Abstract
Purpose of review
Molecular pathways in colorectal carcinogenesis involve several complex genetic and epigenetic modulations that cause normal colonic mucosa to metamorphose into a benign polyp and subsequently into a malignant tumor. Our purpose is to recapitulate historical and recent genomic research in order to augment the understanding of colorectal cancer pathogenesis.
Recent Findings
In 2015, the molecular classification for colorectal cancers was unified into one system with four distinct groups, also called as consensus molecular subtypes. This led to an enhanced understanding of molecular and immune signatures which has implications on predicting the clinical behavior as well as response to different therapeutic agents.
Summary
In this review, we expound on the current literature as well as draw on our own experience to present the important molecular pathogenesis pathways, key genetic mutations, differences in pathogenesis of left versus right sided tumors as well as the molecular classification of colorectal cancers.
Keywords: Molecular Pathogenesis, Colorectal Carcinoma, Molecular Subtypes
Introduction
Colorectal cancer (CRC) is the third most frequently diagnosed cancer in the US, estimated to cause 53,200 deaths and 147,950 new cases in 2020. [1]. Sporadic colorectal cancer accounts for nearly 70% of the cases. Only 5% of the new colorectal cancer diagnosis are related to hereditary conditions such as the Lynch syndrome or familial adenomatous polyposis (FAP). 20–30% of the cases have a familial disposition with no associated or known germline mutation [2].
Over the last few decades, our comprehension of the diverse genomic events in the pathogenesis of invasive colorectal cancer has improved significantly. Herein, we will describe the pathogenesis of colorectal cancer while touching upon the role of immune and stromal components in tumorigenesis as well as discuss the recently proposed molecular classification based on transcriptomics.
Molecular Pathogenesis
Adenoma-carcinoma sequence
An accruing body of evidence continues to propound that most CRC arise from pre-cancerous lesions or adenomatous polyps. Pathogenesis of CRC is multi-phasic, starting from the earliest dysplastic lesion called aberrant crypt focus to adenomatous polyp to invasive cancer. On the molecular level, Vogelstein and colleagues proposed that development of carcinogenesis depends on progressive accumulation of changes beneficial to tumor growth over time leading eventually to an invasive malignancy. This is called the adenoma-carcinoma sequence [3]. APC gene truncation is usually the inciting event, followed by KRAS and TP53 mutations later in the sequence.
Serrated polyp pathway
This represents an alternative pathway to the evolution of colorectal cancer and phenotypically present as heterogenous outgrowths such as hyperplastic polyps, sessile serrated adenomas or mixed hyperplastic polyps/serrated adenoma. [4] BRAF mutations are the most frequent initial insult compounded by epigenetic CpG island methylator phenotype, which is reviewed in detail below. Interestingly, dysbiosis in gut microbiome, especially overgrowth of Fusobacterium nucleatum has been implicated in progression of serrated polyp to adenocarcinoma [5,6]. Prognosis is variable, and a combination of high degree of CIMP (CIMP-H), microsatellite stability (MSS) and BRAF mutation carries the worst prognosis [7].
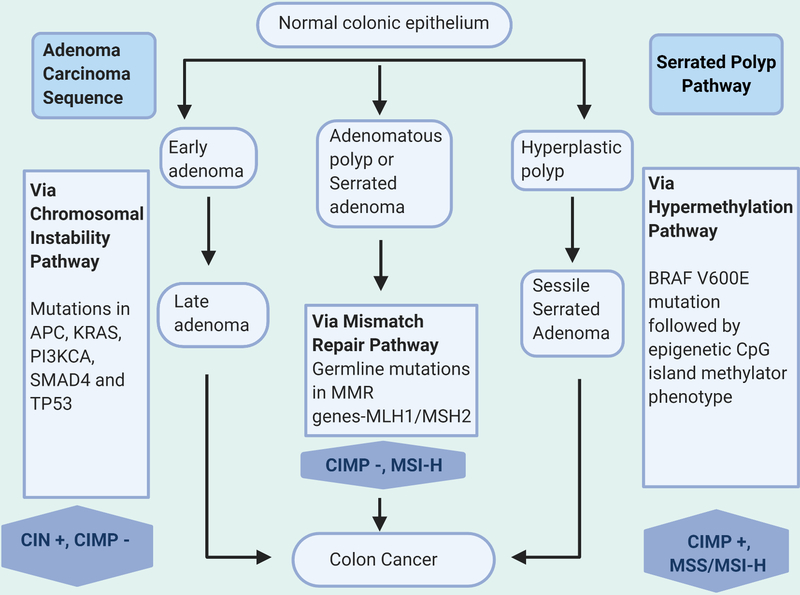
The above sequences are actualized by three major pathways, namely the CIN or chromosomal instability pathway, the MSI or microsatellite instability pathway and CIMP (CpG island methylator phenotype) hypermethylation which are discussed below and outlined in Figure 1. At a molecular level, the adenoma-carcinoma sequence pathway is enriched with KRAS mutations and somatic copy number alterations via CIN whereas BRAF mutations followed by gene promoter hypermethylation drive the serrated polyp pathway
Figure 1.
Key molecular pathways in development of colorectal cancer
Chromosomal instability pathway/APC pathway
Mutations in colon cancer can be traced to two modes of genomic instability: chromosomal instability (CIN) and microsatellite instability. Chromosomal instability can be observed in about 70% of CRC cases [8]. According to the classical adenoma to carcinoma model, mutation in Apc gene instigates progression to carcinoma via chromosomal instability, which involves various numerical chromosomal aberrations, most commonly chromosome 18, sub chromosomal aberrations, and loss of heterozygosity [9]. One significant feature of CIN is that it causes the deletion of tumor suppressor genes. Key genes involved in CIN include APC, TP53, KRAS, PI3KCA, etc [8]. Colon cancers emanating from CIN have worse outcomes than those with microsatellite instability [10].
Mismatch Repair (MMR)
DNA replication is a high-fidelity process and over the course of evolution, several mechanisms have developed to avoid errors in this process. One of these is the mismatch repair (MMR) mechanism, which was initially discovered in prokaryotes [11]. In humans, 9 different homologues have been described [12]. Dysfunction of these mismatch repair proteins as a result of inheritance of one germline mutated allele forms the basis for Lynch syndrome. The other allele may be inactivated through loss of heterozygosity (LOH), epigenetic silencing or mutation. Regions of short segments of DNA repeated hundreds of thousands of time in the genome are called microsatellites. Cells with MMR deficiency are likely to accumulate DNA errors by many folds. A deficient mismatch protein system leads to expansion or contraction of these microsatellites, thus called microsatellite instability [13].
