Abstract
Novel porcine circovirus type 3 (PCV3), first identified in the United States, has been detected in many other countries. Porcine circovirus is associated with postweaning multisystemic wasting syndrome, reproductive failure, congenital tremors, and other clinical symptoms. In this study, we established a double polymerase chain reaction assay for detecting both porcine circovirus type 2 (PCV2) and PCV3. This is the first study to detect and characterize the PCV3 genome in the Tianjin region of North China. We collected a total of 169 tissue samples from seven farms between 2016 and 2018. The PCV3-positive rate of all tissue samples was 37.3% (63/169) and the rate of PCV2 and PCV3 coinfection was 14.8% (25/169). PCV2 and PCV3 coinfections with more serious clinical symptoms were found in only three farms. We sequenced three PCV3 strains selected from tissue samples that were positively identified. The complete genome sequences of the three strains shared 97.6–99.4% nucleotide identities with the PCV3 strains in GenBank. Our results showed the extent of PCV3’s spread in Tianjin, and the need to further study PCV3’s pathobiology, epidemiology, isolation, and coinfection.
Keywords: Porcine circovirus type 3, Double PCR, Coinfection, Complete genome sequences
Introduction
Porcine circovirus (PCV), a member of the genus Circovirus, is a single-stranded DNA virus that can autonomously replicate. PCV has a genome size of 1,700 bp and two basic open reading frames (ORFs) in its DNA sequence called rep and cap. The rep and cap ORFs mainly code for the replicase and capsid proteins, respectively (Hamel, Lin & Nayar, 1998; Harms et al., 2001; Meng, 2013). Porcine circovirus type 1 (PCV1) was first found in 1974 as a nonpathogenic contaminant in PK-15 cells (Tischer et al., 1986). Porcine circovirus type 2 (PCV2) is a variant of PCV1 first found in piglets in 1998 (Allan et al., 1998; Tischer et al., 1995). Previous studies have shown that the nucleotide sequence homology between PCV2 and PCV1 is only about 68%, and that the two viruses have different antigens and phenotypes (Allan et al., 1994; Hamel, Lin & Nayar, 1998; Meehan et al., 1998). PCV2 is associated with many diseases including reproductive disorders, enteric diseases, respiratory diseases, and postweaning multisystemic wasting syndrome (PMWS). These diseases are known as porcine circovirus associated diseases (PCVAD) in the United States, and porcine circovirus diseases (PCVD) in Europe (Harms et al., 2001; Krakowka et al., 2000; Opriessnig, Meng & Halbur, 2007; Segales, 2012).
In 2016, novel porcine circovirus type 3 (PCV3) was first reported in the United States. Palinski et al. (2017) observed that PCV3’s putative cap ORF encoded a 214-aa protein that was 36–37% identical to PCV2. PCV3 is associated with cardiac, multisystemic (Phan et al., 2016), and reproductive failure, as well as porcine dermatitis and nephropathy syndrome (Palinski et al., 2017). PCV3’s complete sequence size is 2,000 bp with three major ORFs (Palinski et al., 2017; Phan et al., 2016). PCV3 has been detected in pigs across many countries (Faccini et al., 2017; Hayashi et al., 2018; Klaumann et al., 2019; Stadejek et al., 2017; Sukmak et al., 2019; Ye et al., 2018). Although there have been some reports of PCV3 in South China, no cases of PCV3 in Tianjin, North China have been reported (Chen et al., 2017; Fan et al., 2017; Ku et al., 2017; Liu et al., 2019; Qi et al., 2019; Shen et al., 2018; Wen et al., 2018; Xu et al., 2018; Zheng et al., 2017; Zou et al., 2018).
This is the first investigation of the spread of PCV3 in Tianjin, North China. We looked at seven farms in seven of its regions (Jizhou, Baodi, Wuqing, Ninghe, Dongli, Jinghai, and Binhai) between 2016 and 2018. Since pig breeding and farming are major industries in Tianjin, it is crucial to study the epidemic nature of PCV3 in North China.
Materials and Methods
Sample collection and clinical symptoms
We collected a total of 169 pig tissue samples (including hearts, livers, spleens, lungs, and kidneys) from seven farms (in Jizhou, Baodi, Wuqing, Ninghe, Dongli, Jinghai, and Binhai) in Tianjin, North China between December 2016 and May 2018. All samples were stored at −80 °C. We selected 10 pigs from each farm for a total of 70 pigs. The selected pig’s major clinical symptoms included fever, cough, anorexia, depression, an increase in mortality rate, and a decrease in conception rate. We compared the clinical symptoms from each farm to determine the distinction between mild and severe cases. In this study, Animal committee of Shanghai Veterinary Research Institute provided full approval for this research (permit number SHVRI-Pig-20161206-05), field experiments were approved by the Scientific Research Office of Shanghai Veterinary Research Institute (permit number SHVRI-Pig-160417-19).
