had an excellent capability for metabolically discriminating the GC sera from the CON sera with high AUC values: all, 0.9552; glycine, 0.9437; lactate, 0.8369; and choline, 0.8182 (Figure S7D).
Additionally, the multivariate ROC analyses based on the above‐described potential gastric tissue biomarkers also showed good capabilities for metabolic discrimination among the 4 pathological groups of sera. The biomarker model established on taurine and glutamine gave AUC values of 0.6364 for GS sera vs LGD sera, 0.7883 for GS sera vs HGD sera, and 0.8003 for GS sera vs GC sera (Figure S8A). Similarly, the biomarker model based on taurine, creatine, and acetate gave AUC values of 0.7768 for HGD sera vs GS sera, 0.6106 for HGD sera vs LGD sera, and 0.8261 for HGD sera vs GC sera (Figure S8C). The combination of glycine, lactate, and choline acting as potential biomarkers for GC gastric tissues vs CON gastric tissues, also displayed high capacities for metabolically discriminating the GC sera from the precancerous sera with the following AUC values: 0.9352 for GC vs GS, 0.7860 for GC vs LGD, and 0.8522 for GC vs HGD (Figure S8D). Regrettably, glutamate could not be used as a potential marker for the LGD state, because these AUC values were mostly low than 0.7 (Figure S8B).
4. DISCUSSION
Gastric carcinogenesis is a multistep process related to several pathological stages, involving distinctly changed metabolic profiles and significantly disturbed metabolic pathways. Molecular mechanisms of GCG remain elusive. Previously, we established a rat model of GCG and carried out NMR‐based metabolomic analyses of sera derived from the rat model. 7 Until now, no metabolomic analyses of gastric tissues have been undertaken to address the underlying molecular mechanisms during the progression of GCG. Given that the physiological state of a complex tissue is reflected in the full complement of various metabolites from its constituent cells, metabolic profiling of gastric tissues would be beneficial to comprehensively understand molecular mechanisms underlying GCG. Here, we undertook the NMR‐based metabolomic analysis of gastric tissues derived from the same rat model of GCG. 7 We compared distinctly altered metabolic profiles, dramatically changed metabolite levels, and significantly disturbed metabolic pathways associated with the 4 pathological stages of GCG.
4.1. Glutamate‐related metabolism is disordered during GCG
We found that 2 metabolic pathways were continually disordered in the multistage process of GCG (glutamine and glutamate metabolism, and alanine, aspartate, and glutamate metabolism). Our previous metabolomic analysis of sera derived from the rat model of GCG also revealed the 2 identical disordered pathways. 7 Compared with the CON rats, the serum levels of aspartate, glutamate and glutamine were increased in the MODEL rats. 7 Previously, Sharma et al also got a consistent result by studying the gastrointestinal mucosa in celiac disease with inflammation. 34 It is well known that glutamate, glutamine, and aspartate are major sources of energy for small intestinal mucosa. 35 Once undergoing deamination in their carbon skeleton, these changed amino acids are converted into intermediate metabolites of anaplerotic reactions of the Krebs cycle. 36 In addition, glutamate and glutamine metabolism also participate in oxidation stress. 37 It has been suggested that significantly altered metabolic pathways related to glutamate might respond to oxidation stress, which are probably associated with chronic inflammation and malignant transformation during GCG. 38
Our work indicated that oxidation stress continuously influenced gastric tissues in the 4 typical pathological stages of GCG. Moreover, glutamine was identified as a potential biomarker in the GS stage, and glutamate was identified as a potential biomarker in the LGD stage. Thus, these results suggest that oxidative stress is the main disordered metabolic pathway in the GS and LGD stages of GCG.