Patients with Lynch syndrome have an onset of invasive cancer at an earlier age, more likely to have proximal site or right sided colon cancer, poorly differentiated histology, mucinous subtype, marked lymphocytic infiltrate but surprisingly have a better prognosis in stage II disease as compared to mismatch proficient colon cancers [13].
Hypermethylation BRAF pathway
A subgroup of sporadic colorectal tumors is typified by microsatellite instability and V600 mutant BRAF. The pathogenetic hallmark is epigenetic silencing of mismatch repair proteins mediated via CpG sequence hypermethylation. This hypermethylation process specifically occurs in the promoter region of genes that code for mismatch repair proteins, resulting in mismatch repair enzyme deficiency. These tumors are termed as CIMP+ (CpG island methylator phenotype) tumors [14]. Activating BRAF mutations including BRAF V600 is an exclusive feature of these tumors [15]. This mutation is also more commonly associated with the proximal or right colon cancers but portends a worse prognosis. Presence of CIMP has been studied as a biomarker of response to fluoropyrimidine containing cytotoxic regimens such as FOLFOX or FOLFIRI, however, its role remains debatable [16].
Molecular Abnormalities
Oncogenes
Gain of function mutations in oncogenes leads to constitutive activation and uncontrolled growth leading to cancer. Several oncogenes have been incriminated in colorectal tumorigenesis. Some of the more important ones are discussed below.
1. RAS
RAS proteins encode for small GTP hydrolases that function as a growth switch which is tightly regulated. In the active form, this pathway triggers stimulation of distal mechanisms including the MAPK and PI3K pathways [17]. Three cellular variants are known to exist, of which, KRAS is the most frequent to undergo mutation in colon cancer. Point mutations in KRAS gene result in constitutive stimulation of Ras protein and uncontrolled growth [18]. This is postulated to be a pilot event in CRC pathogenesis. Mutated KRAS gene results in constant activation of the RAS-RAF-MEK-ERK pathway and this has shown to confer resistance to anti epidermal growth factor receptor therapies such as cetuximab or panitumumab [19].
2. BRAF
The RAF proteins belong to the family of serine-threonine kinases located just downstream to RAS, which are responsible for stimulating the RAF/MEK/MAPK sequence leading to cellular growth and proliferation [20]. Mutant BRAF has been described in roughly 10% of sporadic CRC, more commonly on the right side [21]. V600E is the most common mutation encountered in clinical practice [22]. BRAF mutation can confer resistance to EGFR directed therapy and usually portends worse survival outcomes [23].
3. HER2
The HER2 transmembrane glycoprotein receptor is classified within the EGFR receptor cohort, with inherent tyrosine kinase functionality. HER2 protein is responsible for the activation of downstream pathways controlling epithelial cell growth [24]. About 5% of metastatic colorectal cancer patients have HER2 gene amplification, short variant modulations, or both[25]. Cancer cells utilize the HER2 pathway as a bypass signal transduction pathway conferring them immunity against anti-EGFR therapies. This resistance mechanism is especially acquired in wild-type RAS and BRAF cancers which would otherwise be a sensitive target for anti-EGFR antibodies [26]. HER-2 is emerging as a therapeutic target with trastuzumab-lapatinib and trastuzumab-pertuzumab combination therapies showing promising results in early clinical trials [27,28].
Tumor suppressor genes
1. Apc gene
According to the adenoma-carcinoma sequence model, as mentioned above, CRC origin is initiated by mutations in the Apc gene. Mutant Apc gene occurs in about 80–90% of CRC. Germline mutations in one allele followed by loss of heterozygosity in Apc gene is the fundamental mechanism in development of familial adenomatous polyposis (FAP), a condition that inevitably causes CRC at an early age, usually under 35 [29]. Apc gene controls several key cellular functions such as differentiation, cellular migration, cell-cell adhesion, cytoskeletal integrity and genomic stability [30]. The significance of Apc gene can be adjudged by the fact that restoring the Apc gene has been shown to cause rapid cell differentiation and tumor regression in mouse models [31].
In the absence of a functional Apc gene, β-catenin accumulates via the Wnt signaling pathway [32]. β-catenin along with T-cell lymphocyte enhancer factor activates central pro-oncogenic cell-cycle controller genes such as c-Myc and cyclin D1 via transcription [33]. Therapeutic agents aimed at disrupting this interaction as well as other steps in the canonical Wnt signaling pathway are currently under investigation [32,34].
2. TP53 gene
TP53 is also known as the “gatekeeper of the genome” and is a pivotal mutation in a multitude of cancers including colon cancer. Its protein product (tumor protein 53) recognizes any form of injury to the DNA and either halts the cell cycle to allow for DNA repair or in case of irreparable damage, it can initiate the process of cellular suicide or apoptosis. Nearly 50% of invasive colon cancers harbor this mutation, and it is often seen in cancers growing in the rectum or distal colon [35]. Role of TP53 in CRC pathogenesis is dichotomous and may involve “loss of function” of TP53 gene resulting from inactivating mutations in one allele and loss of other allele as a result of chromosomal instability or acquisition of missense-type mutations in the DNA binding domain which convert TP53 into a pro-oncogene [36]. This event is theorized to occur later in the adenoma - carcinoma transformation sequence, due to a relatively low rate of presence in precancerous lesions, and a higher presence in cases of invasive cancer [37]. Only exception is colitis associated carcinoma where loss of TP53 is an earlier step in the pathogenesis [38].
3. PTEN
Mutations in the Phosphatase and Tensin homolog (PTEN) gene are linked with CRC in the young (<50 years). Decreased PTEN expression causes Cowden Syndrome, also associated with thyroid and breast cancers. PTEN protein directly inhibits the PI3K/Atk signaling pathway by dephosphorylating a key second messenger and thereby stifles cell cycle progression and induced cellular apoptosis. Promoter hypermethylation stemming from genomic instability is a common reason for PTEN inactivation, which explains the correlation between MSI and PTEN loss [39]. Likewise, PTEN mutant CRCs are seen to arise more commonly in proximal and right colon. Other reasons for PTEN loss in CRC include LOH and somatic mutations. The risk of CRC associated with PTEN is around 9–16% [40]. PTEN inactivated tumors are also known to carry concomitant BRAF and KRAS mutations, whereas TP53 and PTEN mutation do not occur simultaneously [41,42].
