Abstract
Whether germline (g) breast cancer susceptibility gene (BRCA) mutations are located within or outside the ovarian cancer cluster region (OCCR) (1380‐4062 bp for gBRCA1, and between 3249‐5681 bp and 6645‐7471 bp for gBRCA2) may influence risk variations for ovarian cancers. This ad hoc analysis of the CHARLOTTE epidemiological study in Japan assessed the distribution of gBRCA1/2 mutations in patients with newly diagnosed ovarian cancer, and investigated an association between gBRCA1/2 mutation locations and ovarian cancer risk. Differences in patient background and clinical characteristics in subgroups stratified by gBRCA1/2 mutation locations were also evaluated. We analyzed the data of 93 patients (14.7%) from the CHARLOTTE study who were positive for gBRCA1/2 mutations. After excluding 16 cases with L63X founder mutation, 28 (65.1%) of gBRCA1 mutations were within the OCCR. Of 30 gBRCA2 mutations, 15 (50.0%) were within the OCCR. Of 27 patients (one patient excluded for unknown family history) with gBRCA1 mutations located in the OCCR, 11 (40.7%) had a family history of ovarian cancer; the proportion of patients with a family history of ovarian cancer and gBRCA1 mutations outside the OCCR was lower (13.3%). Sixty percent of patients with gBRCA1 mutations outside the OCCR had a family history of breast cancer; the proportion of patients with a family history of breast cancer and gBRCA1 mutations within the OCCR was relatively lower (33.3%). Understanding the mutation locations may contribute to more accurate risk assessments of susceptible individuals and early detection of ovarian cancer among gBRCA mutation carriers.
Keywords: BRCA1 gene, BRCA2 gene, mutation, ovarian cancer, ovarian cancer cluster region
We investigated the prevalence of the gBRCA1/2 mutations according to the mutation location inside or outside the OCCR in patients with newly diagnosed ovarian cancer who were enrolled in the CHARLOTTE study. Among Japanese ovarian cancer patients, approximately half of the gBRCA1/2 mutations were located within the OCCR.

Abbreviations
- BCCR
breast cancer cluster region
- BRCA1
breast cancer susceptibility gene 1
- BRCA2
breast cancer susceptibility gene 2
- FIGO
International Federation of Gynecology and Obstetrics
- g
germline
- m
mutation
- OB
oligonucleotide binding
- OCCR
ovarian cancer cluster region
- SAS
Statistical Analysis Software
1. INTRODUCTION
Ovarian cancer is the 7th most common cancer in women, with an estimated 239 000 new cases and 152 000 deaths worldwide per annum. 1 It has been estimated that approximately 1.3% of women in the general population will develop ovarian cancer at some point in their lives. 2
Breast cancer susceptibility gene 1 (BRCA1) and BRCA2 3 encode proteins BRCA1 and BRCA2, which play important roles in DNA repair, transcriptional regulation in response to DNA damage, maintenance of chromosomal stability, and regulation of the cell cycle and apoptosis. 3 Germline (g) mutations of these genes are associated with a well established high risk of developing breast and ovarian cancer. 4 , 5 , 6 , 7 , 8 , 9 It has been recently estimated that approximately 44% of women who inherit gBRCA1 mutations and approximately 17% of those who inherit gBRCA2 mutations will develop ovarian cancer by 80 y of age. 6 It has also been reported that the spectrum of gBRCA1/2 mutations in breast and ovarian cancer patients also varies across different races and ethnicities. 10 , 11 , 12
The importance of gBRCA mutation testing is growing as the gBRCA1/2 status is not only important for breast/ovarian cancer risk assessment, but also for the selection of therapy. 13 Recent studies 6 , 8 , 14 have shown the importance of estimating cancer risk among individuals with a family history of ovarian or breast cancer by gBRCA testing before the onset of cancer. Data from the SOLO1 study, in which women with BRCA1/2‐mutated advanced ovarian cancer were randomly assigned to receive olaparib or placebo after first‐line chemotherapy, reported a 70% reduction in the hazard risk for disease progression or death with olaparib after a median follow‐up of 41 mo. 15 A clinical benefit of this magnitude is likely to change the clinical treatment algorithm for this patient population, 16 and these data are of critical interest to physicians.
In addition to knowing the gBRCA mutation status, it is important to assess the location of the mutation to estimate the risk for certain cancers. A recent observational study on a large sample of women with ovarian and breast cancer and gBRCA1/2 mutations showed that the risk of developing breast and ovarian cancer varied by the type and location of the mutation. 8 In a previous study, Rebbeck et al identified an ovarian cancer cluster region (OCCR) that was associated with a relative decrease and an increase in the risks of breast and ovarian cancers, respectively. 8
The CHARLOTTE (Characterizing the cross‐sectional approach to ovarian cancer: Genetic testing of BRCA) study evaluated the prevalence of gBRCA1/2 mutations in Japanese patients with ovarian cancer, and reported the prevalence of 9.9% and 4.7% for gBRCA1 and gBRCA2 mutations, respectively, in 634 patients. 14 Additionally, this study has shown that there was a slightly higher prevalence of gBRCA mutations in patients with high‐grade serous carcinoma and a family history of ovarian cancer. However, the prevalence of gBRCA1/2 mutations within the OCCR has never been evaluated in Japanese ovarian cancer patients, highlighting the importance of this analysis. Furthermore, the most common sites of gBRCA1/2 mutations in ovarian cancer patients, and the relationship between family history and gBRCA mutation site, remain to be clarified.
