Abstract
Gastric cancer (GC) is one of the most common malignant tumors. The mechanism of how GC develops is vague, and therapies are inefficient. The function of microRNAs (miRNAs) in tumorigenesis has attracted the attention from many scientists. During the development of GC, miRNAs function in the regulation of different phenotypes, such as proliferation, apoptosis, invasion and metastasis, drug sensitivity and resistance, and stem-cell-like properties. MiRNAs were evaluated for use in diagnostic and prognostic predictions and exhibited considerable accuracy. Although many problems exist for the application of therapy, current studies showed the antitumor effects of miRNAs. This paper reviews recent advances in miRNA mechanisms in the development of GC and the potential use of miRNAs in the diagnosis and treatment of GC.
Keywords: Apoptosis, Diagnosis, Drug resistance, MicroRNAs, Neoplasm metastasis, Neoplastic stem cells, Prognosis, Stomach neoplasms, Treatment
Introduction
Gastric cancer (GC) is one of the most common malignant tumors, with high morbidity and mortality worldwide.[1] It produces a tremendous burden, especially in Asia, Latin America, and central and eastern Europe.[2] GC is second in incidence and mortality of all malignant tumors in China. Approximately 498,000 Chinese people died from GC in 2015.[3–5] Gastrointestinal endoscopic screening reduces the risk of death in Asian countries.[6] However, the economic and medical burden for endoscopic screening is too high.[7] Therefore, it is important and imperative to study and understand the mechanism throughout the development of GC and identify new efficient non-invasive methods for the detection, prognostic prediction, and treatment.
MicroRNAs (miRNAs) are endogenous RNAs, approximately 22 nucleotides (nts), that play regulatory roles in animals and plants. MiRNAs are an abundant class of gene regulators in multi-cellular organisms that may affect the expression of numerous protein-coding genes.[8,9] MiRNAs directly bind to complementary sequences in the 3′-untranslated regions (3′-UTR) of target mRNAs to regulate degradation or translational repression of the target mRNAs. Many miRNAs contribute to cancer development, and the ectopic expression of miRNAs is associated with tumor initiation and progression, such as proliferation, invasion, and other malignant phenotypes.[10,11] During the past decade, studies found that miRNAs were unexpectedly rich in information content with respect to cancer. Analysis of microarray data showed that many miRNAs, long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) were detected during the process of malignant transformation of GC from normal tissues to atrophic gastritis and tumorigenesis.[12,13] The differential expression pattern of miRNAs in GC creates specific miRNA signatures that characterize distinct histologic subtypes, cancer progression, and patient prognosis.[14–16] The study of miRNA characteristics and their regulatory genes will help us better understand the functions and mechanism of the development of GC. The present review summarizes the recent advances of the role and application of miRNAs in GC.
Roles of MiRNAs in the Development of Gastric Cancer
MiRNAs are involved in different cellular processes, like metabolism, differentiation, development, and apoptosis. These miRNAs function as promoters or suppressors depending on the character of the target mRNAs or genes. Specific expression profiles exist in normal and gastric tumor tissues.[17,18] Previous studies showed that miRNAs played a pivotal role in GC progression. MiRNAs function as oncogenes or tumor-suppressive genes to regulate many aspects of GC, such as proliferation, apoptosis, metastasis, chemo-sensitivity and chemo-resistance, and the stemness of cancer stem cells (CSCs) [Figure 1].
Figure 1.