While multiple reports point towards a positive association between PTEN loss and decreased life expectancy, local recurrence, lymphatic invasion, liver metastasis and advanced TNM staging, many others failed to show such an association [43–45]. In breast cancer, absence of PTEN expression is associated with a lack of response to trastuzumab, however studies analyzing the contribution of PTEN as a predictive biomarker for EGFR directed therapies have yielded discordant results so far [46–50].
4. Transforming Growth Factor β (TGFβ) signaling
TGFβ is a signaling molecule that regulates inhibition of cellular proliferation and induction of apoptosis. It mediates its effects by binding to type I and II TGFβ receptors leading to the activation of several downstream targets including the SMAD proteins [51]. In one study, 75% of the colon cancer cell lines had defective TGFβ mediated signaling caused by mutations in the type II receptor or the downstream molecules [52]. Although, TGFβ has a negative stimulatory effect, there is a paradoxical increased expression of TGF-beta ligand by tumor cells in the mesenchymal molecular subtype [53]. Rise in TGFβ levels may have a pathogenic role in increasing angiogenesis and inducing immunosuppression, leading to worse prognosis in this subtype [54]. TBGβ mutations are more commonly associated with MSI-H tumors.
5. Chromosome 18q: the DCC, SMAD4, and SMAD2 genes
Deleted in colon cancer or DCC is a tumor suppressor gene housed in the long arm of chromosome 18 and is frequently deleted in cases of sporadic colon cancer [55]. Unlike other tumor suppressor genes, DCC only acts conditionally. It is a dependence receptor, which leads to cellular proliferation when bound to netrin-1 whereas in the unbound state, it induces cell death. Role of DCC in prognostication and as a therapeutic target is currently unclear.
- SMAD4
SMAD 4 [Small Mothers Against Decapentaplegic homolog 4], or DPC4 [Deleted in Pancreatic Cancer-4] is another tumor suppressor gene located on 18q [56]. SMAD is a cohort of transcription factor proteins instrumental to the TGF-Beta signaling pathway. Mutation in SMAD4 can release the cancer cell from the growth suppressive effect of TGF-B [57] (13). Inherited mutation in SMAD4 is responsible for Juvenile polyposis syndrome.
- SMAD2
SMAD 2 is a messenger in the TGF-B pathway, which upon binding of TGF-B to its receptor gets phosphorylated and subsequently binds to SMAD4. SMAD4 then translocates into the nucleus and is responsible for transcribing pro-apoptotic proteins. Mutation is SMAD2 is also associated with cases of sporadic colon cancer [58]. Pro-oncogenic mutant form of TP53 can inhibit the phosphorylation of SMAD2/3 disrupting the downstream effects of TGF-B [59]. Interestingly, SMAD proteins and TGF-B have been incriminated in the development of inflammatory bowel disease and may have a role to play in colitis associated cancer [60].
6. Mismatch repair genes- MSI-high versus MSI-low
Microsatellite Instability-high (MSI-H) colorectal tumors, identified on multigene sequencing, originate from dysfunctional mismatch repair genes (MMR). These mismatches can result either from epigenetic silencing (80% of cases) of components of the MMR machinery (such as MLH1, MSH2, PMS2, MSH6), or by germline mutations (20% of cases) [61,62]. Interestingly, MSI-H CRC patients may have a higher life expectancy than microsatellite stable patients [63,64]. Patients with MSI-H CRC have been shown to benefit the most from immunotherapy.
7. MUTYH defects and familial CRC
A biallelic mutation in the MUTY homolog gene causes a hereditary syndrome associated with frequent colorectal cancers, MUTYH-associated polyposis (MAP). MAP is similar in phenotype to attenuated FAP, and results in formation of ten to hundreds of colorectal adenomas, and usually presents at a median age of 46–48 [65]. Roughly 20% cases of adenomatous polyposis which lack Apc gene mutation, harbor MUTYH defects. MUTYH gene product is a DNA glycosylase which serves to repair mismatches caused by oxidative DNA damage via base excision repair [65]. Y179C and G396D, are the most common missense mutations in the MUTYH gene, accounting for ~80% of MAP [66]. Patients with homozygous Y179C mutations are known to suffer from a more severe form of CRC occurring at an earlier age as compared to those with homozygous G396D mutations or compound heterozygotes [67]. Unfortunately, ~50% of these patients have already developed CRC by the time of diagnosis due to lack of vigilant screening given the autosomal recessive nature of inheritance [68]. MAP related CRCs frequently develop in proximal colon, have a mucin rich histology, abundance of lymphocytes as well as a relatively better prognosis than sporadic CRC [67].
Modifier genes
COX-2
COX-2 is an inducible cyclooxygenase, which has been found to be overexpressed in human colon cancer. Its functions include blocking cellular differentiation, reduction of apoptosis and promoting angiogenesis [69]. In patients with FAP, an NSAID called sulindac was associated with regression of adenomas [70]. One of the plausible reasoning for this effect could be reduction in COX-2 levels leading to increased apoptosis. More recently, randomized controlled trials of aspirin and selective COX2 have also shown a decreased incidence of adenomas and colorectal cancers in average risk patients [71].
PPAR gene
PPAR gene product is a nuclear hormone receptor that assists in inhibition of cellular growth and promotion of cellular differentiation. This receptor is distal to the Wnt/APC/β-Catenin pathway and may also be an effector of the COX-2 pathway. Mutations can lead to sporadic CRC [72].
Inflammatory bowel disease related CRC
Inflammatory bowel disease poses a high risk of dysplasia and invasive CRC at an earlier age [73]. The risk is increased with the duration, intensity and expanse of inflammation within the colon. Colitis-associated cancers have been found to have distinct genetic features. They usually harbor TP53, IDH1 and MYC mutations whereas mutant APC and KRAS are not as common. [74–77]. Also, LOH for TP53 is an earlier event in colitis associated colon cancer.
Other rare ones like CHEK2
CHEK2 is a tumor suppressor gene, which through its interaction with TP53, halts cell division and induces apoptosis when the cell sustains irrevocable damage to its DNA [78]. Breast malignancies more often carry mutant CHEK2 but it has been linked with a higher likelihood of developing colon cancer particularly in patients with a family history of colon cancer [79].
