Abstract
Deep learning is a powerful tool for predicting transcription factor binding sites from DNA sequence. Despite their high predictive accuracy, there are no guarantees that a high-performing deep learning model will learn causal sequence-function relationships. Thus a move beyond performance comparisons on benchmark datasets is needed. Interpreting model predictions is a powerful approach to identify which features drive performance gains and ideally provide insight into the underlying biological mechanisms. Here we highlight timely advances in deep learning for genomics, with a focus on inferring transcription factors binding sites. We describe recent applications, model architectures, and advances in local and global model interpretability methods, then conclude with a discussion on future research directions.
Keywords: Deep learning, transcription factor binding, motifs, neural networks, interpretability
1. Introduction
Deep learning is a machine learning paradigm that is represented as a multi-layer, i.e. deep, neural network, composed of layers that enable hierarchical representations to be learned automatically from the data through training on one or more tasks. The popularity of deep learning in -omics applications has exploded in recent years [1]. One major reason for this rise is the democratization of deep learning code through high-level APIs, such as Pytorch [2] and Tensorflow [3], which make it possible to seamlessly build and train deep neural networks (DNNs) on graphical processing units in just a few lines of code. Another reason is the big data boom in genomics, enabled by high-throughput experiments and next generation sequencing [4]. Deep learning is thriving in this big data regime and its applications are extending to many areas in genomics [5, 6, 7, 8, 9, 10, 11]. Here, we highlight timely advances in applications for deep learning in genomics, with a focus on inferring transcription factors binding sites. We highlight recent applications and advances in model interpretability and then conclude with a discussion on future research directions.
Modeling sequence-function relationships with deep learning
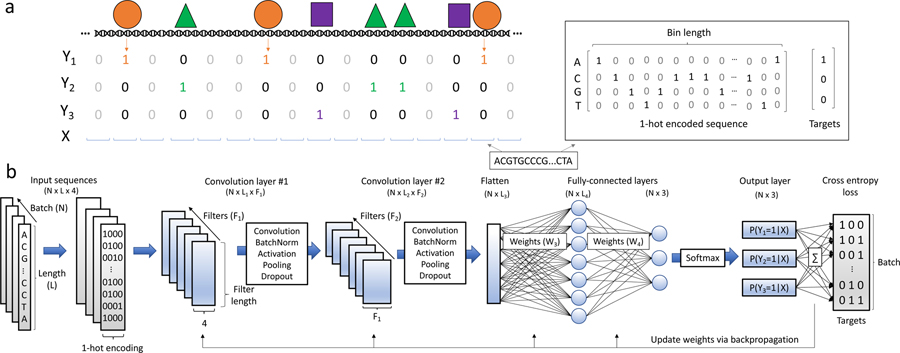
The computational task for inferring TF binding sites from DNA sequence is framed as a single-class or multi-class binary classification problem (for an overview, see Fig. 1a). The 2017 ENCODE-DREAM challenge exemplifies this task, as competitors were ranked on their ability to accurately predict in vivo TF binding on held out test cells and TFs (https://www.synapse.org/#!Synapse:syn6131484). The processed data consists of DNA sequences (as a one-hot representation) that are input to the model and corresponding binary labels (peak or no peak). Convolutional neural networks (CNNs) are particularly adept at modeling regulatory genomic sequences (see Fig. 1b for details of CNNs). A more detailed review of the computational task and CNNs can be found in Ref. [12]. The primary focus of the following sections will be in the context of CNNs, however many of the techniques described, (e.g. interpretation) are extendable to other classes of DNNs. Moreover, these methods extend naturally to other data modalities that describe sequence-function relationships, such as inferring chromatin accessibility sites and RNA-protein interaction sites.
Figure 1:
Overview of TF binding site prediction task. a) Transcription factors bind to regions of the genome based on sequence specificities and modulate various biological functions. ChIP-seq experiments enrich for short DNA sequences that are interacting with the TF under investigation. The resultant DNA sequences (so-called reads) are aligned to a reference genome and a peak calling tool is employed to find read distributions that are statistically significant compared to background levels. Upon binning the full genome into bins of length L, it is possible to then associate each bin with a binary label denoting the presence (Yi = 1) or absence (Yi = 0) of TF i based on sufficient overlap between the peaks and the bin. The DNA within each bin is represented by a 1-hot encoded matrix and the associated label vectors are used to train a model as a single-class or multi-class supervised learning task. b) Convolutional neural networks are powerful methods to learn sequence-function relationships directly from DNA sequence. A CNN is comprised of a number of first layer filters (F1) which learn features directly from the N input sequences by computing the cross-correlation between each set of filter weights and the 1-hot encoded sequence. The resultant scans, so-called feature maps, intuitively represent the match between each pattern being learned in a given filter and the input sequence. The feature map then undergoes a series of functional (e.g. batch normalization, non-linear activation) and spatial transformations (e.g. pooling) resulting in a truncated length (L1). This tensor is then fed into deeper convolutional layers which discriminate higher-order relationships between the learned features. Two convolutional blocks are depicted however this feed-forward process may be repeated any number of times, after which a flattening operation is utilized to reshape the tensor into a N × L3 matrix. Fully-connected layers perform additional matrix multiplications and ultimately output a probability of class membership for each target. Loss is calculated between the predicted values and the targets, and the weights are updated with a learning rule that uses backpropagation to calculate gradients throughout network.