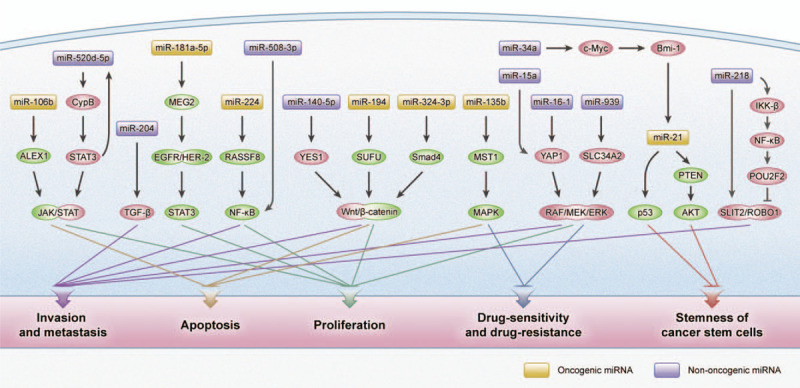
Common signaling pathway with miRNAs involved in the different malignant phenotypes of gastric cancer. Akt: Protein kinase B; ALEX1: Arm protein lost in epithelial cancers on chromosome X 1; Bmi-1: B-lymphoma moloney murine leukemia virus insertion region-1; CypB: Cyclophilin B; EGFR: Epidermal growth factor receptor; ERK: Extracellular signal-regulated kinase; HER-2: Human epidermal growth factor receptor 2; IKK-β: IκB kinase-β; JAK: Janus kinase; MAPK: Mitogen-activated protein kinase; MEG2: Protein-tyrosine phosphatase megakaryocyte 2; MEK: Mitogen-activated protein kinase; MST1: Mammalian sterile 20-like 1; NF-κB: Nuclear factor-κB; POU2F2: POU class 2 homeobox 2; PTEN: Phosphatase and tensin homolog; RASSF8: Ras-association domain family 8; SLC34A2: Solute carrier 34 A2; SLIT2/ROBO1: Slit guidance ligand 2/a ligand of the roundabout 1; Smad4: Drosophila protein, mothers against decapentaplegic homolog 4; STAT3: Signal transducer and activator of transcription 3; SUFU: Suppressor of fused homolog; TGF-β: Transforming growth factor-β; YAP1: YES-associated protein 1; YES1: YES proto-oncogene 1.
Proliferation
Several miRNAs may contribute to the proliferation of GC via influencing the growth of cancer cells. Previously, miR-21 was closely related to tumorigenesis. Zhang et al[19] found that miR-21 was markedly over-expressed in human GC tissues and cancer cell lines, and it repressed reversion-inducing cysteine-rich protein with Kazal motifs (RECK) expression, which resulted in the proliferation and development of GC. Another frequently reported miRNA related to GC cell proliferation is Let-7. The Let-7 miRNA family works as a negative regulator of the target high mobility group AT-hook 2 (HMGA2).[20] Hayashi et al[21] showed that CagA induced the epigenetic silence of let-7 expression, which triggered the up-regulation of Ras expression in Helicobacter pylori-related tumorigenesis of GC. Li et al[22] also found that miR-503 played a negative regulatory role in the proliferation of GC cells via the targeting of the 3′-UTR regions of HMGA2 mRNA. A couple of miRNAs related to GC cell proliferation target classic signaling pathways, such as the nuclear factor-kB (NF-κB) pathway. For example, miR-508-3p directly targets NF-κB1 and RELA (p65), and the inactivation of miR-508-3p may activate the canonical NF-κB signaling pathway in GC.[23] MiR-411 regulates the NF-κB signaling pathway via the targeting of SET domain containing 6.[24]
Recent studies showed that several miRNAs acted as onco-miRs that were pro-proliferative factors in GC. Protein-tyrosine phosphatase megakaryocyte 2 (MEG2), which is a target of miR-181a-5p, suppressed the proliferation and migration of tumor cells via the dephosphorylation of epidermal growth factor receptor (EGFR) and human epidermal growth factor receptor 2, and indirect inhibition of EGF-induced signal transducer and activator of transcription 3 (STAT3) activation.[25,26] Wu et al[27] demonstrated that miR-17-5p may play a pro-proliferative role in GC. It directly targeted suppressor of cytokine signaling 6 and promoted the proliferation of GC cells. MiR-92b also activated the phosphatidylinositol 3 kinase (PI3K)/protein kinase B (AKT) signaling pathway. Ni et al found that miR-92b played a supporting role by targeting DOC-2/DAB2 interacting protein in GC cell proliferation.[28] Wang et al[29] showed that the up-regulation of miR-664a-3p remarkably enhanced the proliferation and invasion via the Hippo pathway in vitro and in vivo. MiR-192 and miR-215 target adenomatous polyposis coli, which is a well-known negative regulator of Wnt signaling function during the tumorigenesis of colorectal cancer to play an oncogenic in GC proliferation and migration.[30] MiR-371a-3p is also a positive force in tumorigenesis via the targeting of transducer of human epidermal growth factor receptor 2, 1 (TOB1).[31] Myeloid-derived suppressor cells, which are marked by the myeloid differentiation factor Schlafen 4, showed increasing expression of miR-130b, which stimulated epithelial cell proliferation, promoted the induction of metaplasia, and advanced xenograft tumor growth.[32]