Sidedness of colon cancer
Over the years, there has been convincing evidence that right and left sided malignancies behave as two clinically disparate entities [80]. Various theories have been presented in order to explain this disparity including the differences in embryological development, immune cell infiltration, differences in gut microbiota as well as delays in diagnosis in right sided colon cancer [80,81]. The right half, up to the proximal 2/3rd of the transverse colon originates from the mid-gut whereas the embryonic hindgut gives rise to the rest of the colon and rectum. This fundamental difference in pre-natal origin has led to numerous dissimilarities in these sites with >200 genes significantly differing in their expression when compared between the dextral and sinistral side of the colon. [82]
At a molecular level, right sided colon cancers (RCRC) are more frequently associated with high microsatellite instability, mismatch repair deficiency, CpG island methylation, and BRAF mutations [3, 82–84]. Except for microsatellite instability, the rest of these features are associated with poor outcomes [85]. Left sided colon/rectal cancers (LCRC) more frequently emanate from the chromosomal instability pathway and harbor TP53, KRAS, PI3KCA, SMAD2 &4 and APC mutations [84]. MSI-H nature also makes right sided tumors highly immunogenic owing to the extensive mutational and neoantigen load promoting T- lymphocyte infiltration. On the other hand, left sided tumors have a cold uninflamed tumor microenvironment. This could explain the better response of RCRC to immune checkpoint blockade. KRAS and BRAF mutational status has also been linked to survival differences, with double wild type RCRCs carrying a worse DFS as compared to LCRC [85].
Left and right sided tumors also starkly differ in terms of morphology and histology. LCRC usually present as tubular or villous adenocarcinomas whereas RCRC appear as sessile serrated mucinous adenocarcinomas. RCRCs are frequently “flat” or laterally growing tumors making them difficult to be picked up at an early stage by routine colonoscopic screening and end up being diagnosed at an advanced stage. LCRC are usually polypoid in appearance allowing for easy detection. KRAS mutation has been implicated in causing exophytic tumor growth and absence of mutant KRAS in RCRC can explain the flat morphology of these tumors [86].
Epidemiologically speaking, younger individuals and males are more likely to have LCRC, whereas RCRC occurs predominantly in the elderly and females. Although, LCRC still remains the most commonly diagnosed cancer site across all age groups, with approximately 70% of all CRC being left sided [87]. LCRC on an average correlates with a 19% absolute risk reduction in terms of mortality when compared with RCRC [83]. Several studies have shown that stage 4 RCRC carries worse prognosis than stage 4 LCRC. Furthermore, a metanalysis showed that right-sided location had poorer outcomes irrespective of mucinous histology or mutant BRAF [88].
Classification
Consensus molecular subtypes of colon cancer
Transcriptomics and analysis of gene expressions can explain heterogeneity in patient outcomes and treatment response. In 2015, the CRC subtyping consortium consolidated all prior molecular classification methodologies into a singular system known as the consensus molecular subtypes or CMS. This includes four unique molecular subtypes described in detail below and summarized in table 1
Table 1:
Consensus molecular subtypes of Colorectal Cancer
| CMS subtypes | CMS 1 (MSI Immune) | CMS 2 (Canonical) | CMS 3 (Molecular) | CMS 4 (Mesenchymal) |
|---|---|---|---|---|
| Frequency | 14% | 37% | 23% | 13% |
| Pathological hallmark | Hypermutation, MSI-H and CIMP-H | Wnt/B-catenin and MYC signaling pathway activation | Metabolic dysregulation of carbohydrate and fatty acid oxidation pathways | Epithelial mesenchymal transformation with prominent stomal infiltration, angiogenesis and TGF-B upregulation |
| Somatic copy number alternations (SCNA) | Low | High | Low-moderate | High |
| Immunological profile | Immune activated-Infiltration of TH1, NK-1 cells as well as PD-1 and CTLA-4, CXCR3/CCR5 chemokine expression | Immune ignorant | Immune ignorant | Immune tolerant inflamed-Infiltration of TH-17 and MDSCs, Complement activation, CCL2 chemokine expression |
| Associated gene mutations | BRAF | TP53, EGFR | KRAS, PIK3CA and IGFBP2 | NOTCH3/ VEGFR2 |
| Site | Proximal right sided colon predominance | Left sided colon and rectal predominance | Relatively more common in right-sided tumors | Relatively more common in left sided and rectal tumors |
| 5-year survival rates | 73% but poor post relapse survival | 77%, higher post relapse survival | 75% | 62%, worst OS and relapse free survival |
| Clinical Implication | May benefit for immune checkpoint inhibitors | May benefit from Anti-EGFR therapies | EGFR resistance | May benefit from angiogenesis inhibitors, no benefit from adjuvant chemotherapy |
1. CMS1
14% of all cases fall in this category with the majority of cases being sporadic while the rest of them are associated with Lynch syndrome [54]. CMS1 is associated with high rates of hypermutation either due to hypermethylation or mutation of promoters of mismatch repair genes. This subtype is not associated with somatic copy number alterations or CIN. BRAF is frequently mutated [89]. Cancers in this subtype often originate in the proximal colon.
One of the other distinguishing features on gene expression profiling in this subtype is a heightened expression of genes mediating immune infiltration and trafficking of T helper 1 and cytotoxic cells, which occurs in conjunction with the activation of immune evasion mechanisms. This likely explains improved outcomes in this subtype in patients treated with immunotherapy [90].
Stage II patients do not need adjuvant chemotherapy and may do worse if treated with adjuvant chemotherapy, likely due to negative effect of chemotherapy on the already activated immune system. On the other hand, stage 3 cancers may benefit from adjuvant systemic therapy. Although the prognosis is generally good, prognosis maybe poor in the subset of CRCs harboring BRAF mutations [91].
2. CMS2
This is the largest molecular subtype comprising 39% of all cases (56). CMS2 is also known as the canonical subtype, characterized by chromosomal instability with somatic copy number alterations with low rates of hypermutation. This subtype is most closely related to adenoma - carcinoma sequence described earlier and is often associated with early loss of APC, followed by KRAS mutation and subsequent late loss of TP53 gene [92]. CMS2 is more commonly associated with left sided tumors with the best prognosis of the 4 subtypes with a 77% 5 year survival rate and an overall survival of close to 3 years in metastatic disease [91].