Several metabolites in amino acid metabolism, such as BCAAs, lysine, phenylalanine, and tyrosine, were identified only in the malignant stage, but not in other pathological stages. These results suggest that these metabolites could be involved in malignancy, rather than in inflammation or antioxidative defense. It has been reported previously that reprogrammed BCAA metabolism can directly regulate cancer development and drive cancer progression. 39
4.2. Antioxidant metabolic pathway is abnormal during GCG
Taurine and hypotaurine metabolism was continually disturbed during GCG. Taurine has been well recognized as an antioxidant both in vitro and in vivo. Previous works showed that taurine could elevate the activities of the antioxidant enzymes superoxide dismutase 40 and glutathione peroxidase. 41 Compared to the CON rats, the level of taurine was profoundly increased in GS rats. The high concentration of taurine probably contribute to the protection of gastric mucosa from oxidation stress. In LGD rats, taurine fell back to the normal control level. With the development of GCG, taurine was decreased in HGD and GC rats. This result indicated that the activity of antioxidation was reduced during the progression of GCG. It is expected that biochemical experiments for testing oxidative stress will provide strong evidence to support the correlation between these metabolites and oxidative stress. Such experiments should be undertaken in the future.
Notably, the metabolic profiling of gastric tissues derived from the rat model of GCG revealed abnormal taurine and hypotaurine metabolism, whereas that of sera derived from the same rat model did not identify this metabolic pathway as a significantly disturbed pathway. 7 This work is indicative of the metabolic distinction between gastric tissues and sera.
Furthermore, the creatine level also displayed an interesting tendency during GCG. Creatine was significantly increased in the gastritis stage, but profoundly decreased in high dysplasia and cancer stages. As is known, creatine is an antiinflammatory metabolite. Such antiinflammatory, antioxidative defense by metabolites might be involved in homeostatic mechanisms in localized tissues. In addition, creatine also displayed potential anticancer effects. 42 Thus, the reduced level of creatine detected in our work might further accelerate tumor progression.
4.3. Energy‐related metabolic pathways are disturbed in HGD and GC stages
Our work showed that several energy‐related metabolic pathways were significantly disturbed in HGD and GC stages, including glycine, serine, and threonine metabolism and Krebs cycle. Obviously, glycine was increased in the 2 stages and identified as a potential biomarker in the GC stage (AUC = 0.9345). These results showed that cancer cell proliferation was promoted by glycine in HGD and GC stages. Both the impaired Krebs cycle and glycine, serine, and threonine metabolism suggest that the anaplerosis of Krebs cycle is activated through the pyruvate metabolism from amino acids, and thus promotes cell proliferation and growth by using Krebs cycle for biosynthesis. 43
Furthermore, the levels of choline, PC, and GPC fluctuated in the 4 pathological stages of GCG. Choline and PC were upregulated in the GS stage but downregulated in HGD and GC stages. Glycerophosphocholine was increased in the GS, HGD, and GC stages. These fluctuations might result from impaired choline phosphorylation, which was previously reported as a common feature of cancer. 44 The enhanced GPC level could promote the biosynthesis of rapidly growing tumor cells with high invasion ability in the HGD stage. 45 More significantly, choline also participated in the glycine, serine, and threonine metabolism in HGD and GC stages, and played important roles in this metabolic pathway, as indicated by its high AUC values (0.8733 and 0.9191). Notably, as described above, choline could be exploited to be a potential biomarker for GC diagnosis.