Researchers also found that miRNAs acted as tumor suppressive genes in GC. Matsuo et al[33] reported that miR-29c up-regulated regulator of chromosome condensation 2 and suppressed the growth of GC cells, and miR-29c antagomirs promoted proliferation. Another study found that miR-29c was lost in the inception phase of gastric tumorigenesis, and further experiments revealed that miR-29c directly targeted integrin β1 to act as a tumor suppressor.[34] MiR-203 over-expression and knockdown of its target PIBF1 suppressed tumor cell proliferation, and the knockdown of miR-203 promoted the growth of tumor cells.[35] YES proto-oncogene 1 (YES1) activates the Wnt/β-catenin pathway to promote cancer cell proliferation. MiR-140-5p negatively regulates YES1 expression and acts as a tumor suppressor in GC.[36,37] MiR-873 and retinoic acid 6 worked in a similar manner to miR-140-5p and YES1.[38]H. pylori infection induced interleukin-6 (IL-6) to influence STAT3 activity and suppressed the expression of miR-520d-5p. The inhibition of miR-520d-5p expression promoted cyclophilin B expression and caused activation of STAT3 and the Janus kinase (JAK)/STAT pathway, which resulted in the proliferation of GC cells.[39] Min et al[40] demonstrated that miR-30a played a tumor-suppressive role via mediation of integrin subunit α 2 in carcinogenesis. Jiang et al[41] showed that miR-1254 inhibited the PI3K/AKT signaling pathway via the targeting of Smurf1, which led to the suppression of tumor cell proliferation, migration, and invasion.
LncRNA and circRNAs participate in the progression of GC. An increasing number of researchers are paying attention to the interaction between miRNAs and other non-coding RNAs. LncRNA TRPM2-AS acts as a miRNA sponge of miR-612. Silence of TRPM2-AS suppressed GC cell proliferation, metastasis, and radio-resistance, and ectopic expression showed an obvious opposite effect.[42] CircRHOBTB3 acted as a sponge for miR-654-3p. Its over-expression produced decreased proliferation, G/S arrest in vitro and xenograft tumor growth inhibition in vivo.[43]
Apoptosis
Anomalous apoptosis is an important reason for cancer occurrence, development, and therapeutic inefficiency. Apoptosis-related genes or miRNAs are novel mechanisms for the study of the development of cancers, and the targeting of these molecules is a potential method for cancer therapeutics.
Zhang et al[44] reported that B-cell lymphoma-2-like protein 11 (BCL2L11) was an important factor mediating apoptosis progress, and miR-24 directly binds to the 3′-UTR of BCL2L11 mRNA to regulate its expression, which enhances GC cell growth and migration and inhibits apoptosis. Duan et al[45] recently found that regenerating gene IV (RegIV) was associated with GC peritoneal metastasis and 5-FU resistance, and it was a target of miR-24. The ectopic expression of miR-24 triggered tumor cell cycle blockade and apoptosis. Except for cancer cell proliferation, miR-17-5p/20a also exerted a pro-apoptotic effect in this experiment. Knockdown of miR-17-5p/20a promoted cancer cell apoptosis and resulted in cell cycle blockade.[46] MiR-1265 suppresses cell autophagy and proliferation and increases apoptosis via the regulation of calcium-binding protein 39 expression and the adenosine monophosphate-activated protein kinase-mammalian target of rapamycin signaling pathway.[47] Early experiments revealed that poor expression of miR-106b enhanced apoptosis of GC cells via inhibition of the JAK1/STAT3 signaling pathway, and arm protein lost in epithelial cancers on chromosome X 1 was a direct target of miR-106b.[48] Low miR-135b expression targeted mammalian sterile 20-like 1 to regulate the mitogen-activated protein kinase signaling pathway, which resulted in apoptosis and inhibition of cisplatin resistance of GC cells.[49] MiR-324-3p over-expression contributed to the loss of drosophila protein, mothers against decapentaplegic homolog 4 and activated the Wnt/β-catenin signaling pathway, which resulted in tumor cell growth and apoptosis inhibition. MiR-19a-3p regulated the expression of pituitary homeobox 1, which targeted genes that were all relating to apoptosis, induced apoptosis and suppressed growth in GC cells.[50] The above references showed that miRNAs target the main apoptotic genes or affect the signaling transduction to regulate cancer cell apoptosis.