3. CMS3
This subtype comprises of about 13% of the cases and is known as the metabolic subtype. Chromosomal instability rates are markedly lower than CMS2 and CMS4 subtypes. CMS3 is characterized by MSI, although lower than CMS1 but higher than the other two subtypes [54]. KRAS mutations are more common, being positive in 68% of the cases and responsible for higher rates of resistance to anti-EGFR therapies [54]. Overall, 5-year survival is promising at 75% [89].
4. CMS4
This subtype is known as the mesenchymal subtype and comprises of about 23% of the cases [54]. Of all four cancer subtypes, CMS4 tends to be diagnosed at a later stage and carries the worst relapse free survival. This subtype is also known to arise from the serrated pathway, similar to CMS1 but carries a high frequency of somatic copy number alterations and a lower frequency of hypermutation [93]. On gene expression profiling, this subtype was found to have a greater presence of proteins involved in stromal infiltration and mesenchymal overactivation.
Correlation of CMS subtypes with immune signatures
The tumor microenvironment is comprised of cellular components belonging to the epithelium, stroma as well as the adaptive and innate immune system. Organization of the tumor microenvironment can predict tumor aggressiveness and response to therapy. Local tumor environment can have an inhibitory or negative effect on cancer growth through an increased and effective immune response and surveillance or it can have the opposite effect promoting angiogenesis, inflammation, immune evasion or immunosuppression resulting in uncontrolled tumor growth and metastasis [94]. Becht et al did an extensive analysis of composition and functional orientation of tumor microenvironment in a substantial volume of colon cancer specimens and discerned a link between tumor microenvironment and consensus molecular subtypes that explain the clinical nuances and response to current therapies as well as provide the rationale for designing future treatments [95].
The MSI-like subtype or CMS1 is enriched with genes involved in recruitment and activation of T-lymphocytes, which explains the higher cytotoxic T cell and NK cell infiltration. At the same time, CMS 1 subtype is also equipped with immune evasion mechanisms with high expression of immune checkpoint molecules, which explains the good response to anti PD1 therapy in this subtype [96]. Interestingly, CMS4 was also found to have a higher expression of immune signatures, but worse prognosis and poor response to immune based treatment [97]. This can be explained by increased myeloid, angiogenic and immunosuppressive molecular signatures in these tumors. The other two subtypes, CMS2 and CMS3 are characterized by low immune signatures. Although CMS4 is associated with poor prognosis, insights into gene expression profiling and tumor microenvironment suggest designing future therapies focused on anti-angiogenesis and anti-inflammatory agents in combination with immune checkpoint inhibitors.
Conclusions
Advances in gene profiling techniques have paved our way to a better discernment of the discrete as well as interlinked pathways in complex molecular pathogenesis of CRC. These advances have helped in diagnosing the driver mutations in tumorigenesis which form the basis of targeted therapies.
In this review, we outlined the most common pathways in the pathogenesis of CRC. Amongst these, the adenoma-carcinoma model which is initiated by APC mutation and propagated by chromosomal instability causing stepwise accretion of molecular and epigenetic changes accounts for ~80% cases. The remaining 15–20% of CRCs are developed through an alternative pathway, such as defective mismatch repair systems, hypermethylation of CIMP or BRAF activation. We discussed the roles of tumor suppressor genes (Apc, TP53, PTEN, TGFβ, Chromosome 18q, SMAD4), oncogenes (KRAS, BRAF, HER2) as well as tumor modifying genes (COX2, PPAR, CHEK2). Developments in identifying the underlying genetic defects have led to the categorization of most CRCs into one of four different subgroups. Tumors marked by MSI are included in CMS1 subtype, while tumors in CMS2 to CMS4 subtypes emanate from chromosomal instability. The creation of this classification has come as a consensus of several prior studies and marks a pivotal milestone in the study of colorectal cancer. However, the clinical implications of understanding the pathogenomics of CRC is yet to be fully realized, and future studies incorporating gene expression profiles as predictors of response to therapy will pave the way for precision medicine in this domain.
Acknowledgments
The editors would like to thank Dr. Gopichand Pendurti for taking the time to review this article.
Footnotes
Compliance with Ethics Guidelines
Conflict of Interest
Anup Kasi, Shivani Handa, Sajjad Bhatti, Shahid Umar, Ajay Bansal, and Weijing Sun each declare no potential conflicts of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394(10207):1467–80. [DOI] [PubMed] [Google Scholar]
- 2.Kastrinos F, Syngal S. Inherited colorectal cancer syndromes. Cancer journal. 2011;17(6):405–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–67. [DOI] [PubMed] [Google Scholar]
- 4.De Palma FDE, D’Argenio V, Pol J, Kroemer G, Maiuri MC, Salvatore F. The Molecular Hallmarks of the Serrated Pathway in Colorectal Cancer. Cancers. 2019;11(7).• A focused review summarizing the genetic and epigenetic basis of the Serrated polyp pathway
- 5.Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. International Journal of Cancer. 2015;137(6):1258–68. [DOI] [PubMed] [Google Scholar]
- 6.Park CH, Han DS, Oh YH, Lee AR, Lee YR, Eun CS. Role of Fusobacteria in the serrated pathway of colorectal carcinogenesis. Scientific reports. 2016;6:25271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murcia O, Juárez M, Hernández-Illán E, Egoavil C, Giner-Calabuig M, Rodríguez-Soler M, et al. Serrated colorectal cancer: Molecular classification, prognosis, and response to chemotherapy. World Journal of Gastroenterology. 2016;22(13):3516–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nature reviews Cancer. 2009;9(7):489–99. [DOI] [PubMed] [Google Scholar]
- 9.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. The Journal of molecular diagnostics : JMD. 2008;10(1):13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walther A, Houlston R, Tomlinson I. Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut. 2008;57(7):941–50. [DOI] [PubMed] [Google Scholar]