Based on the previously reported OCCR, 8 this ad hoc analysis aimed to assess the prevalence of the gBRCA mutation according to its location inside or outside the OCCR, using data from patients in the Japanese CHARLOTTE study. Additionally, we aimed to evaluate patient background characteristics and their family history stratified by gBRCA mutation locations.
2. MATERIALS AND METHODS
2.1. Study design and data sources
The CHARLOTTE study was a collaborative cross‐sectional study (NCT03229122; UMIN000025597) involving 63 study sites throughout Japan, and the methodology and primary analysis have been described previously. 14 The study was conducted in accordance with the principles of the Declaration of Helsinki, and ethical approval was obtained from the institutional review board of each participating institution. Written informed consent was obtained from all patients for study participation and histopathological analysis.
The present ad hoc analysis was based on anonymized patient data collected during the CHARLOTTE study, 14 including patient demographics and clinical characteristics (medical history, medications, menopausal status, obstetric history, and blood biochemical testing) and ovarian cancer data (date of diagnosis, pathological and histological classification, type and grade on diagnosis, and International Federation of Gynecology and Obstetrics [FIGO] classification).
The procedures conducted during the CHARLOTTE study have been described previously. 14 Briefly, testing of patient blood samples for the presence or absence of gBRCA mutations and mutation locations was performed centrally using BRACAnalysis® by Myriad Genetics, Inc. The histological diagnosis was confirmed centrally by a pathologist assigned by the Japanese Gynecologic Oncology Group using hematoxylin and eosin slide specimens from tumor tissues. Only deleterious or suspected deleterious mutations were defined as gBRCA mutations. All patients had undergone pre‐genetic testing counseling by a trained obstetrician‐gynecologist or oncologist, clinical geneticist, or certified genetic counselor.
2.2. Patients
In the CHARLOTTE study, 14 patients were selected in serial order to avoid selection bias. Japanese women aged ≥20 y with newly diagnosed, histologically confirmed FIGO stage I‐IV ovarian cancer, based on surgically resected specimens, and with histological specimens evaluated by central pathological review were included in the CHARLOTTE study. 14
Grounds for exclusion were acute or chronic medical disease, mental illness, histological classification of the surgically resected specimen, and if participation in the study was judged to be inappropriate by the investigator. The present ad hoc analysis focused on the subgroup of patients with confirmed gBRCA1 and gBRCA2 mutations; patients with exon deletion were not included in this analysis.
2.3. Study outcomes
The study outcomes were the prevalence of gBRCA1/2 mutations in patients with newly diagnosed ovarian cancer by the location of gBRCA mutations within and outside the OCCR, which were between 1380 and 4062 bp for gBRCA1, and between 3249‐5681 bp and 6645‐7471 bp for gBRCA2. 8 The patient background and clinical characteristics in subgroups stratified by the location of gBRCA mutations were also evaluated. Ovarian cancer patients with gBRCA1 founder mutation (L63X) were analyzed as a subgroup. 17
2.4. Additional analysis
A further analysis was conducted to determine the location of gBRCA mutations within and outside the OCCR using previously reported breast cancer prevalence data from 7051 breast cancer patients and 11 241 female controls of Japanese ancestry. 18 In that study, 102 patients were positive for gBRCA1 mutations and 191 patients were positive for gBRCA2 mutations.
2.5. Statistical methods
Details of the statistical analysis have been previously described in the main report of the CHARLOTTE study. 14 Demographic and clinical characteristic data are shown using descriptive statistics, with n (%) for categorical variables and mean ± SD for continuous variables. The statistical software used for the ad hoc analysis was Statistical Analysis Software (SAS) 9.4 (SAS Institute).
3. RESULTS
3.1. Study flow
Of the 666 patients enrolled in the CHARLOTTE study, 665 patients underwent gBRCA testing and 634 patients were evaluated (Figure 1). This ad hoc analysis focused on the data of 93 patients (14.7%); 63 patients were positive for gBRCA1 mutations and 30 patients were positive for gBRCA2 mutations.
FIGURE 1.
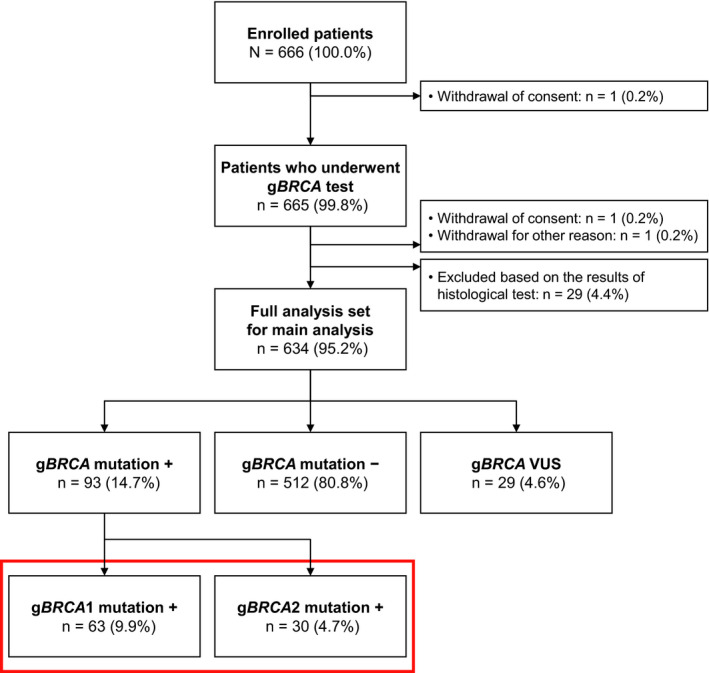
Study flow. BRCA, breast cancer susceptibility gene; g, germline; VUS, variants of uncertain significance. The red box encloses the 2 groups of patients included in the present analysis, corresponding to the patients enrolled in the CHARLOTTE study who were positive for gBRCA1/2 mutations
3.2. Prevalence of gBRCA1/2 mutation by location
Among the 93 patients positive for gBRCA1 or gBRCA2 mutations, the overall prevalence of gBRCA1 mutation was 9.9% and that of gBRCA2 mutation was 4.7% (Figure 1).