1.1. Recent advances in DNN architectures
There have been many advances in DNN architectures over recent years, primarily driven by applications in computer vision and natural language processing (NLP), that have been slowly ported into genomics, including hybrid models, such as CNN-recurrent neural networks (RNNs) [13, 14, 15], dilated convolutions [16], residual connections [17], dense connections [18], and (self)attention [19].
Network modules.
Dilated convolutions are interesting because they provide a mechanism for considering a large sequence context, with receptive fields as large as 10kb without pooling [10, 20, 21]. Dilated convolutions can be combined with other network modules such as residual blocks [10, 21] or dense connections [20], both of which foster gradient flow to lower layers. Notably, dilated residual modules were a key component of Alphafold [22], the top protein folding method in the CASP13 free modeling competition.
Attention.
An interesting direction that is worth serious exploration is attention [23, 24, 25]. Attention provides an intrinsically interpretable mechanism to place focus on regions-of-interest in the inputs. Albeit, recent evidence suggests that attention is not strongly related to explainability [26]. There are many types of attention mechanisms. State-of-the-art language models in NLP employ a multi-head self-attention, also referred to as a scaled-dot-product attention, which are key components of transformer networks like BERT [27] and XLNet [28]. Recently, Ullah et al. demonstrated how self-attention can be employed to extract associations between TFs that reside in accessible chromatin sites [25].
1.2. Incorporating biophysical priors
The salient features in domains such as computer vision or NLP (where most deep learning progress is taking place) are different from genomics, particularly for TF binding, which consists of primary and alternative protein binding sites, cooperative and competitive binding factors, and sequence context (e.g. DNA shape features, GC-content, nucleosome positioning, accessibility and chromatin structure) [29]. In genomics, low-level sequence features, such as motifs, are of particular interest, whereas in images, higher-level features of objects are generally more important. In TF binding prediction tasks, incorporation of biophysical features may provide additional gains in performance. For instance, the top scoring teams [30, 31] in the ENCODE-DREAM challenge report increases in predictive performance through the inclusion of manually-crafted chromatin accessibility features (median gains on the area under the precision-recall curve of 0.252 and 0.0504, respectively). Thus an emerging trend is to design DNNs with biophysical priors, making them more suitable to model genomic features, including reverse-compliment equivariance and parameters that capture biophysical properties.
Reverse-compliment equivariance.
Reverse-compliment (RC) awareness can be achieved via data augmentation with RC sequences, incorporating separate inputs for RC sequences [15], and weight tying [32, 33, 34], which is more computationally efficient. These domain-motivated models yield improved predictive performance over standard DNNs, with reported gains on the area under the receiver-operating characteristic curve of around 0.02 [32]. Reverse-compliment pooling can further reduce the number of parameters [34], albeit introducing a strong prior of motif directional invariance. These strategies are particularly important when analyzing data generated via single-stranded sequencing. To enforce positional invariance of a motif within a filter, circular filters have been shown to be effective [35].
Biophysical parameters.
Recasting traditional physics-based models as a neural network is an active area of research [36, 37, 38]. Tareen & Kinney recently showed that biophysical models of TF binding can be represented as a neural network [37], where edges represent meaningful biophysical quantities, such as free energies. In parallel, [38] has also demonstrated how DNNs can be designed with strong biophysical priors. These networks are highly-constrained, but provide interpretable biophysical parameters. They offer starting points which can be embellished upon with machine learning tricks-of-the-trade using deep learning frameworks [2, 3].