- 11.Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mechanisms of ageing and development. 2008;129(7–8):391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen SA, Turner EH, Beightol MB, Jacobson A, Gooley TA, Salipante SJ, et al. Frequent PIK3CA Mutations in Colorectal and Endometrial Tumors With 2 or More Somatic Mutations in Mismatch Repair Genes. Gastroenterology. 2016;151(3):440–7.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138(6):2044–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nature genetics. 2006;38(7):787–93. [DOI] [PubMed] [Google Scholar]
- 15.Jones JC, Renfro LA, Al-Shamsi HO, Schrock AB, Rankin A, Zhang BY, et al. (Non-V600) BRAF Mutations Define a Clinically Distinct Molecular Subtype of Metastatic Colorectal Cancer. Journal of Clinical Oncology. 2017;35(23):2624–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jover R, Nguyen TP, Perez-Carbonell L, Zapater P, Paya A, Alenda C, et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology. 2011;140(4):1174–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends in biochemical sciences. 2011;36(6):320–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imamura Y, Morikawa T, Liao X, Lochhead P, Kuchiba A, Yamauchi M, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clinical Cancer Research. 2012;18(17):4753–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saeed O, Lopez-Beltran A, Fisher KW, Scarpelli M, Montironi R, Cimadamore A, et al. RAS genes in colorectal carcinoma: pathogenesis, testing guidelines and treatment implications. Journal of Clinical Pathology. 2019;72(2):135–9. [DOI] [PubMed] [Google Scholar]
- 20.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26(22):3291–310. [DOI] [PubMed] [Google Scholar]
- 21.Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61(6):847–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakamoto N, Feng Y, Stolfi C, Kurosu Y, Green M, Lin J, et al. BRAF(V600E) cooperates with CDX2 inactivation to promote serrated colorectal tumorigenesis. Elife. 2017;6:e20331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Safaee Ardekani G, Jafarnejad SM, Tan L, Saeedi A, Li G. The prognostic value of BRAF mutation in colorectal cancer and melanoma: a systematic review and meta-analysis. PloS one. 2012;7(10):e47054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kavuri SM, Jain N, Galimi F, Cottino F, Leto SM, Migliardi G, et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discovery. 2015;5(8):832–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richman SD, Southward K, Chambers P, Cross D, Barrett J, Hemmings G, et al. HER2 overexpression and amplification as a potential therapeutic target in colorectal cancer: analysis of 3256 patients enrolled in the QUASAR, FOCUS and PICCOLO colorectal cancer trials. The Journal of Pathology. 2016;238(4):562–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greally M, Kelly CM, Cercek A. HER2: An emerging target in colorectal cancer. Current Problems in Cancer. 2018;42(6):560–71. [DOI] [PubMed] [Google Scholar]
- 27.Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. The Lancet Oncology. 2016;17(6):738–46. [DOI] [PubMed] [Google Scholar]
- 28.Meric-Bernstam F, Hurwitz H, Raghav KPS, McWilliams RR, Fakih M, VanderWalde A, et al. Pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer (MyPathway): an updated report from a multicentre, open-label, phase 2a, multiple basket study. The Lancet Oncology. 2019;20(4):518–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brannon AR, Vakiani E, Sylvester BE, Scott SN, McDermott G, Shah RH, et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biology. 2014;15(8):454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson S, Näthke IS. Interactions and functions of the adenomatous polyposis coli (APC) protein at a glance. Journal of Cell Science. 2013;126(4):873–77. [DOI] [PubMed] [Google Scholar]
- 31.Dow LE, O’Rourke KP, Simon J, Tschaharganeh DF, van Es JH, Clevers H, et al. Apc Restoration Promotes Cellular Differentiation and Reestablishes Crypt Homeostasis in Colorectal Cancer. Cell. 2015;161(7):1539–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narayan S, Roy D. Role of APC and DNA mismatch repair genes in the development of colorectal cancers. Molecular cancer. 2003;2:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lepourcelet M, Chen YN, France DS, Wang H, Crews P, Petersen F, et al. Small-molecule antagonists of the oncogenic Tcf/beta-catenin protein complex. Cancer Cell. 2004;5(1):91–102. [DOI] [PubMed] [Google Scholar]
- 34.Hashimoto K, Yamada Y, Semi K, Yagi M, Tanaka A, Itakura F, et al. Cellular context-dependent consequences of Apc mutations on gene regulation and cellular behavior. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(4):758–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iacopetta B TP53 mutation in colorectal cancer. Human mutation. 2003;21(3):271–6. [DOI] [PubMed] [Google Scholar]
- 36.Nakayama M, Oshima M. Mutant p53 in colon cancer. Journal of Molecular Cell Biology. 2019;11(4):267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikolaev SI, Sotiriou SK, Pateras IS, Santoni F, Sougioultzis S, Edgren H, et al. A single-nucleotide substitution mutator phenotype revealed by exome sequencing of human colon adenomas. Cancer Research. 2012;72(23):6279–89. [DOI] [PubMed] [Google Scholar]
- 38.Schwitalla S, Ziegler PK, Horst D, Becker V, Kerle I, Begus-Nahrmann Y, et al. Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell. 2013;23(1):93–106. [DOI] [PubMed] [Google Scholar]
- 39.Goel A, Arnold CN, Niedzwiecki D, Carethers JM, Dowell JM, Wasserman L, et al. Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability-high sporadic colorectal cancers. Cancer Research. 2004;64(9):3014–21. [DOI] [PubMed] [Google Scholar]
- 40.Chang CC, Lin PC, Lin CC, Lan YT, Lin HH, Lin CH, et al. Molecular and Clinicopathological Differences by Age at the Diagnosis of Colorectal Cancer. International Journal of Molecular Sciences. 2017;18(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danielsen SA, Lind GE, Bjornslett M, Meling GI, Rognum TO, Heim S, et al. Novel mutations of the suppressor gene PTEN in colorectal carcinomas stratified by microsatellite instability- and TP53 mutation- status. Human Mutation. 2008;29(11):E252–62. [DOI] [PubMed] [Google Scholar]
- 42.Custodio A, Feliu J. Prognostic and predictive biomarkers for epidermal growth factor receptor-targeted therapy in colorectal cancer: beyond KRAS mutations. Critical Reviews in Oncology/Hematology. 2013;85(1):45–81. [DOI] [PubMed] [Google Scholar]