Figure 2A,B show the prevalence of gBRCA 1 and gBRCA 2 mutations according to their locations, ie, inside or outside the OCCR. For gBRCA1, the mutations were largely concentrated in the middle of the OCCR. For gBRCA2, the most frequent mutation was the R2318* nonsense mutation within the OCCR.
FIGURE 2.
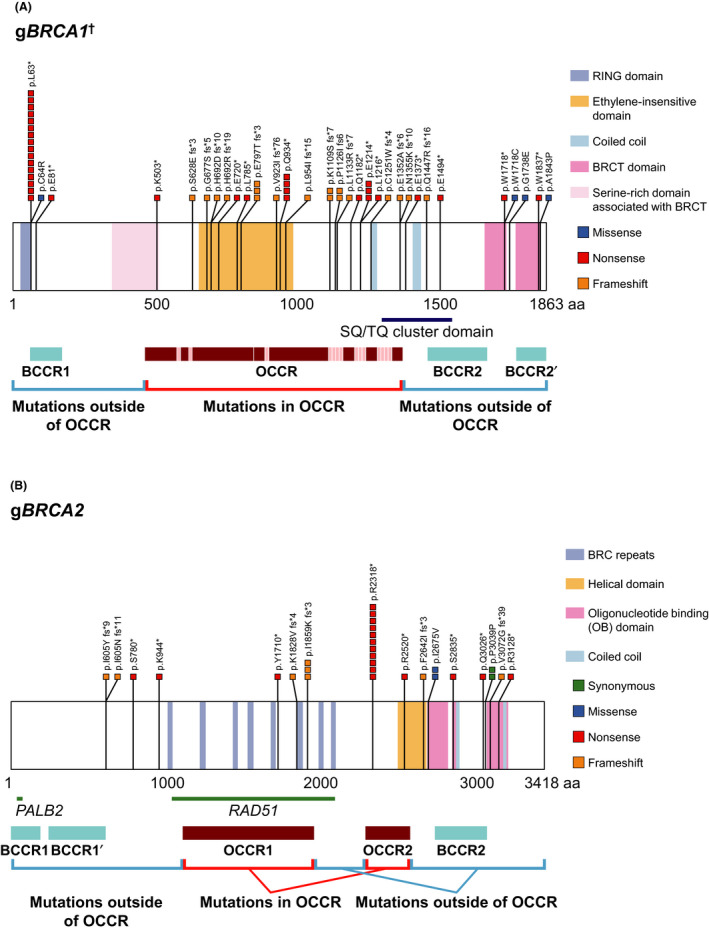
Prevalence of (A) gBRCA1 † and (B) gBRCA2 mutations by location. Adapted from Rebbeck et al. JAMA. 2015;313:1347‐1361. †Four patients with silent gBRCA1 mutations (exon deletion) are not indicated in the figure. aa, amino acid; BCCR, breast cancer cluster region; BRCA, breast cancer susceptibility gene; BRCT, BRCA1 C‐terminal; g, germline; OB, oligonucleotide binding; OCCR, ovarian cancer cluster region; PALB2, partner and localizer of BRCA2; RAD51, RAD51 recombinase; RING, really interesting new gene
Among ovarian cancer patients with gBRCA1 mutation, when excluding 16 patients with L63X founder mutation and 4 with silent mutations, 65.1% (28/43) were distributed within the OCCR and 34.9% (15/43) were distributed outside of the OCCR (Figure 3A). When including patients with the founder mutation (27.1%, 16/59), the prevalence of gBRCA1 mutations within the OCCR was 47.5% (28/59) and outside the OCCR was 25.4% (15/59) (Figure S1a). Among ovarian cancer patients with gBRCA2 mutation, 50.0% were distributed within the OCCR and 50.0% were distributed outside of the OCCR (Figure 3B).
FIGURE 3.
