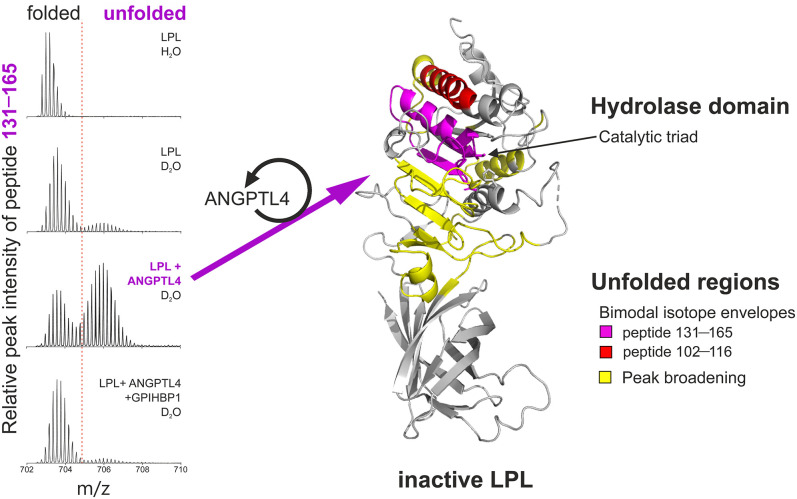
The ANGPTL proteins regulate the catalytic activity of lipoprotein lipase (LPL) by catalyzing the unfolding of LPL’s N-terminal hydrolase domain. Intravascular processing of triglyceride-rich lipoproteins depends on LPL and GPIHBP1 (an endothelial cell protein that binds LPL and transports it to the capillary lumen). A deficiency of either protein severely impairs triglyceride hydrolysis, resulting in markedly elevated plasma triglyceride levels and a substantial risk of acute pancreatitis. Angiopoietin-like proteins (ANGPTL) 3, 4, and 8 regulate LPL activity. Biophysical studies, but in particular hydrogen–deuterium exchange mass spectrometry (HDX-MS), revealed that the ANGPTL proteins inactivate LPL activity by catalyzing the unfolding of LPL’s hydrolase domain (1, 2). HDX-MS quantifies the mass increase resulting from protium(1H)−deuterium(2H) exchange in the presence of deuterium oxide (D2O). Deuterium exchange is faster in protein regions that are exposed to solvent; thus, HDX-MS is useful for defining regional differences in solvent exposure within a protein. Changes in protein conformation alter solvent exposure and hence the extent of deuterium uptake. Regional changes in protein conformation are identified by changes in uptake of deuterium into proteolytic fragments (generated by in-line pepsin digestion). The presence of bimodality or peak broadening in the isotope envelopes for individual peptic peptides provides an unambiguous signature for protein unfolding. Shown here is a bimodal isotope envelope for an LPL peptic peptide spanning amino acids 131–165 after 5 min in D2O. Unfolding of LPL is confined to the catalytic domain of LPL (depicted in red, purple, and yellow). The unfolding of LPL is catalyzed by sub-stoichiometric amounts of ANGPTL4. Importantly, the binding of GPIHBP1 to LPL protects LPL from unfolding (evident by the absence of a bimodal isotope envelope). In these studies, the unfolding of LPL is mirrored by a loss of LPL catalytic activity. These studies demonstrate that ANGPTL4 regulates LPL activity by catalyzing LPL unfolding and that ANGPTL4-mediated LPL unfolding is abrogated by GPIHBP1 binding. The ability of GPIHBP1 to protect LPL from unfolding serves to focus LPL activity on the surface of capillary endothelial cells where it is needed for lipoprotein processing. Recent studies have shown that the GPIHBP1’s intrinsically disordered acidic domain is crucial for protecting against ANGPTL4-catalyzed LPL unfolding (3, 4).
EQUIPMENT: UPLC-ESI-MS with a quadrupole time-of-flight mass spectrometer (Synapt G2; Waters).
REAGENTS: Purified bovine LPL from fresh milk, a soluble human GPIHBP1 (residues 1-131) expressed in Drosophila S2 cells, and a truncated version of ANGPTL4 (1-159) expressed in Escherichia coli
REFERENCES
- 1.Mysling S., Kristensen K. K., Larsson M., Beigneux A. P., Gårdsvoll H., Fong L. G., Bensadouen A., Jørgensen T. J., Young S. G., Ploug M.. 2016. The acidic domain of the endothelial membrane protein GPIHBP1 stabilizes lipoprotein lipase activity by preventing unfolding of its catalytic domain eLife . 5: e12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mysling S., Kristensen K. K., Larsson M., Kovrov O., Bensadouen A., Jørgensen T. J., Olivecrona G., Young S. G., Ploug M.. 2016. The angiopoietin-like protein ANGPTL4 catalyzes unfolding of the hydrolase domain in lipoprotein lipase and the endothelial membrane protein GPIHBP1 counteracts this unfolding. eLife. 5: e20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kristensen K. K., Midtgaard S. R., Mysling S., Kovrov O., Hansen L. B., Skar-Gislinge N., Beigneux A. P, Kragelund B. B., Olivecrona G., Young S. G., et al. 2018. A disordered acidic domain in GPIHBP1 harboring a sulfated tyrosine regulates lipoprotein lipase. Proc. Natl. Acad. Sci. USA. 115: E6020–E6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young S. G., Fong L. G., Beigneux A. P., Allan C. P., He C., Jiang H., Nakajima K., Meiyappan M., Birrane G., Ploug M.. 2019. GPIHBP1 and lipoprotein lipase, partners in plasma triglyceride metabolism. Cell Metab. 30: 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]