During the progression of GCG, the changes of creatine and 3‐hydroxybutyrate levels are also worth attention. Creatine was decreased in HGD and GC stages, indicative of reduced creatine biosynthesis. Similar results were previously reported by Manju Ray et al. 46 They found that arginine, glycine, and related metabolites were regulated to support polyamine and methionine synthesis in cancer cells, rather than supporting creatine biosynthesis. Furthermore, our work also identified creatine to be a potential biomarker in the HGD stage. Compared to the other 4 pathological stages, the GC state showed significantly increased 3‐hydroxybutyrate, which was a metabolite in ketone bodies’ metabolism, implying that the fatty acid metabolism was regulated to meet the body’s energy needs. Previously, Yeh et al showed that fatty acid metabolism had high rates of overexpression in colorectal carcinogenesis. 47
In the GC stage, the metabolic pathway of valine, leucine, and isoleucine biosynthesis was also significantly disturbed, and the related metabolites were profoundly upregulated. This result showed that the metabolism of amino acids was promoted in this stage for tumor cell proliferation. Furthermore, the GC stage showed markedly disturbed glycolysis, together with the highest level of lactate and the lowest level of glucose. In addition, lactate was identified as a potential biomarker in the GC stage (AUC = 0.9226). These results indicate that glycolysis is the main disordered metabolic pathway in the GC stage, which requires more energy for the rapid growth of tumor cells. Similar phenomena have been observed in tumor cells. 48
We undertook NMR‐based metabolomic analyses of gastric tissues derived from a rat model of GCG. We characterized distinctly changed metabolic profiles, identified significant metabolites with dramatically altered levels, and identified significantly disturbed metabolic pathways associated with the 4 pathological stages of GCG. The progression of GCG shows 3 continually disturbed metabolic pathways (taurine and hypotaurine metabolism, glutamine and glutamate metabolism, and alanine, aspartate, and glutamate metabolism). Moreover, both the HGD and GC stages show 2 extra impaired metabolic pathways (glycine, serine, and threonine metabolism, and Krebs cycle), which might contribute to the supply of more energy for tumor cell proliferation and growth. Furthermore, the GC stage displays 2 unique impaired pathways (glycolysis, and valine, leucine, and isoleucine biosynthesis) due to increased energy requirements for the rapid growth of tumor cells. More significantly, the GC stage also shows more remarkably altered metabolite levels relative to the precancerous stages, indicative of severe metabolic disorder. In this work, we have not measured expression levels or activities of regulatory enzymes involved in the identified significantly disturbed metabolic pathways. Such work should be carried out in the future to confirm these significant pathways associated with the progression of GCG. In addition, we identified potential gastric tissue biomarkers for metabolically discriminating the 4 pathological stages of GCG from the normal CON stage: taurine and glutamine for GS; glutamate for LGD; creatine, taurine, and acetate for HGD; and glycine, lactate, and choline for GC. These potential biomarkers, identified for pathological gastric tissues, also showed good discriminant capabilities for their serum counterparts. Further works are required to comprehensively evaluate the potencies of these potential biomarkers for clinical diagnoses based on large‐scale gastric tissue and serum samples. Our results provide new insights into the metabolic mechanisms underlying the 4 pathological stages of GCG and could be beneficial to exploit potential biomarkers for clinically diagnosing and monitoring GC progression.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Supporting information
Appendix S1
ACKNOWLEDGMENTS
The work was supported by grants from the National Natural Science Foundation of China (Nos. 31971357 and 81574080), the Xiamen Ocean Economic Innovation and Development Demonstration Project (No.16PZP001SF16), the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (No. 21521004), and the Natural Science Foundation of Zhejiang Province, China (No. LQ18B050003).
Gu J, Huang C, Hu X, Xia J, Shao W, Lin D. Nuclear magnetic resonance‐based tissue metabolomic analysis clarifies molecular mechanisms of gastric carcinogenesis. Cancer Sci. 2020;111:3195–3209. 10.1111/cas.14443
Contributor Information
Caihua Huang, Email: huangcaihua@xmut.edu.cn.
Donghai Lin, Email: dhlin@xmu.edu.cn.
REFERENCES
- 1. Stewart BW, Wild CP. Cancer worldwide In: World Cancer Report 2014. World Cancer Reports Lyon, France: IARC Nonserial Publication, 2015. [Google Scholar]