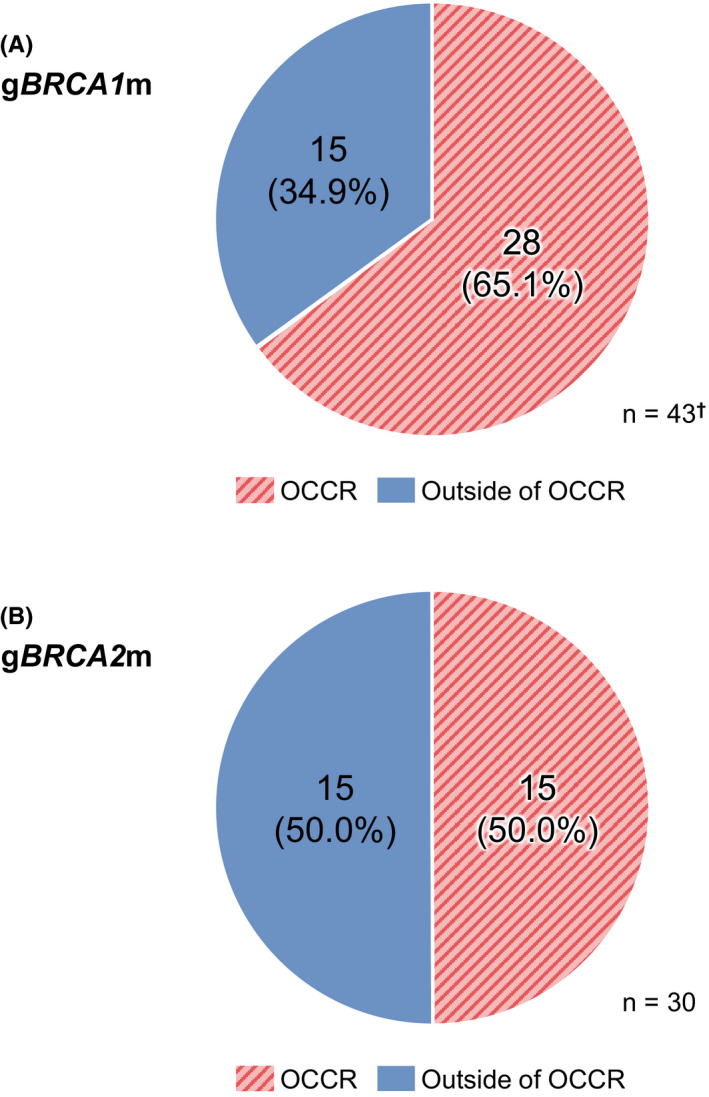
Prevalence of (A) gBRCA1 and (B) gBRCA2 mutations by location in the 93 newly diagnosed Japanese ovarian cancer patients from the CHARLOTTE study who were positive for mutations. †Four patients with silent gBRCA1 mutations (exon deletion) are not indicated in the figure; 16 patients with L63X founder mutation are also excluded. BRCA, breast cancer susceptibility gene; OCCR, ovarian cancer cluster region
3.3. Patient background and clinical characteristics in subgroups stratified by gBRCA mutation locations
Table 1 shows patient background and clinical characteristics in subgroups stratified by gBRCA mutation locations. No marked difference was found in patient age, birth history, disease stage, histopathological classification, history of breast cancer, and family history of prostate and pancreatic cancers, by mutation site. Approximately half of the patients at stage IV had gBRCA mutations located within the OCCR and half had gBRCA mutations outside the OCCR.
TABLE 1.
Prevalence of gBRCA1/2 mutations in subgroups stratified by gBRCA mutation location and patient background and clinical characteristics
| gBRCA mutations | gBRCA1 mutations a | gBRCA2 mutations | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Present | gBRCA1 only | gBRCA2 only | Absent | Inside the OCCR | Outside the OCCR | Founder mutation (L63X) | Inside the OCCR | Outside the OCCR | |
| All, n | 93 | 63 | 30 | 512 | 28 | 15 | 16 | 15 | 15 |
| Age, y | |||||||||
| 20‐29 | 0 | 0 | 0 | 4 (0.8) | 0 | 0 | 0 | 0 | 0 |
| 30‐39 | 3 (3.2) | 3 (4.8) | 0 | 29 (5.7) | 1 (3.6) | 1 (6.7) | 1 (6.3) | 0 | 0 |
| 40‐49 | 24 (25.8) | 19 (30.2) | 5 (16.7) | 113 (22.1) | 8 (28.6) | 5 (33.3) | 6 (37.5) | 3 (20.0) | 2 (13.3) |
| 50‐59 | 22 (23.7) | 16 (25.4) | 6 (20.0) | 153 (29.9) | 8 (28.6) | 3 (20.0) | 3 (18.8) | 3 (20.0) | 3 (20.0) |
| 60‐69 | 37 (39.8) | 20 (31.7) | 17 (56.7) | 136 (26.6) | 7 (25.0) | 6 (40.0) | 5 (31.3) | 8 (53.3) | 9 (60.0) |
| 70‐79 | 5 (5.4) | 3 (4.8) | 2 (6.7) | 66 (12.9) | 3 (10.7) | 0 | 0 | 1 (6.7) | 1 (6.7) |
| ≥80 | 2 (2.2) | 2 (3.2) | 0 | 11 (2.1) | 1 (3.6) | 0 | 1 (6.3) | 0 | 0 |
| ≤40 | 4 (4.3) | 4 (6.3) | 0 | 36 (7.0) | 2 (7.1) | 1 (6.7) | 1 (6.3) | 0 | 0 |
| ≥41 | 89 (95.7) | 59 (93.7) | 30 (100.0) | 476 (93.0) | 26 (92.9) | 14 (93.3) | 15 (93.8) | 15 (100.0) | 15 (100.0) |
| ≤49 | 27 (29.0) | 22 (34.9) | 5 (16.7) | 146 (28.5) | 9 (32.1) | 6 (40.0) | 7 (43.8) | 3 (20.0) | 2 (13.3) |
| ≥50 | 66 (71.0) | 41 (65.1) | 25 (83.3) | 366 (71.5) | 19 (67.9) | 9 (60.0) | 9 (56.3) | 12 (80.0) | 13 (86.7) |
| ≤64 | 63 (67.7) | 46 (73.0) | 17 (56.7) | 368 (71.9) | 21 (75.0) | 11 (73.3) | 11 (68.8) | 8 (53.3) | 9 (60.0) |
| ≥65 | 30 (32.3) | 17 (27.0) | 13 (43.3) | 144 (28.1) | 7 (25.0) | 4 (26.7) | 5 (31.3) | 7 (46.7) | 6 (40.0) |
| History of childbirth | |||||||||