Polymerase chain reaction assay
The collected tissue samples were cut into small pieces (0.1 cm × 0.1 cm) and packed into two mL Eppendorf tubes. After three freeze-thaw cycles, the small pieces of tissue were ground thoroughly with one mL of Dulbecco’s modified Eagle’s medium (DMEM; Hyclone, Beijing, China) and centrifuged at 3,000×g for 10 min at 4 °C. The supernatants were stored at −80 °C. We extracted nucleic acid using a DNeasy® Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
A double PCR assay was established to detect PCV2 and PCV3. Using bioinformatics analysis, we designed and synthesized two pairs of specific primers to detect PCV2 and PCV3 (Table 1) from a commercial source (GENEWIZ). Amplicon products were cloned into a PLB vector (Tiangen, Beijing, China) and sequenced by Sangon Biotech Company (Shanghai, China). The complete PCV2 and PCV3 sequences were amplified using the primer pairs PCV2-F (5″-ATCCACGGAGGAAGGGGGCCAGTT-3″) and PCV2-R (5″-GTGGATTGTTCTGTAGCATTCTTCCA-3″) (Guo et al., 2010), PCV3-1F (5″-CACCGTGTGAGTGGATATAC-3″) and PCV3-1R (5″-CAAACCCACCCTTAACAG-3″), PCV3-2F (5″-GTCGTCTTGGAGCCAAGTG-3″) and PCV3-2R (5″-CGACCAAATCCGGGTAAGC-3″), PCV3-3F (5″-TGTTGTACCGGAGGAGTG-3″) and PCV3-3R (5″-TGCCGGGTAATACTAGCC-3″), and PCV3-4F (5″-GAAGTTGCGGAGAAGATG-3″) and PCV3-4R (5″-TCCAAGACGACCCTTATG-3″) (Palinski et al., 2017). The amplicon products were also cloned into a PLB vector (Tiangen, Beijing, China) and sequenced by Sangon Biotech Company (Shanghai, China).
Table 1. Primers for PCV2 and PCV3 specific target gene amplification by the double PCR.
| Virus | Primer sequence (5′ → 3′) |
Product length (bp) | Annealing temperature (°C) |
|---|---|---|---|
| PCV2 | F: ATAGGGGTCATAGGTTAGGGCATT | 305 | 56 |
| R: GAAAAATGGCATCTTCAACACCCG | |||
| PCV3 | F: GGAGGTTCACTAAGGTTGTTTGTT | 767 | 58 |
| R: ACCACTTCATTACCCGCCTAAACGA |
We carried out a single PCR for PCV2 and PCV3 in a 50 μl mixture containing 25 μl Takara La Taq™ DNA polymerase (5 U/μl), one μl forward primer (10 mmol/μl), one μl reverse primer (10 mmol/μl), one μl DNA template, and 22 μl distilled water. The DEPC-treated water was used as a template for negative control reactions. The reaction was performed under the following conditions: initial denaturation at 94 °C for 3 min; 30 cycles of 94 °C for 30 s, 55 °C (PCV2) or 58 °C (PCV3) for 30 s, and 72 °C for 1 min; and the final extension at 72 °C for 10 min. We detected PCR products by electrophoresing through 1% agarose gel in 1× TAE buffer. Each specific viral target fragment was cloned into the plasmid PLB vector (Tiangen, Beijing, China) and sequenced (Invitrogen, Carlsbad, CA, USA).
We optimized the double PCR reaction by changing a single condition and leaving the other conditions unchanged. We also experimentally optimized the Takara La Taq™ DNA polymerase from 1 to 5 U, each primer from 2 to 20 pmol, the annealing temperature from 50 to 70 °C, and the number of cycles from 25 to 40. We detected PCR products by electrophoresing through 1% agarose gel in 1× TAE buffer. To ensure the sensitivity of the double PCR assay and to determine the limits of detection, we diluted the recombinant plasmid of the complete DNA sequences of the two viruses using a 10-fold series. The following formula was used to calculate the number of gene copies per μl in each dilution: copies/μl = (6.02 × 1023) × (Plasmid concentration (ng/μl) × 10−9)/(DNA length (bp) × 660) (Yue et al., 2009). To confirm the specificity of the double PCR assay, we also tested for pseudorabies virus (PRV), porcine reproductive and respiratory syndrome (PRRSV), classical swine fever virus (CSFV), Japanese encephalitis virus (JEV), porcine epidemic diarrhea virus (PEDV), swine influenza virus (SIV), porcine parvovirus (PPV), and transmissible gastroenteritis virus (TGEV) using the primers listed earlier in Table 1.
Detecting viruses in clinical samples using PCR assays
We extracted the DNA from the 169 collected samples using the methods described earlier. The samples were tested for PCV2 and PCV3 using single PCR and double PCR. Additionally, we prepared the extracted nucleic acid of the collected samples and used PCR to detect other common porcine viruses PCV1, PRV, PRRSV, CSFV, JEV, PEDV, SIV, PPV, and TGEV. The primers for PRV, PRRSV, CSFV, JEV and PPV were referred to Zeng et al. (2014), the primers for PEDV and TGEV were referred to Ogawa et al. (2009), the primers for SIV were referred to Poddar (2002) and the primers for PCV1 were referred to Yu et al. (2018). All the primers were synthesized from a commercial source (GENEWIZ, South Plainfield, NJ, USA).
Virus isolation
Virus isolation was carried out in a PK-15 cell lines were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were resuspended in 15 ml of DMEM (Hyclone, Beijing, China) containing 10% fetal bovine serum (FBS) (Hyclone, Beijing, China) and inoculated with 500 μl filtered tissue sample supernatants. These cultures were then incubated at 37 °C for 18 h in a 10% CO2 atmosphere, and the resulting semiconfluent monolayers were treated with 300 mM D-(+)-glucosamine hydrochloride (Sigma, St. Louis, MO, USA) and incubated for an additional 48–72 h at 37 °C (Allan et al., 1998). All PCV2- and PCV3-positive samples were used to isolate the viruses. After three passages, we used the freeze-thawed third generation PK-15 cell culture to extract DNA and the DNA was detected by using the established double PCR assay.
Phylogenetic analysis
Phylogenetic trees were reconstructed using the maximum-likelihood method with a MEGA 6.0 software bootstrap analysis of 1,000 replicates.