- 2. Correa P, Piazuelo MB, Camargo MC. The future of gastric cancer prevention. Gastric Cancer. 2004;7:9‐16. [DOI] [PubMed] [Google Scholar]
- 3. Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230‐233. [DOI] [PubMed] [Google Scholar]
- 4. Lazebnik Y. What are the hallmarks of cancer? Nat Rev Cancer. 2010;10:232‐233. [DOI] [PubMed] [Google Scholar]
- 5. Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646‐674. [DOI] [PubMed] [Google Scholar]
- 6. Chan AW, Gill RS, Schiller D, Sawyer MB. Potential role of metabolomics in diagnosis and surveillance of gastric cancer. World J Gastroenterol. 2014;20:12874‐12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gu JP, Hu XM, Shao W, et al. Metabolomic analysis reveals altered metabolic pathways in a rat model of gastric carcinogenesis. Oncotarget. 2016;7:60053‐60073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Griffin JL, Nicholls AW. Metabolomics as a functional genomic tool for understanding lipid dysfunction in diabetes, obesity and related disorders. Pharmacogenomics. 2006;7:1095‐1107. [DOI] [PubMed] [Google Scholar]
- 9. Song H, Wang L, Liu HL, et al. Tissue metabolomic fingerprinting reveals metabolic disorders associated with human gastric cancer morbidity. Oncol Rep. 2011;26:431‐438. [DOI] [PubMed] [Google Scholar]
- 10. Sreekumar A, Poisson LM, Rajendiran TM, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910‐914. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 11. Hirayama A, Kami K, Sugimoto M, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time‐of‐flight mass spectrometry. Can Res. 2009;69:4918‐4925. [DOI] [PubMed] [Google Scholar]
- 12. Nicholson JK, Lindon JC, Holmes E. 'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181‐1189. [DOI] [PubMed] [Google Scholar]
- 13. Wang HJ, Zhang HL, Deng PC, et al. Tissue metabolic profiling of human gastric cancer assessed by 1H‐NMR. BMC Cancer. 2016;16:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang HL, Cui LZ, Liu W, et al. 1H‐NMR metabolic profiling of gastric cancer patients with lymph node metastasis. Metabolomics. 2018;14:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jung J, Jung Y, Bang EJ, et al. Noninvasive diagnosis and evaluation of curative surgery for gastric cancer by using NMR‐based metabolomic profiling. Ann Surg Oncol. 2014;21:736‐742. [DOI] [PubMed] [Google Scholar]
- 16. Wu H, Xue R, Tang Z, et al. Metabolomic investigation of gastric cancer tissue using gas chromatography/mass spectrometry. Anal Bioanal Chem. 2009;396:1385‐1395. [DOI] [PubMed] [Google Scholar]
- 17. Chen J‐L, Tang H‐Q, Hu J‐D, Fan J, Hong J, Gu J‐Z. Metabolomics of gastric cancer metastasis detected by gas chromatography and mass spectrometry. World J Gastroenterol. 2010;16:5874‐5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin C, Wu H, Tjeerdema R, Viant M. Evaluation of metabolite extraction strategies from tissue samples using NMR metabolomics. Metabolomics. 2007;3:55‐67. [Google Scholar]
- 19. Beckonert O, Keun HC, Ebbels TMD, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protocols. 2007;2:2692‐2703. [DOI] [PubMed] [Google Scholar]
- 20. Fonville JM, Maher AD, Coen M, Holmes E, Lindon JC, Nicholson JK. Evaluation of Full‐Resolution J‐Resolved 1H NMR Projections of Biofluids for Metabonomics Information Retrieval and Biomarker Identification. Anal Chem. 2010;82:1811‐1821. [DOI] [PubMed] [Google Scholar]
- 21. Ding L, Hao F, Shi Z, et al. Systems biological responses to chronic perfluorododecanoic acid exposure by integrated metabonomic and transcriptomic studies. J Proteome Res. 2009;8:2882‐2891. [DOI] [PubMed] [Google Scholar]
- 22. Zhang L, Ye Y, An Y, Tian Y, Wang Y, Tang H. Systems Responses of Rats to Aflatoxin B1 Exposure Revealed with Metabonomic Changes in Multiple Biological Matrices. J Proteome Res. 2011;10:614‐623. [DOI] [PubMed] [Google Scholar]
- 23. Savorani F, Tomasi G, Engelsen SB. icoshift: A versatile tool for the rapid alignment of 1D NMR spectra. J Magn Reson. 2010;202:190‐202. [DOI] [PubMed] [Google Scholar]
- 24. Trygg J, Holmes E, Lundstedt T. Chemometrics in Metabonomics. J Proteome Res. 2007;6:469‐479. [DOI] [PubMed] [Google Scholar]