- 43.Colakoglu T, Yildirim S, Kayaselcuk F, Nursal TZ, Ezer A, Noyan T, et al. Clinicopathological significance of PTEN loss and the phosphoinositide 3-kinase/Akt pathway in sporadic colorectal neoplasms: is PTEN loss predictor of local recurrence? American Journal of Surgery. 2008;195(6):719–25. [DOI] [PubMed] [Google Scholar]
- 44.Sawai H, Yasuda A, Ochi N, Ma J, Matsuo Y, Wakasugi T, et al. Loss of PTEN expression is associated with colorectal cancer liver metastasis and poor patient survival. BMC Gastroenterology. 2008;8:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kara O, Duman BB, Kara B, Erdogan S, Parsak CK, Sakman G. Analysis of PTEN, VEGF, HER2 and P53 status in determining colorectal cancer benefit from bevacizumab therapy. Asian Pacific Journal of Cancer Prevention. 2012;13(12):6397–401. [DOI] [PubMed] [Google Scholar]
- 46.Negri FV, Bozzetti C, Lagrasta CA, Crafa P, Bonasoni MP, Camisa R, et al. PTEN status in advanced colorectal cancer treated with cetuximab. British Journal of Cancer. 2010;102(1):162–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loupakis F, Pollina L, Stasi I, Ruzzo A, Scartozzi M, Santini D, et al. PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. Journal of Clinical Oncology. 2009;27(16):2622–9. [DOI] [PubMed] [Google Scholar]
- 48.Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Research. 2009;69:1851–1857. [DOI] [PubMed] [Google Scholar]
- 49.Tol J, Dijkstra JR, Klomp M, Teerenstra S, Dommerholt M, Vink-Börger ME, et al. Markers for EGFR pathway activation as predictor of outcome in metastatic colorectal cancer patients treated with or without cetuximab. European Journal of Cancer. 2010;46(11):1997–2009. [DOI] [PubMed] [Google Scholar]
- 50.Ulivi P, Capelli L, Valgiusti M, Zoli W, Scarpi E, Chiadini E, et al. Predictive role of multiple gene alterations in response to cetuximab in metastatic colorectal cancer: a single center study. Journal of Translational Medicine. 2012;10:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iacopetta BJ, Welch J, Soong R, House AK, Zhou XP, Hamelin R. Mutation of the transforming growth factor-beta type II receptor gene in right-sided colorectal cancer: relationship to clinicopathological features and genetic alterations. The Journal of pathology. 1998;184(4):390–5. [DOI] [PubMed] [Google Scholar]
- 52.Jung B, Staudacher JJ, Beauchamp D. Transforming Growth Factor β Superfamily Signaling in Development of Colorectal Cancer. Gastroenterology. 2017;152(1):36–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fischer JM, Calabrese PP, Miller AJ, Munoz NM, Grady WM, Shibata D, et al. Single cell lineage tracing reveals a role for TgfbetaR2 in intestinal stem cell dynamics and differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(43):12192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nature medicine. 2015;21(11):1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehlen P, Fearon ER. Role of the dependence receptor DCC in colorectal cancer pathogenesis. Journal of Clinical Oncology. 2004;22(16):3420–8. [DOI] [PubMed] [Google Scholar]
- 56.Fleming NI, Jorissen RN, Mouradov D, Christie M, Sakthianandeswaren A, Palmieri M, et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Research. 2013;73(2):725–35. [DOI] [PubMed] [Google Scholar]
- 57.Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell. 2018;33(1):125–36.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andre T, Lonardi S, Wong KYM, Morse M, McDermott RS, Hill AG, et al. Combination of nivolumab (nivo)+ ipilimumab (ipi) in the treatment of patients (pts) with deficient DNA mismatch repair (dMMR)/high microsatellite instability (MSI-H) metastatic colorectal cancer (mCRC): CheckMate 142 study. Journal of Clinical Oncology. 2017; 35(15):supp_3531–3531. [Google Scholar]
- 59.Kalo E, Buganim Y, Shapira KE, Besserglick H, Goldfinger N, Weisz L, et al. Mutant p53 attenuates the SMAD-dependent transforming growth factor beta1 (TGF-beta1) signaling pathway by repressing the expression of TGF-beta receptor type II. Molecular and Cellular Biology. 2007;27(23):8228–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chandrasinghe P, Cereser B, Moorghen M, Al Bakir I, Tabassum N, Hart A, et al. Role of SMAD proteins in colitis-associated cancer: from known to the unknown. Oncogene. 2018;37(1):1–7.• A detailed review linking SMAD protein to Colitis associated Cancer
- 61.Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. The Lancet Oncology. 2017;18(9):1182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Phipps AI, Limburg PJ, Baron JA, Burnett-Hartman AN, Weisenberger DJ, Laird PW, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology. 2015;148(1):77–87.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sinicrope FA, Shi Q, Smyrk TC, Thibodeau SN, Dienstmann R, Guinney J, et al. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology. 2015;148(1):88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nielsen M, Morreau H, Vasen HF, Hes FJ. MUTYH-associated polyposis (MAP). Critical Reviews in Oncology/Hematology. 2011;79(1):1–16. [DOI] [PubMed] [Google Scholar]
- 65.Sinicrope FA, Gill S. Role of cyclooxygenase-2 in colorectal cancer. Cancer Metastasis Reviews. 2004;23(1–2):63–75. [DOI] [PubMed] [Google Scholar]
- 66.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, et al. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nature Genetics. 2002;30(2):227–32. [DOI] [PubMed] [Google Scholar]
- 67.Nielsen M, Morreau H, Vasen HF, Hes FJ. MUTYH-associated polyposis (MAP). Critical Reviews in Oncology/Hematology. 2011;79(1):1–16. [DOI] [PubMed] [Google Scholar]
- 68.Sieber OM, Lipton L, Crabtree M, Heinimann K, Fidalgo P, Phillips RK, et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. The New England Journal of Medicine. 2003;348(9):791–9. [DOI] [PubMed] [Google Scholar]
- 69.Wang D, Dubois RN. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene. 2010;29(6):781–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mohammed A, Yarla NS, Madka V, Rao CV. Clinically Relevant Anti-Inflammatory Agents for Chemoprevention of Colorectal Cancer: New Perspectives. International Journal of Molecular Sciences. 2018;19(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vallée A, Lecarpentier Y. Crosstalk Between Peroxisome Proliferator-Activated Receptor Gamma and the Canonical WNT/β-Catenin Pathway in Chronic Inflammation and Oxidative Stress During Carcinogenesis. Frontiers in Immunology. 2018;9:745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beaugerie L, Itzkowitz SH. Cancers complicating inflammatory bowel disease. The New England Journal of Medicine. 2015;372(15):1441–52. [DOI] [PubMed] [Google Scholar]