| Yes | 72 (77.4) | 50 (79.4) | 22 (73.3) | 351 (68.6) | 22 (78.6) | 13 (86.7) | 11 (68.8) | 9 (60.0) | 13 (86.7) |
| No | 3 (3.2) | 2 (3.2) | 1 (3.3) | 12 (2.3) | 1 (3.6) | 0 | 1 (6.3) | 1 (6.7) | 0 |
| Unknown | 18 (19.4) | 11 (17.5) | 7 (23.3) | 149 (29.1) | 5 (17.9) | 2 (13.3) | 4 (25.0) | 5 (33.3) | 2 (13.3) |
| Histological classification | |||||||||
| Low‐grade serous cancer | 1 (1.1) | 1 (1.6) | 0 | 4 (0.8) | 0 | 1 (6.7) | 0 | 0 | 0 |
| High‐grade serous cancer | 78 (83.9) | 53 (84.1) | 25 (83.3) | 181 (35.4) | 24 (85.7) | 13 (86.7) | 13 (81.3) | 12 (80.0) | 13 (86.7) |
| Mucinous cancer | 0 | 0 | 0 | 19 (3.7) | 0 | 0 | 0 | 0 | 0 |
| Endometrioid cancer | 8 (8.6) | 7 (11.1) | 1 (3.3) | 104 (20.3) | 4 (14.3) | 1 (6.7) | 1 (6.3) | 1 (6.7) | 0 |
| Clear cell carcinoma | 4 (4.3) | 2 (3.2) | 2 (6.7) | 178 (34.8) | 0 | 0 | 2 (12.5) | 1 (6.7) | 1 (6.7) |
| Others | 2 (2.2) | 0 | 2 (6.7) | 22 (4.3) | 0 | 0 | 0 | 1 (6.7) | 1 (6.7) |
| Classification of advanced stage (FIGO 2014) | |||||||||
| I | 8 (8.6) | 3 (4.8) | 5 (16.7) | 221 (43.2) | 2 (7.1) | 0 | 0 | 4 (26.7) | 1 (6.7) |
| II | 7 (7.5) | 7 (11.1) | 0 | 60 (11.7) | 3 (10.7) | 1 (6.7) | 3 (18.8) | 0 | 0 |
| III | 62 (66.7) | 42 (66.7) | 20 (66.7) | 170 (33.2) | 16 (57.1) | 13 (86.7) | 11 (68.8) | 8 (53.3) | 12 (80.0) |
| IV | 16 (17.2) | 11 (17.5) | 5 (16.7) | 58 (11.3) | 7 (25.0) | 1 (6.7) | 2 (12.5) | 3 (20.0) | 2 (13.3) |
| Others | 0 | 0 | 0 | 3 (0.6) | 0 | 0 | 0 | 0 | 0 |
| Geographic location | |||||||||
| East | 58 (62.4) | 40 (63.5) | 18 (60.0) | 268 (52.3) | 18 (64.3) | 5 (33.3) | 15 (93.8) | 9 (60.0) | 9 (60.0) |
| West | 35 (37.6) | 23 (36.5) | 12 (40.0) | 244 (47.7) | 10 (35.7) | 10 (66.7) | 1 (6.3) | 6 (40.0) | 6 (40.0) |
| History of breast cancer | |||||||||
| Yes | 14 (15.1) | 11 (17.5) | 3 (10.0) | 16 (3.1) | 3 (10.7) | 1 (6.7) | 6 (37.5) | 2 (13.3) | 1 (6.7) |
| No | 79 (84.9) | 52 (82.5) | 27 (90.0) | 496 (96.9) | 25 (89.3) | 14 (93.3) | 10 (62.5) | 13 (86.7) | 14 (93.3) |
Data are given as n (%).
Abbreviations: BRCA, breast cancer susceptibility gene; FIGO, International Federation of Gynecology and Obstetrics; OCCR, ovarian cancer cluster region.
Four patients with silent gBRCA1 mutations are not included.
Among patients with gBRCA1 mutations, 40.7% and 33.3% of patients with mutations within the OCCR had a family history of ovarian cancer and breast cancer, respectively. Meanwhile, 13.3% and 60.0% of patients with mutations outside of the OCCR had a family history of ovarian cancer and breast cancer, respectively (Table 2). Among patients with gBRCA1 mutations and a family history of ovarian cancer, the percentage of patients with mutations within the OCCR was larger (40.7%) compared with that outside the OCCR (13.3%). Among patients with gBRCA1 mutations and a family history of breast cancer, the percentage of mutations within the OCCR was relatively smaller (33.3%) than that outside the OCCR (60.0%).