Results
Optimizing double PCR
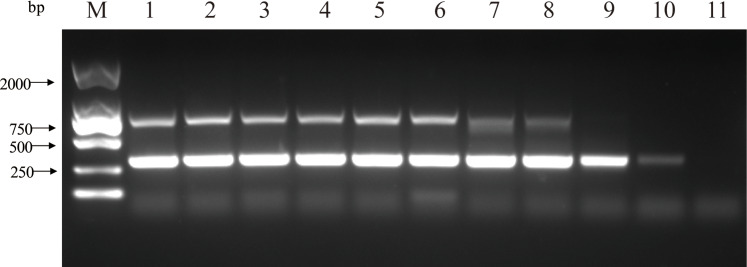
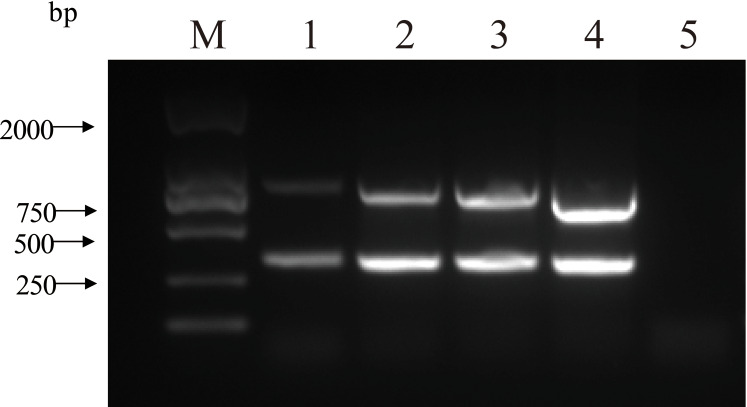
The double PCR using two pairs of primers for PCV2 and PCV3 produced specific amplicon lengths of 305 bp and 767 bp, respectively. The optimal annealing temperature was 58.8 °C (Fig. 1) and the optimal number of cycles was 30 (Fig. 2). Other optimal double PCR conditions were 4.5 U of La Taq™ DNA polymerase and 10 pmol of primer. Under optimized conditions, the double PCR effectively amplified the two viruses.
Figure 1. Influence of a range of annealing temperatures in the efficiency of amplification for two close related viruses by double PCR.
M: DL2000 DNA Marker; 1: 50 °C; 2: 52 °C; 3: 54 °C; 4: 56.4 °C; 5: 58.8 °C; 6: 61.2 °C; 7: 63.6 °C; 8: 66 °C; 9: 68 °C; 10: 70 °C; 11: negative control.
Figure 2. Influence of a range of number of cycles in the efficiency of amplification for two close related viruses by double PCR.
M: DL2000 DNA Marker; 1: 40 cycles; 2: 35 cycles; 3: 30 cycles; 4: 25 cycles; 5: negative control.
Sensitivity and specificity of single and double PCR
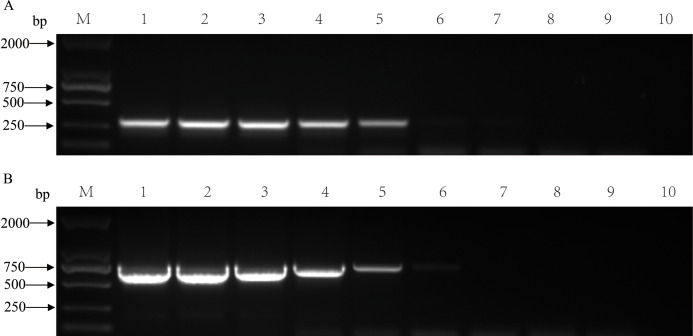
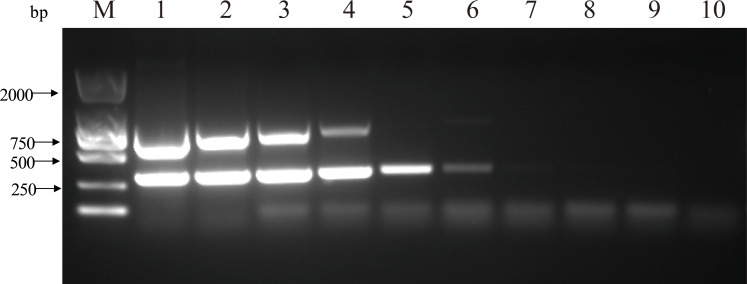
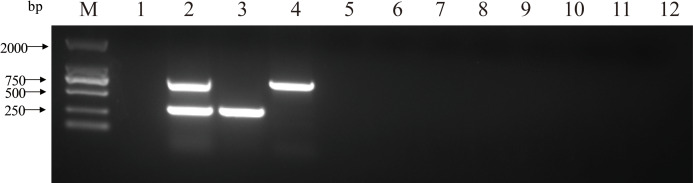
The nucleic acids of the two viruses were diluted 10-fold and mixed to form templates for single and double PCR sensitivity assays. The single PCR detection limit copy numbers were 1.44 × 105 and 2.23 × 105 for PCV2 and PCV3, respectively (Fig. 3). The detection limits of the double PCR were 1.44 × 104 and 2.23 × 106 copies for PCV2 and PCV3, respectively (Fig. 4). We evaluated the specificity of the two pairs of primers using double PCR. As shown in Fig. 5, the double PCR was specific to the target viral agent. No amplicons were produced with the other agents (PRV, PRRSV, CSFV, JEV, PEDV, SIV, PPV or TGEV) or with ddH2O. All mentioned amplicons and plasmids were sequenced to control for potential false positive results. The double PCR amplicon sequences corresponded to 305 bp for PCV2 and 767 bp for PCV3.
Figure 3. Sensitivity of single PCR for detecting single viral nucleic acid of PCV2 or PCV3 by diluting the nucleic acid using a 10-fold series.
(A) Single PCR sensitivity for PCV2. M: DL2000 DNA Marker; 1: 109 copies; 2: 108 copies; 3: 107 copies; 4: 106 copies; 5: 105 copies; 6: 104 copies; 7: 103 copies; 8: 102 copies; 9: 101 copies; 10: negative control. (B) Single PCR sensitivity for PCV3. M: DL2000 DNA Marker; 1: 109 copies; 2: 108 copies; 3: 107 copies; 4: 106 copies; 5: 105 copies; 6: 104 copies; 7: 103 copies; 8: 102 copies; 9: 101 copies; 10: negative control.