Invasion and Metastasis
The main reason cancers cause high mortality is because the cells easily invade and metastasize to adjacent tissues or blood vessels. The mechanisms of invasion and metastasis are not clear. Studies of the mechanisms of tumor invasion and metastasis are urgent for early diagnosis. MiRNAs are related to the invasion and metastasis of GC and may be used as a tool for the detection and therapeutic targeting of the metastasis of cancers.
Many recent studies found that miRNAs played a pivotal role in the metastasis of GC cells, which involved many signaling pathways. The Wnt signaling pathway also produced a marked effect.[51] Peng et al[52] determined that miR-194 targeted suppressor of fused homolog (SUFU), which is an important negative regulator of Wnt and Hedgehog signaling. The anti-SUFU siRNAs eliminated the suppression of the antagonists and promoted tumor migration and proliferation. MiR-29c-3p mediated an inner ear-specific protein, cell migration inducing hyaluronan binding protein (KIAA1199 or CEMIP), expression and activated the fibroblast growth factor receptor 4/Wnt/β-catenin and EGFR signaling pathways, which induced metastasis behavior.[53] Kang et al[54,55] recognized that miR-15a and miR-16-1 acted as tumor suppressors that improved the metastasis and proliferation of GC via the targeting of YES-associated protein 1 and activation of the rapidly accelerates RAF/mitogen-activated protein kinase (MEK)/extracellular signal-regulated kinase (ERK) signaling pathway. Wang et al[56] found that the tumor-suppressive miRNA miR-218 inhibited metastasis via the POU class 2 homeobox 2-oriented network, including simultaneously mediating multiple molecules. The network also regulated the NF-κB and slit guidance ligand 2/a ligand of the roundabout 1 signaling pathways and modulated the metastasis of GC cells. The high expression of miR-27a was closely related to the metastatic characteristic of the patients’ clinical statistics. MiR-27a plays a pro-metastatic effect via inhibition of the PH domain and leucine-rich repeat protein phosphatase 2 and activation of the protein kinase B/glycogen synthase kinase 3 β (AKT/GSK3β) pathway.[57] MiR-939 suppressed the migration and invasion phenotype and decreased the incidence of lung metastasis in vivo via targeting of the solute carrier 34 A2 (SLC34A2) to inhibit the SLC34A2/Raf/MEK/ERK pathway.[58] Many studies clearly indicated the different targets of miRNAs in tumor cells. Wu et al[59] found that miR-19a/b, Max dimerization protein 1 and c-Myc created a positive feedback loop that facilitated the malignant phenotype metastasis. MiR-589 participated in an atypical miR-589-leukemia inhibitory factor receptor-PI3K/AKT-c-Jun feedback loop as a facilitating factor, which induced tumor cell metastasis and invasion.[60] MiR-647 suppressed GC cell migration and invasion via the serum response factor/myosin heavy chain 9 axis.[61] Down-regulation of miR-142-5p directly promoted the expression of the cysteine-rich angiogenic inducer 61 and promoted cell metastasis.[62] The silencing of miR-34a also up-regulated insulin-like growth factor 2 mRNA-binding protein 3 expression and promoted metastasis.[63] Blockade of autophagy-mediated glutaminolysis contributed to GC growth and metastasis, and Zhang et al[64] found that miR-133a-3p targeted gamma-aminobutyric acid type A receptor-associated protein-like 1 (GABARAPL1) to trigger this process. He et al[65] found that hypoxia-inducible miR-224 acted as an oncogene to promote the migration and invasion phenotype via targeting and lowering Ras-association domain family 8 (RASSF8) expression. Knockdown of RASSF8 promoted the transcriptional activity of NF-κB and p65 translocation. Mousa et al[66] showed that miR-4316 inhibited vascular endothelial growth factor A (VEGF-A) expression and induced the suppression of migration and colony formation in tumor cell lines. β-Elemene extracted from Chinese herbal medicine showed a good effect on the inhibition of metastasis in nude mice. Deng et al[67] confirmed that β-elemene suppressed miR-1323 and up-regulated casitas B lymphoma-b (Cbl-b) expression to achieve these results.