Model interpretability is key to moving forward
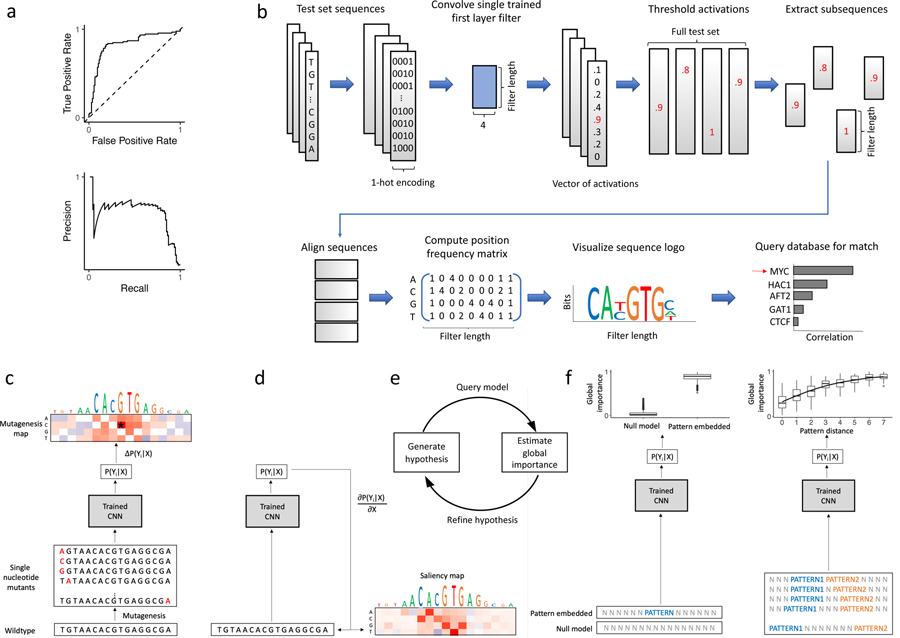
Biological experiments are noisy but often treated as ground truth for both training and testing. Improved predictions on unvalidated experimental benchmark datasets may not necessarily serve as a reliable way of comparing model performance (Fig. 2a). Interpreting models can therefore help to elucidate whether a DNN has learned new biology not captured by previous methods or has gained an advantage by learning correlated features that are indirectly related, such as technical biases of an experiment. Since binary classification tasks require discrimination of sequences between the positive and negative class, interpretability can also help to diagnose whether the DNN has learned poor features that directly result from a poor choice of negative sequences. In genomics, the main approaches to interpret a CNN are through visualizing convolutional filters [5, 7], attribution methods [39, 40, 41], and more recently in silico experiments [21, 42].
Figure 2:
Overview of model evaluation and interpretability. a) Model performance is assessed using the receiver-operating characteristic curve (top) or precision-recall curve (bottom). b) Visualizing CNN filters helps to understand learned representations. This can be achieved by scanning each filter across test set sequences, extracting subsequences (the length of the filter) centered on sufficiently large activations (above some threshold), aligning the subsequences, from which a position frequency matrix can be constructed and visualized as a sequence logo. Motif comparison search tools, such as Tomtom, can compare motif similarity against a database of previously-annotated motifs. c) In silico mutagenesis provides a single-nucleotide resolution map consisting of an importance score for each nucleotide variant at each position by calculating the difference in predicted values between a given wildtype sequence and new sequences with all possible single nucleotide variants. d) Gradient-based attribution methods analogously provide a single-nucleotide resolution map by calculating the derivative of the output (or logits) of a given class with respect to the inputs. e) A CNN can be used to generate and refine biological hypotheses by querying the model with a set of carefully chosen sequence models and estimating the global importance. f) Given a representative null background model (light gray N nucleotides) the global importance of a pattern (left panel) or spacing between patterns (right panel) may be estimated by querying the trained CNN with a sufficiently-large corpus of randomized, null sequences, each with an instance containing the feature as well as a matched instance without the feature. Such a method allows practitioners to quantitatively test a variety of biological hypotheses while controlling for unwanted confounders.
1.3. Filter visualization
First layer filters can be directly visualized as sequence logos via activation-based alignments (Fig. 2b). This representation makes it possible to compare filter representations against known databases of motifs, such as JASPAR [43], using Tomtom [44], a motif comparison search tool. Filter visualization has been a popular interpretability approach to support that a CNN has learned meaningful biology [5, 7, 11, 12, 13, 45, 46, 47]. There are many drawbacks to filter interpretation, including the challenge in quantifying the importance of the feature and how to relate the features to model prediction. Due to the complex dependencies with other filters within and across layers, off-the-shelf CNNs may not necessarily learn complete motif representations in first layer filters. Representations learned by CNNs are strongly influenced by many factors, including inductive biases provided by architectural constraints [48, 49], activation functions [50], and training procedure [51]. Hence, filter analysis should only be employed when a model is explicitly trained to learn interpretable motif representations. A more thorough discussion of the benefits and drawbacks to visualizing first layer filters can be found in [48, 49].