Figure 4. Sensitivity of double PCR for detecting PCV2 and PCV3 by diluting the nucleic acid using a 10-fold series.
M: DL2000 DNA Marker; 1: 109 copies; 2: 108 copies; 3: 107 copies; 4: 106 copies; 5: 105 copies; 6: 104 copies; 7: 103 copies; 8: 102 copies; 9: 101 copies; 10: negative control.
Figure 5. Specificity of double PCR for detecting PCV2, PCV3, PRV, PRRSV, CSFV, JEV, PEDV, SIV, PPV and TGEV.
M: DL2000 DNA Marker; 1: negative control; 2: PCV2 and PCV3; 3: PCV2; 4: PCV3; 5: PRV; 6: PRRSV; 7: CSFV; 8: JEV; 9: PEDV; 10: SIV; 11: PPV; 12: TGEV.
Detecting clinical samples using PCR assays and clinical symptom records
In this study, we analyzed a total of 169 collected samples for PCV2 and PCV3. Sixty-three tissue samples were positive for PCV3, with a 37.3% positive rate. Forty-eight tissue samples were positive for PCV2, with a 28.4% positive rate. Additionally, 25 tissue samples were coinfected with PCV2 and PCV3, and the coinfection rate was 14.8%. We found no other common porcine viruses among the 169 tissue samples. The specific clinical symptoms were different across the seven farms (Table 2) and coinfection was only found in Farm-D, Farm-E, and Farm-G.
Table 2. List of clinical symptoms at the seven farms in Tianjin.
| Farm name | Fever | Cough | Anorexic | Depression | Increase in mortality rate | Decrease in conception rate |
|---|---|---|---|---|---|---|
| Farm-Aa | + | – | + | – | + | + |
| Farm-Bb | + | – | + | – | + | – |
| Farm-Ca | – | – | – | + | – | + |
| Farm-Dc | + | – | + | ++ | ++ | + |
| Farm-Ec | ++ | + | + | ++ | + | ++ |
| Farm-Fa | + | – | + | – | – | + |
| Farm-Gc | ++ | ++ | + | ++ | ++ | ++ |
Notes:
Only PCV3 infection was detected on the farm.
Only PCV2 infection was detected on the farm.
Both PCV2 and PCV3 were detected on the farm.
+/−, positive or negative appearance of clinical symptoms; ++, degree of severity of the demonstrated clinical symptoms.
Virus isolation
After three passages through the PK-15 cell culture, the cell culture DNA was extracted and detected using single and double PCR. We isolated six strains of the virus, all of which were PCV2 strains.
Phylogenetic analysis
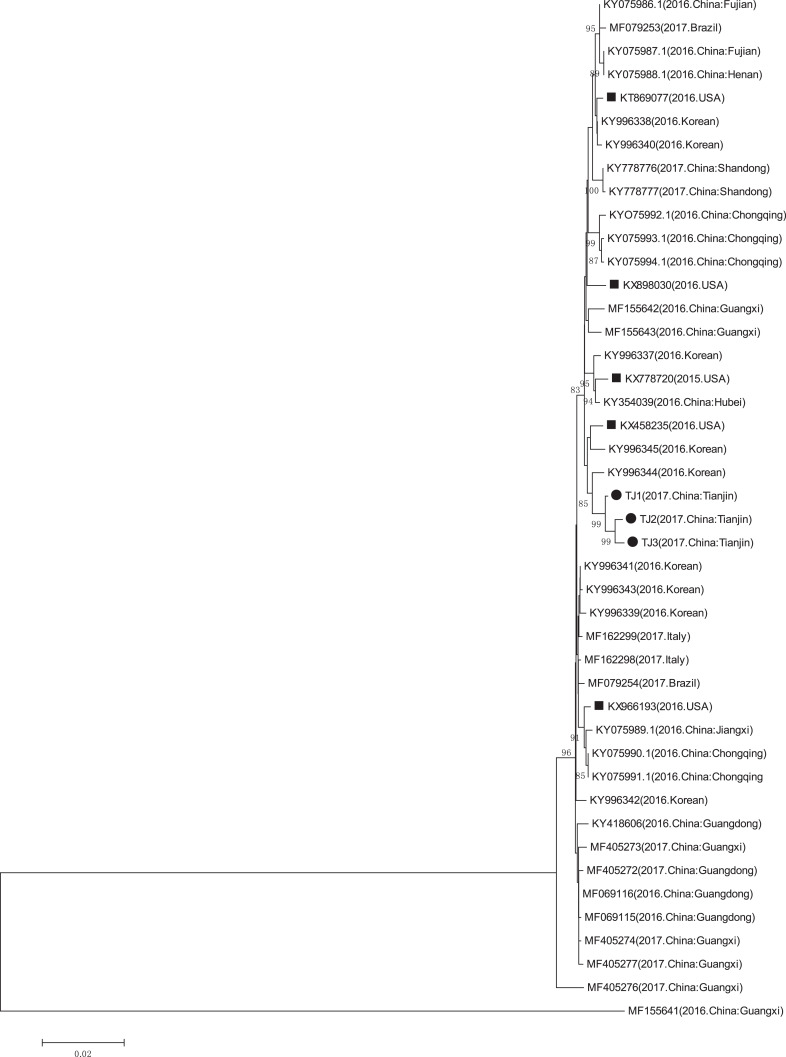
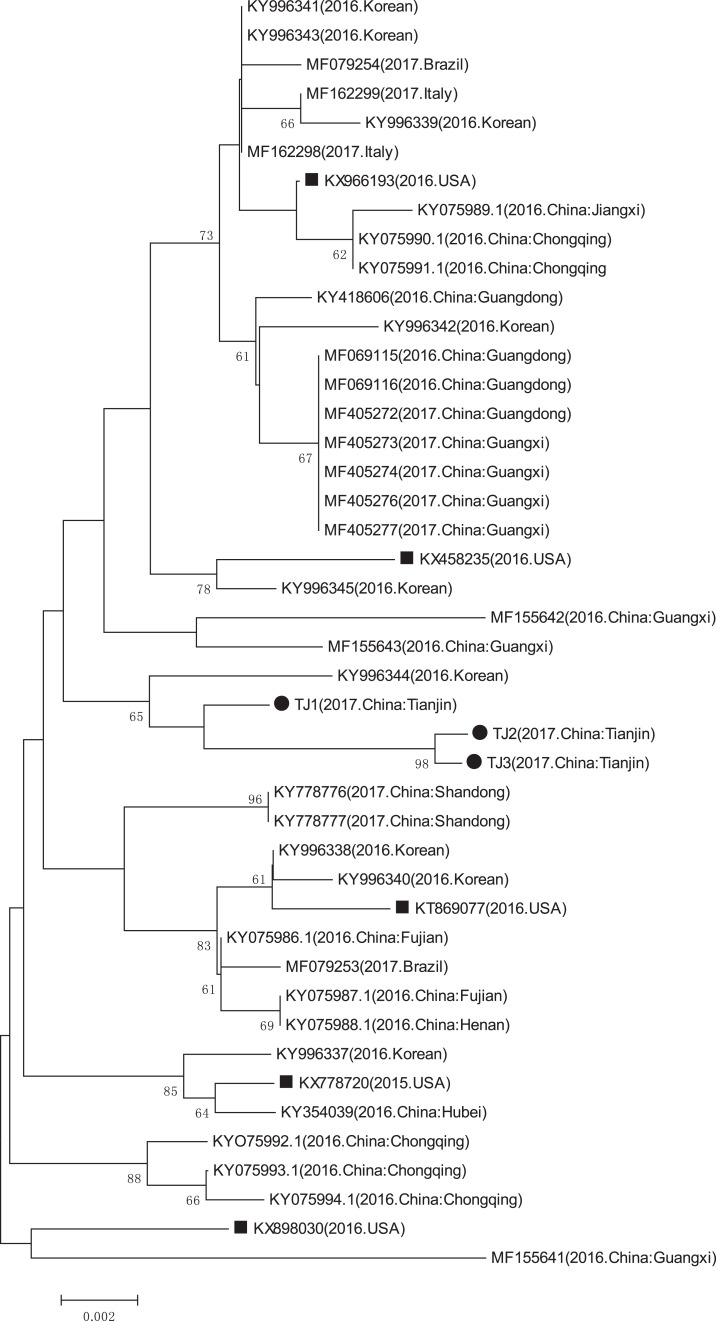
The GenBank accession numbers of the three PCV3 strains identified in this study are MN790774, MN790775, and MN790776. The genomic homology of the complete genome and the cap gene of the three identified PCV3 strains with that of the other PCV3 strains (except the PCV3-China/GX2016-1 strain which had a deletion (Wen et al., 2018)), were in the 97.6–99.4% and 97.1–99.1% ranges, respectively. The complete genome and cap gene shared 99.5–99.6% and 99.1–99.8% homology between the three identified PCV3 strains, respectively. The phylogenetic analyses of the complete genome and the cap gene showed that the three PCV3 strains from Tianjin were closely related to the PCV3/KU-1608 (KY996344) strain first reported in South Korea (Figs. 6 and 7) (Kwon et al., 2017) and the genomic homology of the complete genome and the cap gene of the three identified PCV3 strains with PCV3/KU-1608 (KY996344) were up to 99.4% and 99.1%. Additionally, the cap gene length of all three PCV3 strains measured 645 bp.