Angiogenesis is critical for the progression of invasion and metastasis. MiR-21 played an important role during tumor angiogenesis via the targeting of RECK and regulation of matrix metalloproteinases.[19] Ye et al[68] discovered that the low expression of miR-7 activated p65-mediated abnormal NF-κB expression and promoted metastasis, which ultimately led to a poor prognosis. The delivery of miR-7 in BALB/c mice produced therapeutic effects via the inhibition of angiogenesis, lymph node metastases and inflammation. Zhang et al[69] demonstrated that low miR-29a/c expression promoted the release of VEGF in GC cells, which enhanced vascular cell growth and angiogenesis. Exosomal miR-155 caused tumor angiogenesis via the down-regulation of c-Myb and up-regulation VEGF, but it also suppressed the expression of Forkhead box O3 protein.[70,71] MiR-23a derived from exosomes promoted tumor angiogenesis via suppression of phosphatase and tensin homolog (PTEN) expression.[72]
Interaction with other non-coding RNAs also affects invasion behavior. LncRNA RGMB-AS1 functioned as a miR-574 sponge and up-regulated histone deacetylase 4 to promote aggressiveness behavior.[73] LncRNA UCA1 is a competing endogenous RNA of miR-495 that up-regulates phosphatase of regenerating liver-3 expression to accelerate the development of GC.[74] MiR-150-5p and its sponge circLMTK2 showed a positive effect on proliferation and metastasis in GC via the regulation of c-Myc expression.[75] The above studies demonstrated the indispensable roles of miRNAs in the process of migration, invasion, and metastasis of GC.
Drug Sensitivity and Drug Resistance
Chemotherapy is the standard therapy for advanced GC. Chemotherapy is useful in the treatment of advanced malignant tumor patients and extends their survival time. However, multi-drug resistance generally leads to treatment failure and a poor prognosis of patients. Drug sensitivity and drug resistance are common in diverse cancer cells, and many factors influence the drug response. Commonly administered chemotherapeutic agents include platinum drugs and 5-fluorouracil. The roles of miRNAs in the modulation of chemo-sensitivity and chemo-resistance have received increased attention.
MiRNAs have the capacity to affect drug sensitivity via specific genes or signaling pathways. Wang et al[76] found that cisplatin (DDP) and docetaxel (DOC) restored the expression of miR-29 family members. Via suppression of catenin-δ and Ras homolog gene family member A (RhoA) signaling, MiR-29 family members mediate a significant part of the efficacious benefits of patients via suppression of catenin-δ and Ras homolog gene family member A (RhoA) signaling, especially patients who experienced recurrence after chemotherapy. MiR-148a-3p conferred mitochondrial fission and suppressed cyto-protective autophagy by enhancing cisplatin cytotoxicity in experiments.[77] Chen et al[78] found a decreased concentration of miR-143-3p in gastric tumor tissues, and miR143-3p transgenic expression in cell lines diminished proliferation and regenerated chemosensitivity to cisplatin via the targeting of bromodomain-containing protein 2. Suppression of miR-633 expression significantly enhanced doxorubicin (DOX)/DDP-induced GC cell apoptosis in vitro by increasing Fas-associated protein with death domain protein levels, and miR-633 antagomirs in combination with DOX prominently decreased tumor growth and sensitized tumors cells to chemotherapy.[79] A positive feedback loop played an important role in the chemoresistance to oxaliplatin and metastasis via epithelial-mesenchymal transition (EMT) and stemness-like properties. The over-expression of miR-577 suppressed SDPR expression and released a complex that included serum deprivation protein response protein to activate the ERK-NF-κB pathway, which up-regulated the transcription of miR-577.[80] MiR-508-5p also influenced multi-drug resistance. High expression of miR-508-5p targeted P-glycoprotein and zinc ribbon domain-containing 1 and changed the chemo-resistant phenotype of tumor cells.[81] Shao et al[82] found that a high level of miR-135b-5p suppressed apoptosis and induced cisplatin resistance. Zheng et al[83] demonstrated that E2F transcription factor 1 (E2F1) decreased miR-34c expression to promote resistance to paclitaxel combined with cisplatin in GC cells. Liu et al[84] created nanoparticles (NPs) that were stimulated by gelatinase to co-transport miR-200c and docetaxel, and the results showed that the released miR-200c had a significantly synergistic effect of docetaxel. Exosome-delivered anti-miRs also have chemo-sensitive and chemo-resistant functions. For example, miR-374a-5p over-expression promoted a chemo-resistance characteristic of GC cells, and miR-374a-5p knockdown had an opposite effect. Exosome-delivered anti-miR-374a-5p promoted cell apoptosis and reversed oxaliplatin resistance via enhancing neurogenic differentiation 1 levels.