1.4. Attribution methods
In genomics, attribution methods – such as in silico mutagenesis [6, 7], saliency maps [39], integrated gradients [52], DeepLift [41], and DeepSHAP [53] – provide a single-nucleotide resolution map consisting of an importance score for each nucleotide variant at each position that are directly linked to predictions (Figs. 2, c–d). In practice, attribution methods have been utilized to validate that a model has learned representations that resemble known motifs in TF binding [20, 21], chromatin accessibility [5, 6, 7], RNA-protein interactions [54]. There are other interpretability methods that have been developed for genomics, including maximum entropy-based sampling [55] and occlusion experiments [21, 40], as well as many other methods that have not yet been thoroughly explored in genomics [40, 56, 57, 58].
Limitations.
Attribution methods are local interpretability methods that provide feature importance of individual nucleotides for a single sequence. Hence many attribution maps have to be observed on an individual basis to deduce what features the network has learned globally at a population-level. This can be challenging, because attribution methods tend to produce noisy representations with spurious importance scores for seemingly arbitrary nucleotides. TF-MoDISco aims to simplify this process by clustering attribution scores [59]. Even still, attribution methods are unable to quantify the effect that a whole putative motif (not just one nucleotide) has on model predictions. Ongoing research is exploring to what extent we can trust attribution methods [60, 61, 62, 63].
Second-order interactions.
The previously described attribution methods are first-order interpretability methods, revealing the independent contribution of single nucleotide variants in a sequence. There has been growing interest in uncovering interactions between two nucleotide positions, including second-order in silico mutagenesis [42], integrated Hessians [64], self-attention networks [25], filter visualization in deeper layers [47], and other gradient-based methods [11, 65, 66].
1.5. Global importance analysis
Global importance analysis (GIA) provides a framework to quantify the effect size of such putative motifs as well as the ability to map specific functions learned by a DNN [67]. GIA performs in silico experiments where synthetic sequences are designed with embedded hypothesis patterns while the other positions are randomized by sampling a null sequence model (Fig. 2f). By averaging the predictions of these synthetic sequences, GIA quantifies the average effect of the embedded patterns while marginalizing out the contributions of the other positions. Important to this approach is an appropriate null sequence model that minimizes distributional shift between the synthetic sequences and the experimental data. Prior knowledge is critical to determine the null model. For instance, Koo et al. employed GIA to find that the number of motifs, spacing between motifs, relative positions, and aspects of RNA secondary structure were significant learned features in their DNN [42]. More recently, Avsec et al. employed GIA to understand motif syntax, including cooperative associations and positional periodicity [21]. We envision GIA will play a critical role in testing hypotheses of what DNNs have learned, moving beyond speculation from observing putative features in attribution maps and individual filters.
Conclusion
The timely advances in deep learning and genomics have made research at this intersection progress at a rapid pace. Improvements to architecture and interpretability have been key to the synergy. Yet there are many pressing avenues that are beginning to emerge, including end-to-end models, generative modeling, causal inference, variant effect prediction, and robustness properties.
End-to-end models.
Framing TF binding as a binary classification task is limiting, because peak calling is noisy and the read distributions themselves can be informative of the underlying biological signals. Recent applications have by-passed the peak calling preprocessing step altogether, directly predicting read distributions from sequence [20, 21]. This allows the DNN to learn how to discriminate peaks. Interpreting these so-called end-to-end DNNs may help to isolate biological signals from experimental noise.
Generative modeling.
In contrast to supervised representation learning, which are informed only through the task they are trained on, unsupervised representations learned with deep generative models, such as generative adversarial networks [68] and variational autoencoders [69], can reveal latent structure of the data on a low dimensional manifold. Deep generative models are an active research area in protein sequence modeling [70, 71] but is largely lagging for regulatory genomic sequences. Applications for proteins demonstrate that deep generative models could potentially help to study evolution of sequences across phylogenies [72] and design new sequences with desired properties [73].
Causal inference.
A fundamental assumption in the field of causal inference is ignorability, for which domain-knowledge is employed to build structural causal graphs which capture relevant data dependencies and explicitly formulate model assumptions to ensure there are no unmeasured confounders. On the other hand, highly-parameterized DNNs which estimate complex functions from rich functional classes run counter to such explicit formulations. A hallmark technique to ensure ignorability is the randomized control trial (RCT). Experiments performed in regulatory genomics, such as massively parallel reporter assays [4], are by design RCTs given a sufficiently large library. While costly, such experiments provide valuable insight into the underlying causal mechanism dictating sequence-function relationships. An alternative to physically performing these experiments is to simulate them in silico, namely by performing global importance analysis. To do so, however, requires robust models which accurately learn the functional relationships under consideration. We therefore prioritize the collaboration between bench scientists and computational scientists such that hypotheses generated in silico may be validated in vivo and a feedback loop may be utilized to develop better models (Fig. 2e). DNNs that accurately model the true causal effects are more robust to distribution shifts and improve generalizability [74]. The same may be said when integrating multiple data modalities. For instance, adjusting for confounders such as chromatin accessibility is critical for learning a generalizable function across cell types. Subsequent improved design of models will reduce costs associated with experimental validation, accelerate hypothesis generation and refinement, and provide more accurate discovery of causal biological mechanisms.