TABLE 2.
Family history of cancer per subgroups stratified by gBRCA1/2 mutation locations
| gBRCA1 mutations a | gBRCA2 mutations | |||
|---|---|---|---|---|
| Inside the OCCR | Outside the OCCR | Inside the OCCR | Outside the OCCR | |
| n | 27 b | 15 | 15 | 15 |
| Family history | ||||
| Ovarian cancer | ||||
| Yes | 11 (40.7) | 2 (13.3) | 1 (6.7) | 2 (13.3) |
| No | 16 (59.3) | 13 (86.7) | 14 (93.3) | 13 (86.7) |
| Breast cancer | ||||
| Yes | 9 (33.3) | 9 (60.0) | 5 (33.3) | 6 (40.0) |
| No | 18 (66.7) | 6 (40.0) | 10 (66.7) | 9 (60.0) |
| Prostate cancer | ||||
| Yes | 1 (3.7) | 2 (13.3) | 1 (6.7) | 1 (6.7) |
| No | 26 (96.3) | 13 (86.7) | 14 (93.3) | 14 (93.3) |
| Pancreatic cancer | ||||
| Yes | 2 (7.4) | 2 (13.3) | 3 (20.0) | 1 (6.7) |
| No | 25 (92.6) | 13 (86.7) | 12 (80.0) | 14 (93.3) |
Data are given as n (%).
Data of 16 patients with founder mutation (L63X) are not shown.
One patient with unknown family history was excluded.
Figure S2 shows the geographic distribution of patients by the location of their mutations within the OCCR. Almost all cases with gBRCA1 L63X founder mutation (15/16) were treated at centers in eastern Japan. For gBRCA2 mutation, the proportions of patients with mutations outside and inside the OCCR were the same in both the East and West of Japan.
3.4. Prevalence of gBRCA1/2 mutations by location in 7051 Japanese breast cancer patients from the study by Momozawa et al 18
Figure 4A shows the prevalence of gBRCA1 mutations within the OCCR and outside the OCCR in a large sample of Japanese breast cancer patients. Most gBRCA1 mutations (62.3%) were outside the OCCR. When accounting for 25 cases (24.5%) with L63X founder mutation, 28.4% and 47.1% of mutations were located inside and outside of the OCCR, respectively (Figure S1b). Figure 4B shows the prevalence of gBRCA2 mutations within the OCCR and outside the OCCR. The distribution of gBRCA2 mutations within the OCCR and outside the OCCR was similar, with a slightly higher prevalence of gBRCA2 mutations outside the OCCR (52.4%).
FIGURE 4.
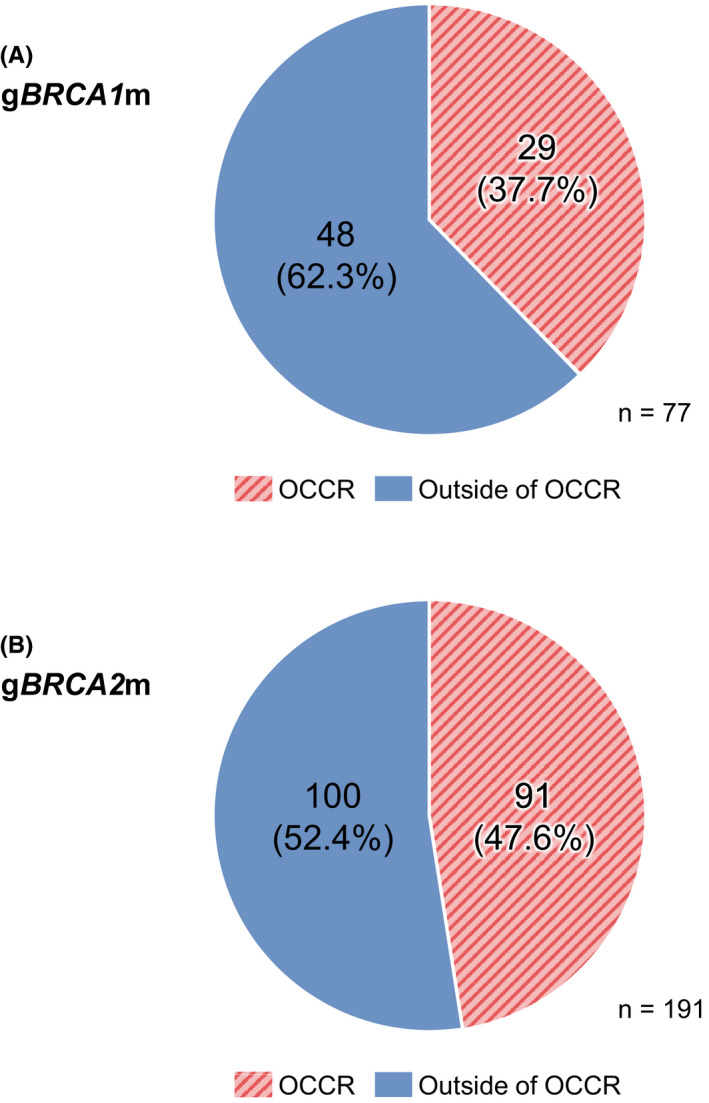
Prevalence of (A) gBRCA1 and (B) gBRCA2 mutations by location in 7051 patients studied in Momozawa et al. 18 BRCA, breast cancer susceptibility gene; OCCR, ovarian cancer cluster region
4. DISCUSSION
The main purpose of this ad hoc analysis was to report the prevalence of gBRCA1 and gBRCA2 mutations within and outside the OCCR using data from newly diagnosed Japanese ovarian cancer patients from the CHARLOTTE study. 14 To the best of our knowledge, this is the first study to report the prevalence of gBRCA1 and gBRCA2 mutations with respect to the location in and outside the OCCR 8 in Japanese ovarian cancer patients.