[85] Wang et al[86] also reported that exosome-mediated delivery of a miR-214 inhibitor reversed a DDP-resistant phenotype. They found that gradually injecting exo-anti-214 in nude mice reduced GC cell growth and restored sensitivity to DDP. Exosomal transfer of the oncogenic miRNA miR-501 inhibited cell death inducer expression, inactivated caspases-9/-3 and induced phosphorylation of AKT, which led to a rapid growth rate of tumors and resistance to DOX.[87]
Chemotherapy with fluorouracil is also widely used in the clinic for GC treatment. Li et al[88] demonstrated that treatment with 5-fluorouracil increased the expression of miR-204 and inhibited the transforming growth factor-β (TGF-β) pathway, and high miR-204 expression may reverse the chemotherapy resistance via the targeting of TGF-β receptors 1. Zhang et al[89] found that miR-567 exerted its chemo-sensitive function to 5-fluorouracil and oxaliplatin in tumor cells primarily by a miR-567-PIK3AP1-PI3K/AKT-c-Myc feedback loop. Another report showed that miR-939 enhanced 5-fluorouracil-induced chemo-sensitivity via inhibition of the SLC34A2/Raf/MEK/ERK pathway, which was activated in research studies.[58] For TNF-related apoptosis-inducing ligand (TRAIL), which is a novel and low-toxic antitumor drug, one study showed that GC miR-494 expression level was associated with a TRAIL-sensitive phenotype. The over-expression of miR-494 decreased survivin levels and established an axis for a potential mechanism to promote TRAIL-induced mitochondrial collapse and apoptosis pathway.[90] Notably, Wei et al[91] found that a well-known neurotransmitter related to stress, isoproterenol, also led to chemo-resistance in GC cells, and transfection with miR-373 inhibited the effects of isoproterenol on drug sensitivity. Interestingly, different non-coding RNAs, such as miRNAs, lncRNAs, and circRNAs, are critically involved in GC drug-resistant phenotype development.[92] Apatinib is a novel and highly selectively targeted agent of anti-angiogenesis, and it showed great prospects as an advanced GC patient treatment. The silencing of an endogenous sponge for miR-3657, circRACGAP1, suppressed apatinib-induced autophagy. MiR-3657 rescued this effect, which may increase safety.[93] For example, circAKT3, as a sponge for miR-198, increased predominant regulatory isoform of PI3K expression to confer DDP resistance and suppress apoptosis of tumor cells.[94] Similarly, there some cirRNAs contribute to DDP resistance via interaction with miRNAs, such as circFN1 and circPVT1.[95,96]
Treatments with these miRNAs may be a novel approach to reverse drug resistance. The above research achievements are meaningful in the clinical application of drug-resistant diagnosis and further chemotherapy drug sensitization treatments, especially for advanced patients.
Stemness of Cancer Stem Cells
CSCs are an important origin for cancer occurrence and recurrence. CSCs are the origin of cancer and targeting CSCs may be of significant clinical value in the treatment of GC. One miRNA may regulate CSCs via various target genes and have an amplifying effect in the regulation of CSC properties.
CD44 is a CSC marker. Mounting evidence shows that the ectopic expression of CD44 also influences the stemness of cancer cells.[97] MiR-34a, a key regulator of tumor suppression, decreases CD44 level.[98,99] Jang et al[100] designed nanovesicles containing poly L-lysine-graft-imidazole (PLI)/miR, PLI-condensing miRs, and found that the application of this PLI/miR-34a-containing nanovesicle to nude mice suppressed the expression of CD44. They further found that in CD44-positive tumor-bearing mouse models, this PLI/miR-34a-containing nanovesicle also significantly delayed tumor growth in CD44-positive tumor-bearing mouse models. MiR-200c NPs and miR-200c/DOC NPs also negatively regulated CD44 expression and elevated E-cadherin levels and were further reported to inhibit CSC stemness and reverse EMT.[84] Similarly, Li et al[101] also found that miR-149 expression was markedly down-regulated in cancer-associated fibroblasts, which promoted stem-like properties and EMT of tumor cells. B-lymphoma Moloney murine leukemia virus insertion region-1 (Bmi-1), another known stem cell marker, regulated stemness phenotype in various CSCs. Wang et al[102] found that miR-34a regulated stem cell-like properties in GC via the regulation of p53 and PTEN-AKT signaling through the c-Myc/Bmi-1/miR-21 pathway.