Robustness and interpretability.
By learning sequence-function relationships, a trained DNN can be used to score the effect that disease-associated variants have on the phenotype that it was trained on [5, 7, 9, 10, 11, 20, 75]. This of course assumes that the model has learned an invariant causal representation which is generalizeable beyond the data that it was trained on. Demonstration of out-of-distribution generalization performance has been limiting due to a lack of reliable benchmark datasets with ground truth. In other domains, it has been shown that small, targeted perturbations to the inputs, so-called adversarial examples [76], generated by an adversary whose sole mission is to trick the classifier, can result in highly unreliable predictions. This has resurrected the field of robust machine learning which focuses on the trustworthiness of model predictions [77]. Counterintuitively, high performing DNNs do not necessarily yield reliable attribution scores [78, 79], even in genomics [63]. This raises a red flag that we should not blindly trust model predictions on variant effects just because they generalize well on held-out test data generated from the same distribution, which share the same biases. It has been demonstrated that adversarial training, which incorporates adversarial examples during training, not only leads to improved robustness properties but also improved interpretability [51, 63]. Although adversarial examples is not a meaningful phenomenon in genomics, their potential for improving the robustness and interpretability properties of DNNs through adversarial training makes them an exciting area of exploration. A thorough evaluation and understanding of how training procedure, incorporation of biophysical priors, and the various advances in DNN architectures all influence model robustness and interpretability is an avenue for future research.
Beyond validation – discovering new biology.
Deep learning offers a new paradigm for data analysis in genomics. As powerful function approximators, DNNs can be employed to challenge our underlying assumptions made by traditional (non-deep learning) models. To make meaningful contributions, however, we need to move beyond performance comparisons on benchmark datasets. Through model interpretation, we can identify what novel features drive performance gains. In practice, we believe that a combination of interpretability methods – such as first-order and second-order attribution methods and filter visualization – can collectively help to generate hypotheses of putative features and their syntax. This strategy should compensate for the failures of any individual approach. As a follow up, global importance analysis can be employed to quantify the effect size of putative features and also tease out specific functional relationships of the features, including positional dependence, sequence context, and higher-order interactions. We recommend training various DNNs – ranging from models designed to be highly expressive to models designed to learn interpretable representations – to identify features that are robust across models and initializations. Averaging an ensemble of models is a powerful approach to improve performance and it can also be extended to improve interpretability. Interpreting model predictions is a powerful approach to suggest biological insights and generate hypotheses. The patterns they learn are not proof of biological mechanisms, so any new insights should be followed with experimental validation.
Acknowledgements
This work was supported in part by funding from the NCI Cancer Center Support Grant (CA045508) and the Simons Center for Quantitative Biology at Cold Spring Harbor Laboratory. MP was supported by NIH NCI RFA-CA-19-002. The authors would like to thank Ammar Tareen and Justin Kinney for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
Nothing declared.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Peter K. Koo, Simons Center for Quantitative Biology, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA
Matt Ploenzke, Department of Biostatistics, Harvard University, Cambridge, MA, USA.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- [1].Eraslan G, Avsec Z, Gagneur J, Theis F: Deep learning: new computational modelling techniques for genomics. Nature Reviews Genetics 2019, 20(7):389–403. [DOI] [PubMed] [Google Scholar]
- [2].Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, et al. : Pytorch: An imperative style, high-performance deep learning library, in: Advances in Neural Information Processing Systems 32, Curran Associates, Inc, 2019, pp. 8024–8035. URL http://www.https://pytorch.org/ [Google Scholar]
- [3].Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M, et al. : 2016. Tensorflow: Large-scale machine learning on heterogeneous distributed systems, arXiv 2016, 1603.04467.