The present results show that gBRCA1 mutations, excluding the founder mutation and silent mutations, in this study cohort were more frequently found within the OCCR (65.1%) than outside the OCCR (34.9%). In the case of gBRCA2 mutations, the proportions were evenly split: 50.0% each for within and outside the OCCR. A previous study reported that 71% of gBRCA1 mutations of ovarian cancer patients with or without additional ovarian cancer family history were within the gBRCA1 OCCR, while 29% were located in non‐BRCA1 OCCRs. 19 Interestingly, our analysis of the data from a recent large‐scale Japanese study 18 showed that gBRCA1 mutations in breast cancer patients were more frequently located outside the OCCR (62.3%, 48/77). In the case of gBRCA2 mutations, the proportions were similar inside and outside the OCCR, but slightly more mutations were outside the OCCR (52.4%, 100/191).
In the present study, for gBRCA1 mutations, patients with mutations located within the OCCR had a higher frequency of family history of ovarian cancer compared with patients with mutations located outside of the OCCR. Furthermore, patients with gBRCA1 mutations outside the OCCR had a higher frequency of family history of breast cancer than patients with mutations within the OCCR. This finding is consistent with a previous report in which ovarian cancer patients with a family history of breast cancer had significantly fewer mutations located within the OCCR. 19 Conversely, there were no apparent differences between gBRCA2 mutations located inside or outside the OCCR for patients with either a family history of ovarian cancer or a family history of breast cancer. This finding is not consistent with previously reported results, as significant differences in patterns of distribution have been reported for patients with gBRCA2 mutations located within the OCCR. 19 Moreover, a study in a different population reported that gBRCA2 mutations located within the OCCR conferred different risks of developing breast, ovarian, and prostate cancer compared with other mutations. 20
Based on the present results, no marked differences were observed in age, histopathological classification, stages, history of breast cancer, or family history of prostate or pancreatic cancers by mutation location. We did not expect the mutation location to affect factors such as stage or histopathological classification. Regarding the history of breast cancer and family history of prostate cancer, the small sample size may have precluded finding any differences according to the location of the mutation in relation to the OCCR. As the incidence of pancreatic cancer is generally low, it may be that family history of pancreatic cancer is not an influential factor in terms of the mutation location in relation to the OCCR.
When analyzed according to the geographic distribution, most patients with the L63X founder mutation were identified in testing centers located in eastern Japan. This tendency was similar to that reported in the study by Sekine et al. 17
This ad hoc study had some limitations such as small sample size, limited power, and potential biases. As it is based on data from the main CHARLOTTE study, 14 the limitations pertaining to that study also apply here, including that Japanese clinical practices may have limited comparability with those in other countries, and that the varying levels of enrollment across the different sites may confound the regional analyses performed. Another important limitation of the study was the varying number of patients included from different sites; no upper limit was set for the number of patients registered at individual sites, whereas few patients were registered at some sites.
Among Japanese ovarian cancer patients, approximately half of the gBRCA1/2 mutations were located within the OCCR, with some regional differences in eastern vs western Japan. Patients with gBRCA1 mutations located within the OCCR had a higher frequency of family history of ovarian cancer, while those with gBRCA1 mutations located outside the OCCR had a higher frequency of family history of breast cancer. The analysis of gBRCA mutation location is important, as this information may contribute to more accurate risk assessments of susceptible individuals and early detection of ovarian cancer among gBRCA mutation carriers.
DISCLOSURE
Takayuki Enomoto has received lecture fees, honoraria, or other fees from Chugai Pharmaceutical Co., Ltd. and AstraZeneca K.K. Daisuke Aoki has received lecture fees, honoraria, or other fees from AstraZeneca K.K. and Chugai Pharmaceutical Co., Ltd., and scholarship endowments from Sanofi K.K. Hyoe Inomata, Kana Hattori, and Masahisa Jinushi are employees of AstraZeneca K.K. All other authors have nothing to declare.
Supporting information
Figure S1
Figure S2
ACKNOWLEDGMENTS
This study was sponsored by AstraZeneca K.K.
The authors wish to thank Keyra Martinez Dunn, MD, of Edanz Medical Writing for providing medical writing services, which were funded by AstraZeneca K.K. in accordance with Good Publication Practice guidelines (http://www.ismpp.org/gpp3).
The authors would also like to thank Dr. Masami Arai (genetic testing), Dr. Yuko Sasajima, Dr. Reiko Watanabe (Central Pathological Diagnosis Committee members), Dr. Hirofumi Nakaoka (National Institute of Genetics, statistical analysis), and the non‐profit organization Japanese Gynecologic Oncology Group.