Figure 6. Phylogenetic tree based on the complete genome sequences of PCV3 obtained from GenBank database and three sequences detected in this study.
The Phylogenetic tree was constructed using the p-distance-based neighbor-joining method in MEGA 6.0 software. Bootstrap values were calculated with 1,000 replicates. Black solid circles indicate the strains from Tianjin. Black solid squares indicate the strains from the United States. Scale bars indicate nucleotide substitutions per site.
Figure 7. Phylogenetic tree based on the cap gene sequences of PCV3 obtained from GenBank database and three sequences detected in this study.
The phylogenetic tree was based on the partial capsid gene sequences. The phylogenetic tree was constructed using the p-distance-based neighbor-joining method in MEGA 6.0 software. Bootstrap values were calculated with 1,000 replicates. Black solid circles indicate the strains from Tianjin. Black solid squares indicate the strains from the United States. Scale bars indicate nucleotide substitutions per site.
Discussion
PCV3 cases have been reported in many countries. This new virus is associated with reproductive failure, cardiac, multisystemic inflammation, congenital tremors, and other porcine diseases (Chen et al., 2017; Palinski et al., 2017; Phan et al., 2016; Wen et al., 2018). PCVAD presents an immense economic burden to the global swine industry (Meng, 2013), creating a high demand for rapid and precise diagnostic methods. Additionally, PCV1 and PCV2 diagnostic methods have been established and are widely used (Li et al., 2013). In this study, we established a double PCR assay to detect PCV2 and PCV3 in individual reaction systems. The double PCR assay can increase the efficiency of sample detection, help monitor incident rates, and confirm the prevalence of the two viruses. The rapid development of the pig breeding industry calls for a better understanding of PCV3 prevalence. We collected a total of 169 tissue samples from Tianjin. The PCV3 positive rate was 37.3%, similar to the national average in China (Chen et al., 2017; Ku et al., 2017; Wen et al., 2018; Zheng et al., 2017). The phylogenetic tree showed that the three PCV3 strains were all in the same clade as another PCV3 strain first found in Korea (Kwon et al., 2017) and the genomic homology of the complete genome and the cap gene of the three identified PCV3 strains with PCV3/KU-1608 (KY996344) were up to 99.4% and 99.1%, perhaps due to geographic distribution.