- [4].Kinney JB, McCandlish DM: Massively parallel assays and quantitative sequencefunction relationships. Annual Review of Genomics and Human Genetics 2019, 20:99–127. [DOI] [PubMed] [Google Scholar]
- [5].Alipanahi B, Delong A, Weirauch MT, Frey BJ: Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nature Biotechnology 2015, 33(8):831–838. [DOI] [PubMed] [Google Scholar]
- [6].Zhou J, Troyanskaya OG: Predicting effects of noncoding variants with deep learning-based sequence model. Nature Methods 2015, 12(10):931–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kelley DR, Snoek J, Rinn JL: Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Research 2016, 26(7):990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tunney R, McGlincy NJ, Graham ME, Naddaf N, Pachter L, Lareau LF: Accurate design of translational output by a neural network model of ribosome distribution. Nature Structural & Molecular Biology 2018, 25(7):577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhou J, Theesfeld CL, Yao K, Chen KM, Wong AK, Troyanskaya OG: Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk. Nature Genetics 2018, 50(8):1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jaganathan K, Panagiotopoulou S, McRae S, and Darbandi JF, Knowles D, Li Y, Kosmicki J, Arbelaez J, Cui W, Schwartz G, Chow E: Predicting splicing from primary sequence with deep learning. Cell 2019, 176(3):535–548.** Study that trains a DNN, called SpliceAI, to predict splice sites and then investigates features that it has learned. Impressively, it is able to predict noncoding cryptic splice mutations in patients with rare genetic diseases and is followed up with experimental validation.
- [11].Bogard N, Linder J, Rosenberg AB, Seelig G: A deep neural network for predicting and engineering alternative polyadenylation. Cell 2019, 178(1):91–106.** A DNN, called APARENT, is trained to predict alternative polyadenlyation sites. They interpret their network by visualizing filters and gradient-based methods, and then use APARENT to investigate human variants.
- [12].Angermueller C, Pärnamaa T, Parts L, Stegle O: Deep learning for computational biology. Molecular Systems Biology 2016, 12(7):878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Quang D, Xie X: Danq: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Research 2016, 44(11):107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shen WB, Zhen D-S Huang: Recurrent neural network for predicting transcription factor binding sites. Scientific Reports 2018, 8(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Quang D, Xie X: Factornet: a deep learning framework for predicting cell type specific transcription factor binding from nucleotide-resolution sequential data. Methods 2019, 166:40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yu F, Koltun V: Multi-scale context aggregation by dilated convolutions. arXiv 2015, 1511.07122.
- [17].He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2016, 770–778. [Google Scholar]
- [18].Huang G, Liu Z, Van Der Maaten L, Weinberger KQ: Densely connected convolutional networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2017, 4700–4708. [Google Scholar]
- [19].Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser L, Polosukhin I: Attention is all you need. Advances in Neural Information Processing Systems 2017, 5998–6008.
- [20].Kelley DR, Reshef YA, Bileschi M, Belanger D, McLean CY, Snoek J: Sequential regulatory activity prediction across chromosomes with convolutional neural networks. Genome Research 28(5):739–750.** End-to-end DNN, called Bassenji, trained to predict epigenetic profiles, followed by model interpretation using filter visualization and attribuiton methods, and an investigation of variant effect predictions for eQTLs and disease-associated loci.
- [21].Avsec Z, Weilert M, Shrikumar A, Alexandari A, Krueger S, Dalal K, , Fropf R, McAnany C, Gagneur J, Kundaje A, Zeitlinger J: Deep learning at base-resolution reveals motif syntax of the cis-regulatory code. bioRxiv 2019, 737981.** End-to-end CNN, called BPNet, to predict chip-nexus profiles of pluripotency TFs. They thoroughly interpret their model using DeepLIFT, TF-MoDISco and in silico experiments.
- [22].Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Žídek A, Nelson AW, Bridgland A, et al. : Improved protein structure prediction using potentials from deep learning. Nature 2020, 577:706–710. [DOI] [PubMed] [Google Scholar]
- [23].Singh R, Lanchantin J, Sekhon A, Qi Y: Attend and predict: Understanding gene regulation by selective attention on chromatin. Advances in Neural Information Processing Systems 2017, 6785–6795. [PMC free article] [PubMed]
- [24].Chen C, Hou J, Shi X, Yang H, Birchler JA, Cheng J: Interpretable attention model in transcription factor binding site prediction with deep neural networks. bioRxiv 2019, 648691. [DOI] [PMC free article] [PubMed]
- [25].Ullah F, Ben-Hur A: A self-attention model for inferring cooperativity between regulatory features. bioRxiv 2020, 927996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jain S, Wallace B: Attention is not explanation. arXiv 2019, 1902.10186.
- [27].Devlin J, Chang M-W, Lee K, Toutanova K: BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, 1810.04805.
- [28].Yang Z, Dai Z, Yang Y, Carbonell J, Salakhutdinov RR, Le QV: XLNet: Generalized autoregressive pretraining for language understanding. Advances in Neural Information Processing Systems 2019, 5754–5764.