Yoshihara K, Enomoto T, Aoki D, et al. Association of gBRCA1/2 mutation locations with ovarian cancer risk in Japanese patients from the CHARLOTTE study. Cancer Sci. 2020;111:3350–3358. 10.1111/cas.14513
Clinical trial registration: ClinicalTrials.gov, NCT03229122; University hospital Medical Information Network Clinical Trials Registry, UMIN000025597
DATA AVAILABILITY STATEMENT
Data underlying the findings described in this manuscript may be obtained in accordance with AstraZeneca’s data sharing policy described at https://astrazenecagrouptrials.pharmacm.com/ST/Submission/Disclosure.
REFERENCES
- 1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359‐E386. [DOI] [PubMed] [Google Scholar]
- 2. Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2014. Bethesda, MD: National Cancer Institute; https://seer.cancer.gov/csr/1975_2014/, based on November 2016 SEER data submission, posted to the SEER web site, April 2017. [Google Scholar]
- 3. Yoshida K, Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004;95:866‐871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brekelemans CT, Tilanus‐Linthorst MM, Seynaeve C, et al. Tumour characteristics, survival and prognostic factors of hereditary breast cancer from BRCA2‐, BRCA1‐ and non‐BRCA1/2 families as compared to sporadic breast cancer cases. Eur J Cancer. 2007;43:867‐876. [DOI] [PubMed] [Google Scholar]
- 5. Goodwin PJ, Phillips KA, West DW, et al. Breast cancer prognosis in BRCA1 and BRCA2 mutation carriers: an International Prospective Breast Cancer Family Registry population‐based cohort study. J Clin Oncol. 2012;30:19‐26. [DOI] [PubMed] [Google Scholar]
- 6. Kuchenbaecker KB, Hopper JL, Barnes DR, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317:2402‐2416. [DOI] [PubMed] [Google Scholar]
- 7. Pierce LJ, Levin AM, Rebbeck TR, et al. Ten‐year multi‐institutional results of breast‐conserving surgery and radiotherapy in BRCA1/2‐associated stage I/II breast cancer. J Clin Oncol. 2006;24:2437‐2443. [DOI] [PubMed] [Google Scholar]
- 8. Rebbeck TR, Mitra N, Wan F, et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA. 2015;313:1347‐1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rennert G, Bisland‐Naggan S, Barnett‐Griness O, et al. Clinical outcomes of breast cancer in carriers of BRCA1 and BRCA2 mutations. N Engl J Med. 2007;357:115‐123. [DOI] [PubMed] [Google Scholar]
- 10. Kurian AW. BRCA1 and BRCA2 mutations across race and ethnicity: distribution and clinical implications. Curr Opin Obstet Gynecol. 2010;22:72‐78. [DOI] [PubMed] [Google Scholar]
- 11. Begg CB, Haile RW, Borg A, et al. Variation of breast cancer risk among BRCA1/2 carriers. JAMA. 2008;299:194‐201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hall MJ, Reid JE, Burbidge LA, et al. BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast‐ovarian cancer. Cancer. 2009;115:2222‐2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tung NM, Garber JE. BRCA1/2 testing: therapeutic implications for breast cancer management. Br J Cancer. 2018;119:141‐152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Enomoto T, Aoki D, Hattori K, et al. The first Japanese nationwide multicenter study of BRCA mutation testing in ovarian cancer: CHARacterizing the cross‐sectionaL approach to Ovarian cancer geneTic TEsting of BRCA (CHARLOTTE). Int J Gynecol Cancer. 2019;29:1043‐1049. [DOI] [PubMed] [Google Scholar]
- 15. Moore K, Colombo N, Scambia G, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2018;379:2495‐2505. [DOI] [PubMed] [Google Scholar]
- 16. Miller RE, Crusz SM, Ledermann JA. Olaparib maintenance for first‐line treatment of ovarian cancer: will SOLO1 reset the standard of care? Future Oncol. 2019;15:1845‐1853. [DOI] [PubMed] [Google Scholar]
- 17. Sekine M, Nagata H, Tsuji S, et al. Mutational analysis of BRCA1 and BRCA2 and clinicopathologic analysis of ovarian cancer in 82 ovarian cancer families: two common founder mutations of BRCA1 in Japanese population. Clin Cancer Res. 2001;7:3144‐3150. [PubMed] [Google Scholar]
- 18. Momozawa Y, Iwasaki Y, Parsons MT, et al. Germline pathogenic variants of 11 breast cancer genes in 7,051 Japanese patients and 11,241 controls. Nat Commun. 2018;9:4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Singer CF, Tan YY, Muhr D, et al. Association between family history, mutation locations, and prevalence of BRCA1 or 2 mutations in ovarian cancer patients. Cancer Med. 2019;8:1875‐1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thompson D, Easton D; Breast Cancer Linkage Consortium . Variation in cancer risks, by mutation position, in BRCA2 mutation carriers. Am J Hum Genet. 2001;68:410‐419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Data Availability Statement
Data underlying the findings described in this manuscript may be obtained in accordance with AstraZeneca’s data sharing policy described at https://astrazenecagrouptrials.pharmacm.com/ST/Submission/Disclosure.