Previous studies have shown that viral coinfection can cause more damage and/or viral replication in animals (Ellis et al., 2000; Meng, 2013; Opriessnig & Halbur, 2012; Pogranichniy et al., 2002; Ramamoorthy & Meng, 2009). We found similar results in the seven farms in this study. The clinical symptoms of Farm-D, Farm-E, and Farm-G were more severe than the other four farms, and PCV2 and PCV3 coinfection was only found on these three farms. It would be interesting to determine whether coinfection can cause more severe clinical symptoms than single PCV2 or PCV3 infection.
However, our isolation of the PCV3 virus failed. The suspension isolated from PCV3-positive tissue samples was inoculated in PK-15 cells and went through three continuous passages (Tischer et al., 1995). The results of the serial passage tests showed that PCV3 could not be isolated by this method. We conjectured that the PK-15 cell line is not a suitable host for PCV3. To find a new host cell line for PCV3 are need to further research.
Conclusions
We investigated the presence of PCV3 and the rate of PCV2 and PCV3 coinfection on farms in Tianjin, North China using an established double PCR assay. The results indicated the prevalence of PCV3 and PCV2 in swine herds. To better understand the impact of this agriculturally important pathogen, future studies should explore the pathobiology and epidemiology of PCV3.
Supplemental Information
Raw data of full-length image of gel
Raw data of full-length image of gel
Raw data of full-length image of gel
Raw data of full-length image of gel
Raw data of full-length image of gel
Raw data of full-length image of gel
Funding Statement
This study was supported by Shanghai Natural Science Foundation of China (No. 16ZR1444000). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Shuai-Yong Wang conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Ying-Feng Sun conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Qi Wang conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Ling-Xue Yu performed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Shi-Qiang Zhu performed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Xiao-Min Liu performed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Yun Yao performed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Juan Wang performed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Tong-Ling Shan analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Hao Zheng analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Yan-Jun Zhou analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Wu Tong analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Ning Kong analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Guang-Zhi Tong conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Hai Yu conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Animal Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
Animal committee of Shanghai Veterinary Research Institute provided full approval for this research (SHVRI-Pig-20161206-05).
Field Study Permissions
The following information was supplied relating to field study approvals (i.e., approving body and any reference numbers):
Field experiments were approved by the Scientific Research Office of Shanghai Veterinary Research Institute (SHVRI-Pig-160417-19).
Data Availability
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental Files. The DNA sequences are available as Supplemental Files and available at GenBank: MN790774–MN790776.
References
- Allan et al. (1994).Allan GM, Mackie DP, McNair J, Adair BM, McNulty MS. Production, preliminary characterisation and applications of monoclonal antibodies to porcine circovirus. Veterinary Immunology and Immunopathology. 1994;43(4):357–371. doi: 10.1016/0165-2427(94)90157-0. [DOI] [PubMed] [Google Scholar]
- Allan et al. (1998).Allan GM, McNeilly F, Kennedy S, Daft B, Clarke EG, Ellis JA, Haines DM, Meehan BM, Adair BM. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. Journal of Veterinary Diagnostic Investigation. 1998;10(1):3–10. doi: 10.1177/104063879801000102. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2017).Chen GH, Mai KJ, Zhou L, Wu RT, Tang XY, Wu JL, He LL, Lan T, Xie QM, Sun Y, Ma JY. Detection and genome sequencing of porcine circovirus 3 in neonatal pigs with congenital tremors in South China. Transboundary and Emerging Diseases. 2017;64(6):1650–1654. doi: 10.1111/tbed.12702. [DOI] [PubMed] [Google Scholar]
- Ellis et al. (2000).Ellis JA, Bratanich A, Clark EG, Allan G, Meehan B, Haines DM, Harding J, West KH, Krakowka S, Konoby C, Hassard L, Martin K, McNeilly F. Coinfection by porcine circoviruses and porcine parvovirus in pigs with naturally acquired postweaning multisystemic wasting syndrome. Journal of Veterinary Diagnostic Investigation. 2000;12(1):21–27. doi: 10.1177/104063870001200104. [DOI] [PubMed] [Google Scholar]
- Faccini et al. (2017).Faccini S, Barbieri I, Gilioli A, Sala G, Gibelli LR, Moreno A, Sacchi C, Rosignoli C, Franzini G, Nigrelli A. Detection and genetic characterization of porcine circovirus type 3 in Italy. Transboundary and Emerging Diseases. 2017;64(6):1661–1664. doi: 10.1111/tbed.12714. [DOI] [PubMed] [Google Scholar]