- 73.Hussain SP, Amstad P, Raja K, Ambs S, Nagashima M, Bennett WP, et al. Increased p53 mutation load in noncancerous colon tissue from ulcerative colitis: a cancer-prone chronic inflammatory disease. Cancer Research. 2000;60(13):3333–7. [PubMed] [Google Scholar]
- 74.Leedham SJ, Graham TA, Oukrif D, McDonald SA, Rodriguez-Justo M, Harrison RF, et al. Clonality, founder mutations, and field cancerization in human ulcerative colitis-associated neoplasia. Gastroenterology. 2009;136(2):542–50.e6. [DOI] [PubMed] [Google Scholar]
- 75.Kern SE, Redston M, Seymour AB, Caldas C, Powell SM, Kornacki S, et al. Molecular genetic profiles of colitis-associated neoplasms. Gastroenterology. 1994;107(2):420–8. [DOI] [PubMed] [Google Scholar]
- 76.Rashid A, Hamilton SR. Genetic alterations in sporadic and Crohn’s-associated adenocarcinomas of the small intestine. Gastroenterology. 1997;113(1):127–35. [DOI] [PubMed] [Google Scholar]
- 77.Yurgelun MB, Kulke MH, Fuchs CS, Allen BA, Uno H, Hornick JL, et al. Cancer Susceptibility Gene Mutations in Individuals With Colorectal Cancer. Journal of Clinical Oncology. 2017;35(10):1086–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suchy J, Cybulski C, Wokolorczyk D, Oszurek O, Gorski B, Debniak T, et al. CHEK2 mutations and HNPCC-related colorectal cancer. International Journal of Cancer. 2010;126(12):3005–9. [DOI] [PubMed] [Google Scholar]
- 79.Baran B, Mert Ozupek N, Yerli Tetik N, Acar E, Bekcioglu O, Baskin Y. Difference Between Left-Sided and Right-Sided Colorectal Cancer: A Focused Review of Literature. Gastroenterology Research. 2018;11(4):264–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stintzing S, Tejpar S, Gibbs P, Thiebach L, Lenz HJ. Understanding the role of primary tumour localisation in colorectal cancer treatment and outcomes. European Journal of Cancer. 2017;84:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bertotti A, Sassi F. Molecular Pathways: Sensitivity and Resistance to Anti-EGFR Antibodies. Clinical Cancer Research. 2015;21(15):3377–83. [DOI] [PubMed] [Google Scholar]
- 82.Peng Q, Lin K, Chang T, Zou L, Xing P, Shen Y, et al. Identification of genomic expression differences between right-sided and left-sided colon cancer based on bioinformatics analysis. OncoTargets and Therapy. 2018;11:609–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Salem ME, Weinberg BA, Xiu J, El-Deiry WS, Hwang JJ, Gatalica Z, et al. Comparative molecular analyses of left-sided colon, right-sided colon, and rectal cancers. Oncotarget. 2017;8(49):86356–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Petrelli F, Tomasello G, Borgonovo K, Ghidini M, Turati L, Dallera P, et al. Prognostic Survival Associated With Left-Sided vs Right-Sided Colon Cancer: A Systematic Review and Meta-analysis. JAMA oncology. 2017;3(2):211–219.• The first large meta-analysis defining the prognostic implication of primary tumor location
- 85.Taieb J, Kourie HR, Emile JF, Le Malicot K, Balogoun R, Tabernero J, et al. Association of Prognostic Value of Primary Tumor Location in Stage III Colon Cancer With RAS and BRAF Mutational Status. JAMA oncology. 2018;4(7):e173695.• First report analyzingt the differential prognostic impact of RAS and BRAF mutational status on left versus right sided colon cancers.
- 86.Yashiro M, Carethers JM, Laghi L, Saito K, Slezak P, Jaramillo E, et al. Genetic pathways in the evolution of morphologically distinct colorectal neoplasms. Cancer Research. 2001;61(6):2676–83. [PubMed] [Google Scholar]
- 87.Jess P, Hansen IO, Gamborg M, Jess T. A nationwide Danish cohort study challenging the categorisation into right-sided and left-sided colon cancer. BMJ Open. 2013;3(5):e002608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Holch JW, Ricard I, Stintzing S, Modest DP, Heinemann V. The relevance of primary tumour location in patients with metastatic colorectal cancer: A meta-analysis of first-line clinical trials. European Journal of Cancer. 2017;70:87–98. [DOI] [PubMed] [Google Scholar]
- 89.Barras D, Missiaglia E, Wirapati P, Sieber OM, Jorissen RN, Love C, et al. BRAF V600E Mutant Colorectal Cancer Subtypes Based on Gene Expression. Clinical Cancer Research. 2017;23(1):104–15. [DOI] [PubMed] [Google Scholar]
- 90.Gang W, Wang JJ, Guan R, Yan S, Shi F, Zhang JY, et al. Strategy to targeting the immune resistance and novel therapy in colorectal cancer. Cancer Medicine. 2018;7(5):1578–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thanki K, Nicholls ME, Gajjar A, Senagore AJ, Qiu S, Szabo C, et al. Consensus Molecular Subtypes of Colorectal Cancer and their Clinical Implications. International Biological and Biomedical Journal. 2017;3(3):105–11. [PMC free article] [PubMed] [Google Scholar]
- 92.Berg KCG, Sveen A, Høland M, Alagaratnam S, Berg M, Danielsen SA, et al. Gene expression profiles of CMS2-epithelial/canonical colorectal cancers are largely driven by DNA copy number gains. Oncogene. 2019;38(33):6109–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bramsen JB, Rasmussen MH, Ongen H, Mattesen TB, Orntoft MW, Arnadottir SS, et al. Molecular-Subtype-Specific Biomarkers Improve Prediction of Prognosis in Colorectal Cancer. Cell Reports. 2017;19(6):1268–80. [DOI] [PubMed] [Google Scholar]
- 94.Becht E, Giraldo NA, Dieu-Nosjean MC, Sautes-Fridman C, Fridman WH. Cancer immune contexture and immunotherapy. Current Opinion in Immunology. 2016;39:7–13. [DOI] [PubMed] [Google Scholar]
- 95.Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, et al. Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. Journal of Clinical Oncology. 2018;36(8):773–9.• • Long term data from CheckMate-142, the largest single-study report of an immunotherapy combination in dMMR/MSI-H mCRC,
- 96.Roelands J, Kuppen PJK, Vermeulen L, Maccalli C, Decock J, Wang E, et al. Immunogenomic Classification of Colorectal Cancer and Therapeutic Implications. International Journal of Molecular Sciences. 2017;18(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Picard E, Verschoor CP, Ma GW, Pawelec G. Relationships Between Immune Landscapes, Genetic Subtypes and Responses to Immunotherapy in Colorectal Cancer. Frontiers in Immunology. 2020;11:369–. [DOI] [PMC free article] [PubMed] [Google Scholar]