MiR-21 was also up-regulated by chromobox protein homolog 7 and maintained the stem cell-like features of GC.[103] MiR-26a targeted homeobox transcription factors C9 (HOXC9) promoted the stemness phenotype and metastasis of tumor cells.[104] Some classic signaling pathways are also regulated by miRNAs. For example, miR-501-5p directly targeted and inhibited several repressors of the Wnt/β-catenin signaling cascade to enhance a stem-cell-like phenotype.[105] In Drosha-silenced or Drosha low-expressing GC cells, miR-6778-5p up-regulated serine hydroxymethyl transferase 1 to mediate a compensatory activation of cytoplasmic carbon metabolism, which was essential for the maintenance of CSCs in GC.[106] IL-1 signaling up-regulated the expression of miR-135b, promoted stem-cell characteristics and the invasiveness of tumor cells via decreasing of RECK and forkhead box N3 (FOXN3) mRNAs.[107] Because CSCs that endow malignant phenotypes to normal cancer cells are regarded as the initiation of cancer, miRNAs in the regulation of CSCs also regulate other malignant phenotypes, such as growth, metastasis, and chemo-resistance, even in precancerous lesions. For example, the miRNA-17-92 cluster includes seven members: miR-17-5p, miR-17-3p, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92a. Wu et al[108] reported that the over-expression of miR-17-92 cluster members (miR-92a, miR-20a, and miR-19b) maintained the self-renewal ability of gastric CSCs and promoted proliferation of tumor cells, perhaps via the targeting of E2F1 and homeodomain-interacting protein kinase 1 and inhibition of the Wnt/β-catenin signaling pathway. The same group found that a member of the miR-17-92 cluster, miR-17-5p, promoted proliferation and miR-19a/b promoted metastasis in vitro and in vivo.[27,59] They also found that another member of this cluster, miR-92a-1-5p, influenced the formation of gastric intestinal metaplasia via the targeting of Forkhead box D1 and regulation of the NF-κB/caudal-type homeobox 2 axis.[109] LncRNA HCP5 from mesenchymal stem cells enhanced the characteristics of stem cells and induced chemo-resistance. It sequestered miR-3619-5p and increased peroxisomal proliferating-activated receptor gamma co-activator 1 alpha expression, which enhanced fatty acid oxidation in GC cells.[110]
Precancerous Inflammatory Lesions
Inflammation in the stomach caused by H. pylori is a significant risk factor for the occurrence and development of GC. Abundant miRNAs are reported as significant tools and markers in cancer and gastric inflammation, such as miR-21, miR-143, miR-145, and miR-155. These miRNAs play roles in cell cycle growth or tumor invasion. For example, high miR-15 expression down-regulated the expression of some genes that are involved in a defective DNA mismatch repair system, which leads to gastric pathologies.[111] NF-κB played an essential role in the process from H. pylori-mediated inflammation to cancer, such as combination with the miR-223-3p promoter. H. pylori infection activated miR-7 via NF-κB signaling, regulation of RELA expression via direct and indirect signaling, which regulated the process of gastric carcinogenesis.[112] CagA-induced MiR-223-3p directly targeted AT-rich interactive domain 1A to play a role in cell proliferation and migration, which provided an explanation of carcinogenesis.[113] Li et al[114] reported that IL-17A was related to H. pylori infection in the gastric mucosa. IL-17A-induced miR-146a regulated the inflammatory response via NF-κB during the H. pylori-induced inflammation. An inflammasome is forms via activation of the innate immune signaling complex when bacterial infections occur.[115] NOD-like receptor protein 3 (NLRP3) is an essential component of the inflammasome, and it accelerated the proliferation of epithelial cells and induced gastric carcinogenesis. Li et al[116] demonstrated that H. pylori infection inhibited miR-22 expression, which resulted in NLRP3 up-regulation and represented oncogenic effects. Hsa-miR-223-3p and cytokine IL-10 work in tandem to control NLRP3 expression.[117] MiR-3178 targeted TNF receptor-associated factor 3, which activated NF-κB and alleviated inflammation and gastric carcinogenesis.[118] The effect of H. pylori-induced inflammation on the occurrence of GC may be further elucidated via the identification of the miRNAs mechanism of action.