- Fan et al. (2017).Fan S, Ku X, Chen F, Wang Y, Yu X, He Q. Complete genome sequence of a novel porcine circovirus type 3 strain, PCV3/CN/Hubei-618/2016, isolated from China. Genome Announcements. 2017;5(15):e00100-17. doi: 10.1128/genomeA.00100-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo et al. (2010).Guo LJ, Lu YH, Wei YW, Huang LP, Liu CM. Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China. Virology Journal. 2010;7:273. doi: 10.1186/1743-422X-7-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel, Lin & Nayar (1998).Hamel AL, Lin LL, Nayar GPS. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. Journal of Virology. 1998;72(6):5262–5267. doi: 10.1128/JVI.72.6.5262-5267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms et al. (2001).Harms PA, Sorden SD, Halbur PG, Bolin SR, Lager KM, Morozov I, Paul PS. Experimental reproduction of severe disease in CD/CD pigs concurrently infected with type 2 porcine circovirus and porcine reproductive and respiratory syndrome virus. Veterinary Pathology. 2001;38(5):528–539. doi: 10.1354/vp.38-5-528. [DOI] [PubMed] [Google Scholar]
- Hayashi et al. (2018).Hayashi S, Ohshima Y, Furuya Y, Nagao A, Oroku K, Tsutsumi N, Sasakawa C, Sato T. First detection of porcine circovirus type 3 in Japan. Journal of Veterinary Medical Science. 2018;80(9):1468–1472. doi: 10.1292/jvms.18-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaumann et al. (2019).Klaumann F, Correa-Fiz F, Sibila M, Nunez JI, Segales J. Infection dynamics of porcine circovirus type 3 in longitudinally sampled pigs from four Spanish farms. Veterinary Record. 2019;184(20):619. doi: 10.1136/vr.105219. [DOI] [PubMed] [Google Scholar]
- Krakowka et al. (2000).Krakowka S, Ellis JA, Meehan B, Kennedy S, McNeilly F, Allan G. Viral wasting syndrome of swine: experimental reproduction of postweaning multisystemic wasting syndrome in gnotobiotic swine by coinfection with porcine circovirus 2 and porcine parvovirus. Veterinary Pathology. 2000;37(3):254–263. doi: 10.1354/vp.37-3-254. [DOI] [PubMed] [Google Scholar]
- Ku et al. (2017).Ku X, Chen F, Li P, Wang Y, Yu X, Fan S, Qian P, Wu M, He Q. Identification and genetic characterization of porcine circovirus type 3 in China. Transboundary and Emerging Diseases. 2017;64(3):703–708. doi: 10.1111/tbed.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon et al. (2017).Kwon T, Yoo SJ, Park CK, Lyoo YS. Prevalence of novel porcine circovirus 3 in Korean pig populations. Veterinary Microbiology. 2017;207:178–180. doi: 10.1016/j.vetmic.2017.06.013. [DOI] [PubMed] [Google Scholar]
- Li et al. (2013).Li J, Shi JL, Wu XY, Cong XY, Xu SJ, Yuan XY, Wu JQ, Sun WB, Du YJ, Peng Z, Wang JB, Huang BH. Differentiation of PCV1 and PCV2 by a multiplex real-time PCR assay. Veterinary Record. 2013;173(14):346. doi: 10.1136/vr.101686. [DOI] [PubMed] [Google Scholar]
- Liu et al. (2019).Liu Y, Zhang S, Song X, Hou B, Gu X, Zhao B, Yang L, Wang C, Zhou Z. The prevalence of novel porcine circovirus type 3 isolates in pig farms in China. Transboundary and Emerging Diseases. 2019;66(5):2143–2151. doi: 10.1111/tbed.13266. [DOI] [PubMed] [Google Scholar]
- Meehan et al. (1998).Meehan BM, McNeilly F, Todd D, Kennedy S, Jewhurst VA, Ellis JA, Hassard LE, Clark EG, Haines DM, Allan GM. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. Journal of General Virology. 1998;79(Pt. 9):2171–2179. doi: 10.1099/0022-1317-79-9-2171. [DOI] [PubMed] [Google Scholar]
- Meng (2013).Meng XJ. Porcine circovirus type 2 (PCV2): pathogenesis and interaction with the immune system. Annual Review of Animal Biosciences. 2013;1(1):43–64. doi: 10.1146/annurev-animal-031412-103720. [DOI] [PubMed] [Google Scholar]
- Ogawa et al. (2009).Ogawa H, Taira O, Hirai T, Takeuchi H, Nagao A, Ishikawa Y, Tuchiya K, Nunoya T, Ueda S. Multiplex PCR and multiplex RT-PCR for inclusive detection of major swine DNA and RNA viruses in pigs with multiple infections. Journal of Virological Methods. 2009;160(1–2):210–214. doi: 10.1016/j.jviromet.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Opriessnig & Halbur (2012).Opriessnig T, Halbur PG. Concurrent infections are important for expression of porcine circovirus associated disease. Virus Research. 2012;164(1–2):20–32. doi: 10.1016/j.virusres.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opriessnig, Meng & Halbur (2007).Opriessnig T, Meng XJ, Halbur PG. Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. Journal of Veterinary Diagnostic Investigation. 2007;19(6):591–615. doi: 10.1177/104063870701900601. [DOI] [PubMed] [Google Scholar]
- Palinski et al. (2017).Palinski R, Pineyro P, Shang P, Yuan F, Guo R, Fang Y, Byers E, Hause BM. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. Journal of Virology. 2017;91(1):1173. doi: 10.1128/JVI.01879-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan et al. (2016).Phan TG, Giannitti F, Rossow S, Marthaler D, Knutson TP, Li L, Deng X, Resende T, Vannucci F, Delwart E. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virology Journal. 2016;13(1):184. doi: 10.1186/s12985-016-0642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poddar (2002).Poddar SK. Influenza virus types and subtypes detection by single step single tube multiplex reverse transcription-polymerase chain reaction (RT-PCR) and agarose gel electrophoresis. Journal of Virological Methods. 2002;99(1–2):63–70. doi: 10.1016/S0166-0934(01)00380-9. [DOI] [PubMed] [Google Scholar]