- 25. Gao HC, Lu Q, Liu X, et al. Application of 1H NMR‐based metabonomics in the study of metabolic profiling of human hepatocellular carcinoma and liver cirrhosis. Cancer Sci. 2009;100:782‐785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pérez‐Enciso M, Tenenhaus M. Prediction of clinical outcome with microarray data: a partial least squares discriminant analysis (PLS‐DA) approach. Hum Genet. 2003;112:581‐592. [DOI] [PubMed] [Google Scholar]
- 27. Trygg J, Wold S. Orthogonal projections to latent structures (O‐PLS). J Chemom. 2002;16:119‐128. [Google Scholar]
- 28. Shao W, Gu JP, Huang CH, et al. Malignancy‐associated metabolic profiling of human glioma cell lines using 1H‐NMR spectroscopy. Mol Cancer. 2014;13:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nguyen DV, Rocke DM. Tumor classification by partial least squares using microarray gene expression data. Bioinformatics. 2002;18:39‐50. [DOI] [PubMed] [Google Scholar]
- 30. Cloarec O, Dumas ME, Trygg J, et al. Evaluation of the orthogonal projection on latent structure model limitations caused by chemical shift variability and improved visualization of biomarker changes in 1H NMR spectroscopic metabonomic studies. Anal Chem. 2005;77:517‐526. [DOI] [PubMed] [Google Scholar]
- 31. Chong J, Soufan O, Li C, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486‐W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goeman JJ, Bühlmann P. Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics. 2007;23:980‐987. [DOI] [PubMed] [Google Scholar]
- 33. Zweig MH, Campbell G. Receiver‐operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39:561‐577. [PubMed] [Google Scholar]
- 34. Sharma U, Upadhyay D, Mewar S, et al. Metabolic abnormalities of gastrointestinal mucosa in celiac disease: An in vitro proton nuclear magnetic resonance spectroscopy study. J Gastroenterol Hepatol. 2015;30:1492‐1498. [DOI] [PubMed] [Google Scholar]
- 35. Intestinal WUG, Catabolism MAA. Intestinal mucosal amino acid catabolism. J Nutri. 1998;128:1249‐1252. [DOI] [PubMed] [Google Scholar]
- 36. Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277:30409‐30412. [DOI] [PubMed] [Google Scholar]
- 37. Ramond E, Gesbert G, Rigard M, et al. Glutamate utilization couples oxidative stress defense and the tricarboxylic acid cycle in Francisella Phagosomal escape. PLoS Pathog. 2014;10:e1003893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roy K, Wu Y, Meitzler JL, et al. NADPH oxidases and cancer. Clin Sci. 2015;128:863‐875. [DOI] [PubMed] [Google Scholar]
- 39. Hattori A, Tsunoda M, Konuma T, et al. Cancer progression by reprogrammed BCAA metabolism in myeloid leukaemia. Nature. 2017;545:500‐504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vohra BPS, Hui X. Taurine protects against carbon tetrachloride toxicity in the cultured neurons and in vivo. Arch Physiol Biochem. 2001;109:90‐94. [DOI] [PubMed] [Google Scholar]
- 41. Anand P, Rajakumar D, Jeraud M, Felix AJW, Balasubramanian T. Effects of taurine on glutathione peroxidase, glutathione reductase and reduced glutathione levels in rats. Pak J Biol Sci. 2011;14:219‐225. [DOI] [PubMed] [Google Scholar]
- 42. Campos‐Ferraz PL, Gualano B, das Neves W, et al. Exploratory studies of the potential anti‐cancer effects of creatine. Amino Acids. 2016;48:1993‐2001. [DOI] [PubMed] [Google Scholar]
- 43. Solaini G, Sgarbi G, Baracca A. Oxidative phosphorylation in cancer cells. Biochim et Biophys Acta. 2011;1807:534‐542. [DOI] [PubMed] [Google Scholar]
- 44. Marchan R, Lesjak MS, Stewart JD, Winter R, Seeliger J, Hengstler JG. Choline‐releasing glycerophosphodiesterase EDI3 links the tumor metabolome to signaling network activities. Cell Cycle. 2012;11:4499‐4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sabatier J, Gilard V, Malet‐Martino M, et al. Characterization of choline compounds with in vitro 1H magnetic resonance spectroscopy for the discrimination of primary brain tumors. Invest Radiol. 1999;34:230‐235. [DOI] [PubMed] [Google Scholar]
- 46. Bera S, Wallimann T, Ray S, Ray M. Enzymes of creatine biosynthesis, arginine and methionine metabolism in normal and malignant cells. FEBS J. 2008;275:5899‐5909. [DOI] [PubMed] [Google Scholar]
- 47. Yeh C‐S, Wang J‐Y, Cheng T‐L, Juan C‐H, Wu C‐H, Lin S‐R. Fatty acid metabolism pathway play an important role in carcinogenesis of human colorectal cancers by Microarray‐Bioinformatics analysis. Cancer Lett. 2006;233:297‐308. [DOI] [PubMed] [Google Scholar]
- 48. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029‐1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1