- [29].Inukai S, Kock KH, Bulyk ML: Transcription factorDNA binding: beyond binding site motifs. Current Opinion in Genetics & Development 2017, 43:110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Keilwagen J, Posch S, Grau J: Accurate prediction of cell type-specific transcription factor binding. Genome Biology 2019, 20(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li H, Quang D, Guan Y: Anchor: trans-cell type prediction of transcription factor binding sites. Genome Research 2019, 29(2):281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shrikumar A, Greenside P, Kundaje A: Reverse-complement parameter sharing improves deep learning models for genomics. bioRxiv 2017, 103663.
- [33].Bartoszewicz JM, Seidel A, Rentzsch R, Renard BY: DeePaC: predicting pathogenic potential of novel DNA with reverse-complement neural networks. Bioinformatics 2020, 36(1):81–89. [DOI] [PubMed] [Google Scholar]
- [34].Brown RC, Lunter G: An equivariant bayesian convolutional network predicts recombination hotspots and accurately resolves binding motifs. Bioinformatics 2019, 35(13):2177–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Blum C, Kollmann M: Neural networks with circular filters enable data efficient inference of sequence motifs. Bioinformatics 2019, 35(20):3937–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dauparas J, Wang H, Swartz A, Koo P, Nitzan M, Ovchinnikov S: Unified framework for modeling multivariate distributions in biological sequences. arXiv 2019, 1906.02598.
- [37].Tareen A, Kinney JB: Biophysical models of cis-regulation as interpretable neural networks. arXiv 2019, 2001.03560.
- [38].Liu KB, Yi J Reinitz: Fully interpretable deep learning model of transcriptional control. bioRxiv 2019, 655639. [DOI] [PMC free article] [PubMed]
- [39].Simonyan K, Vedaldi A, Zisserman A: Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv 2013, 1312.6034.
- [40].Zeiler MD, Fergus R: Visualizing and understanding convolutional networks. European Conference on Computer Vision 2014, 818–833.
- [41].Shrikumar A, Greenside P, Kundaje A: Learning important features through propagating activation differences. Proceedings of the 34th International Conference on Machine Learning 2017, 3145–3153. [Google Scholar]
- [42].Koo P, Anand P, Paul S, Eddy S: Inferring sequence-structure preferences of RNA-binding proteins with convolutional residual networks. bioRxiv 2018, 418459.** A residual CNN, called ResidualBind, is trained to predict RNA-protein interactions. A thorough interpretation of the model using attribution methods and in silico experiments shows that the model learns to count the number of motifs, consider their spatial positions, and learns secondary structure context.
- [43].Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, B. P. M. et al. : JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Research 2020, 48(D1):D87–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS: Quantifying similarity between motifs. Genome Biology 2007, 8(2):R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cuperus J, Groves B, Kuchina A, Rosenberg A, Jojic N, Fields S, Seelig G: Deep learning of the regulatory grammar of yeast 5 untranslated regions from 500,000 random sequences. Genome Research 2017, 27(12):2015–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hoffman G, Bendl J, Girdhar K, Schadt E, Roussos P: Functional interpretation of genetic variants using deep learning predicts impact on chromatin accessibility and histone modification, Nucleic Acids Research 2019, 47(20):10597–10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Maslova A, Ramirez R, Ma K, Schmutz H, Wang C, Fox C, Ng B, Benoist C, Mostafavi S: Learning immune cell differentiation. bioRxiv 2019, 885814.* A CNN model, called AI-TAC, trained on chromatin accessible sites in immune cells, followed by a thorough investigation into the filter representations at higher layers to identify recurring associations between motifs.
- [48].Koo PK, Eddy SR: Representation learning of genomic sequence motifs with convolutional neural networks. bioRxiv 2018, 362756.** Investigation of how CNN design choice influences the extent that motif representations are learned by convolutional filters; it shows how to design them such that they learn more interpretable representations of motifs in first layer filters.
- [49].Ploenzke M, Irizarry R: Interpretable convolution methods for learning genomic sequence motifs. bioRxiv 2018, 411934.
- [50].Koo P, Ploenzke M: Improving convolutional network interpretability with exponential activations. bioRxiv 2019, 650804.
- [51].Ilyas A, Santurkar S, Tsipras D, Engstrom L, Tran B, Madry A: Adversarial examples are not bugs, they are features. Advances in Neural Information Processing Systems, 2019, 125–136.
- [52].Sundararajan M, Taly A, Yan Q: Axiomatic attribution for deep networks. Proceedings of the 34th International Conference on Machine Learning 2017, 3319–3328. [Google Scholar]
- [53].Lundberg S, Lee S: A unified approach to interpreting model predictions. Advances in Neural Information Processing Systems 2017, 4765–4774.