- Pogranichniy et al. (2002).Pogranichniy RM, Yoon KJ, Harms PA, Sorden SD, Daniels M. Case-control study on the association of porcine circovirus type 2 and other swine viral pathogens with postweaning multisystemic wasting syndrome. Journal of Veterinary Diagnostic Investigation. 2002;14(6):449–456. doi: 10.1177/104063870201400601. [DOI] [PubMed] [Google Scholar]
- Qi et al. (2019).Qi S, Su M, Guo D, Li C, Wei S, Feng L, Sun D. Molecular detection and phylogenetic analysis of porcine circovirus type 3 in 21 Provinces of China during 2015–2017. Transboundary and Emerging Diseases. 2019;66(2):1004–1015. doi: 10.1111/tbed.13125. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy & Meng (2009).Ramamoorthy S, Meng XJ. Porcine circoviruses: a minuscule yet mammoth paradox. Animal Health Research Reviews. 2009;10(1):1–20. doi: 10.1017/S1466252308001461. [DOI] [PubMed] [Google Scholar]
- Segales (2012).Segales J. Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Research. 2012;164(1–2):10–19. doi: 10.1016/j.virusres.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Shen et al. (2018).Shen H, Liu X, Zhang P, Wang L, Liu Y, Zhang L, Liang P, Song C. Genome characterization of a porcine circovirus type 3 in South China. Transboundary and Emerging Diseases. 2018;65(1):264–266. doi: 10.1111/tbed.12639. [DOI] [PubMed] [Google Scholar]
- Stadejek et al. (2017).Stadejek T, Wozniak A, Milek D, Biernacka K. First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transboundary and Emerging Diseases. 2017;64(5):1350–1353. doi: 10.1111/tbed.12672. [DOI] [PubMed] [Google Scholar]
- Sukmak et al. (2019).Sukmak M, Thanantong N, Poolperm P, Boonsoongnern A, Ratanavanichrojn N, Jirawattanapong P, Woonwong Y, Soda N, Kaminsonsakul T, Phuttapatimok S, Wajjwalku W. The retrospective identification and molecular epidemiology of porcine circovirus type 3 (PCV3) in swine in Thailand from 2006 to 2017. Transboundary and Emerging Diseases. 2019;66(1):611–616. doi: 10.1111/tbed.13057. [DOI] [PubMed] [Google Scholar]
- Tischer et al. (1995).Tischer I, Bode L, Peters D, Pociuli S, Germann B. Distribution of antibodies to porcine circovirus in swine populations of different breeding farms. Archives of Virology. 1995;140(4):737–743. doi: 10.1007/BF01309961. [DOI] [PubMed] [Google Scholar]
- Tischer et al. (1986).Tischer I, Mields W, Wolff D, Vagt M, Griem W. Studies on epidemiology and pathogenicity of porcine circovirus. Archives of Virology. 1986;91(3–4):271–276. doi: 10.1007/BF01314286. [DOI] [PubMed] [Google Scholar]
- Wen et al. (2018).Wen S, Sun W, Li Z, Zhuang X, Zhao G, Xie C, Zheng M, Jing J, Xiao P, Wang M, Han J, Ren J, Liu H, Lu H, Jin N. The detection of porcine circovirus 3 in Guangxi. China Transboundary and Emerging Diseases. 2018;65(1):27–31. doi: 10.1111/tbed.12754. [DOI] [PubMed] [Google Scholar]
- Xu et al. (2018).Xu PL, Zhang Y, Zhao Y, Zheng HH, Han HY, Zhang HX, Chen HY, Yang MF, Zheng LL. Detection and phylogenetic analysis of porcine circovirus type 3 in central China. Transboundary and Emerging Diseases. 2018;65(5):1163–1169. doi: 10.1111/tbed.12920. [DOI] [PubMed] [Google Scholar]
- Ye et al. (2018).Ye X, Berg M, Fossum C, Wallgren P, Blomstrom AL. Detection and genetic characterisation of porcine circovirus 3 from pigs in Sweden. Virus Genes. 2018;54(3):466–469. doi: 10.1007/s11262-018-1553-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu et al. (2018).Yu QC, Liu Y, Du JL, Liu YY, Zhang LL, Guo T. Porcine circovirus type 1 was undetected in vaccine but could be cultured in the cell substrate of Lanzhou lamb rotavirus vaccine. Journal of General Virology. 2018;99(1):103–108. doi: 10.1099/jgv.0.000875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue et al. (2009).Yue F, Cui S, Zhang C, Yoon KJ. A multiplex PCR for rapid and simultaneous detection of porcine circovirus type 2, porcine parvovirus, porcine pseudorabies virus, and porcine reproductive and respiratory syndrome virus in clinical specimens. Virus Genes. 2009;38(3):392–397. doi: 10.1007/s11262-009-0333-6. [DOI] [PubMed] [Google Scholar]
- Zeng et al. (2014).Zeng ZY, Liu ZJ, Wang WC, Tang DY, Liang HY, Liu Z. Establishment and application of a multiplex PCR for rapid and simultaneous detection of six viruses in swine. Journal of Virological Methods. 2014;208:102–106. doi: 10.1016/j.jviromet.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Zheng et al. (2017).Zheng S, Wu X, Zhang L, Xin C, Liu Y, Shi J, Peng Z, Xu S, Fu F, Yu J, Sun W, Xu S, Li J, Wang J. The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province. Transboundary and Emerging Diseases. 2017;64(5):1337–1341. doi: 10.1111/tbed.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou et al. (2018).Zou Y, Zhang N, Zhang J, Zhang S, Jiang Y, Wang D, Tan Q, Yang Y, Wang N. Molecular detection and sequence analysis of porcine circovirus type 3 in sow sera from farms with prolonged histories of reproductive problems in Hunan. China Archives of Virology. 2018;163(10):2841–2847. doi: 10.1007/s00705-018-3914-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw data of full-length image of gel
Raw data of full-length image of gel
Raw data of full-length image of gel
Raw data of full-length image of gel
Raw data of full-length image of gel
Raw data of full-length image of gel
Data Availability Statement
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental Files. The DNA sequences are available as Supplemental Files and available at GenBank: MN790774–MN790776.