- [54].Ghanbari M, Ohler U: Deep neural networks for interpreting RNA-binding protein target preferences. Genome Research 2020, 30(2):214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Finnegan A, Song J: Maximum entropy methods for extracting the learned features of deep neural networks. PLoS Computational Biology 2017, 13(10):e1005836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ribeiro MT, Singh S, Guestrin C: Why should I trust you?: Explaining the predictions of any classifier. Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining 2016, 1135–1144. [Google Scholar]
- [57].Selvaraju R, Cogswell M, Das A, Vedantam R, Parikh D, Batra D: Grad-cam: Visual explanations from deep networks via gradient-based localization. Proceedings of the IEEE International Conference on Computer Vision 2017, 618–626. [Google Scholar]
- [58].Erion G, Janizek JD, Sturmfels P, Lundberg S, Lee S-I: Learning explainable models using attribution priors. arXiv 2019, 1906.10670.
- [59].Shrikumar A, Tian K, Shcherbina A, Avsec Z, Banerjee A, Sharmin M, Nair S, Kundaje A: Tf-modisco v0. 4.4. 2-alpha. arXiv 2018, 1811.00416.
- [60].Adebayo J, Gilmer J, Muelly M, Goodfellow I, Hardt M, Kim B: Sanity checks for saliency maps. Advances in Neural Information Processing Systems 2018, 9505–9515.
- [61].Adebayo J, Gilmer J, Goodfellow I, Kim B: Local explanation methods for deep neural networks lack sensitivity to parameter values. arXiv 2018, 1810.03307.
- [62].Sixt L, Granz M, Landgraf T: When explanations lie: Why modified bp attribution fails. arXiv 2019, 1912.09818.
- [63].Koo P, Qian S, Kaplun G, Volf V, Kalimeris D: Robust neural networks are more interpretable for genomics. bioRxiv 2019, 657437.
- [64].Janizek SP, J.D, Lee S: Explaining explanations: Axiomatic feature interactions for deep networks. arXiv 2020, 2002.04138.
- [65].Greenside P, Shimko T, Fordyce P, Kundaje A: Discovering epistatic feature interactions from neural network models of regulatory DNA sequences. Bioinformatics 2018, 34(117):i629–i637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Liu ZH, G, Gifford D: Visualizing complex feature interactions and feature sharing in genomic deep neural networks. BMC Bioinformatics 2019, 20(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Koo P, Ploenzke M: Interpreting Deep Neural Networks Beyond Attribution Methods: Quantifying Global Importance of Genomic Features. bioRxiv 2020, 956896.* An overview of local and global interpretability methods, including a detailed explanation of global importance analysis and its applications, followed by its relation to causality.
- [68].Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y: Generative adversarial nets. Advances in neural information processing systems 2014, 2672–2680.
- [69].Kingma DP, Welling M: Auto-encoding variational bayes. arXiv 2013, 1312.6114.
- [70].Rives A, Goyal S, Meier J, Guo D, Ott M, Zitnick CL, Ma J, Fergus R: Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. bioRxiv 2019, 622803. [DOI] [PMC free article] [PubMed]
- [71].Rao R, Bhattacharya N, Thomas N, Duan Y, Chen P, Canny J, Abbeel P, Song Y: Evaluating protein transfer learning with tape. Advances in Neural Information Processing Systems 2019, 9686–9698. [PMC free article] [PubMed]
- [72].Ding X, Zou Z, Brooks CL III: Deciphering protein evolution and fitness landscapes with latent space models. Nature Communications 2019, 10(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Ingraham J, Garg V, Barzilay R, Jaakkola T: Generative models for graph-based protein design. Advances in Neural Information Processing Systems 2019, 15794–15805.
- [74].Schlkopf B: Causality for machine learning. arXiv 2019, 1911.10500.
- [75].Zhou J, Park C, Theesfeld C, Wong A, Yuan Y, Scheckel C, Fak J, Funk J, Yao K, Tajima Y, Packer A: Whole-genome deep-learning analysis identifies contribution of noncoding mutations to autism risk. Nature Genetics 2019, 51(6):973.* Demonstration of how deep learning can be used to score the functional effects of noncoding mutations, for this particular case, patients with autism.
- [76].Szegedy C, Zaremba W, Sutskever I, Bruna J, Erhan D, Goodfellow I, Fergus R: Intriguing properties of neural networks. arXiv 2013, 1312.6199.
- [77].Biggio B, Roli F: Wild patterns: Ten years after the rise of adversarial machine learning. Pattern Recognition 2018, 84:317–331. [Google Scholar]
- [78].Tsipras D, Santurkar S, Engstrom L, Turner A, Madry A: Robustness may be at odds with accuracy. arXiv 2018, 1805.12152.
- [79].Alvarez-Melis D, Jaakkola TS: On the robustness of interpretability methods. arXiv 2018, 1806.08049.