Roles of MiRNAs in the Clinical Application of Gastric Cancer
Diagnostic prediction
Multiple miRNA expression signatures in tissues may contain many information for GC diagnosis. For example, the differential expression of miRNAs in cancer tissues may be used as a representation of different types of GC. Four miRNAs (miR-138, miR-196a, miR-150, and miR-155) were specifically over-expressed and two miRNAs (miR-7 and miR-153) were decreased in the tissues of gastric mucosa-associated lymphoid tissue (MALT) lymphomas patients.[119] Another study in Japan showed that eight miRNAs (miR-145, miR-133a, miR-125b, miR-99a, miR-105, miR-143, miR-199a, and miR-100) were over-expressed in diffuse-type GC, and four miRNAs (miR-202, miR-494, miR-373, and miR-498) were over-expressed in intestinal-type lesions.[14] Studies also showed the outcome prediction value of miRNA expression signatures. Li et al[120] identified a seven-miRNA signature in paraffin-embedded GC tissues, the miR-30a-5p, miR-21, miR-126, miR-338, let-7a, miR-10b, and miR-223 expression signature may be used as a predictor for overall survival (hazard ratio [HR] = 3.046) and relapse-free survival (HR = 3.337) of tumor patients.
The 5-year survival rate of GC is poor, and greater than 80% of patients are diagnosed at an advanced stage rather than an early stage. Most patients with GC are symptomless or present atypical symptoms at the early stage. Therefore, early detection is important and imperative. MiRNAs may serve as a useful tool for the earlier detection of GC. For example, the increased expression of miR-200 family members was associated with early GC (stage IA) in patients.[121] Serum levels of miR-25 improved GC screening with a sensitivity and specificity of 67.3% to 69.4% and 80.4% to 81.0%, respectively.[122] Zhu et al[123] demonstrated that some miRNAs (miR-425-5p, miR-1180-3p, miR-122-5p, miR-24-3p, and miR-4632-5p) in circulating plasma discriminated early stage patients from atrophic gastritis patients.
With the growing number of patients with GC, the lack of non-invasive detection methods for early diagnosis is one of the biggest problems in China. Non-invasive body fluid tests for the detection of cancers are a satisfactory method. However, the present serum tumor biomarkers for GC, such as carcinoembryonic antigen, have a low positive rate.[124] MiRNAs are small and relatively stable in blood, which is easier for testing. Therefore, the detection of circulating tumor-specific miRNAs has become a novel method for the diagnosis and prognosis of tumors. Different miRNAs were found and detected in the blood of patients with carcinoma of the stomach, and some of these markers may be used for further diagnosis and prediction of prognosis.[3] Shin et al[125] recognized a three-miRNA signature (miR-652, miR-629, and miR-627) as a promising classifier for GC using microarray screening. They tested the signature in patient plasma samples and got a sensitivity of 86.7% and specificity of 85.5%. Huang et al[126] also identified a six-miRNA panel in serum, and the area under the receiver-operating characteristic curve was 0.702 in the validation phases.
Except for tissue and blood, detecting miRNA expression in other fluids, such as gastric juice and urine, may also be a useful method for diagnosis. MiR-21 and miR-106a levels in gastric juice were obviously different in patients with GC compared to those with benign gastric diseases. The value of the area under the receiver-operating characteristic curve was up to 0.969 and 0.871, respectively.[127] A meta-analysis revealed that the testing of gastric juice miRNAs was reliable and reproducible and may become a new biologic marker for the screening of gastric carcinoma.[128] MiR-6807-5p and miR-6856-5p expression in urine was higher in patients with GC, their expression levels decreased after curative resection, which supports their use as novel biomarkers for diagnosis.[129]
Prognostic prediction
Except for diagnostic prediction, miRNAs also have prognostic evaluation value in GC. For example, the expression of miR-200 family members provided superior predictive power for overall survival compared to single indicators.[121] Ueda et al[130] verified that low expression of let-7g (HR = 2.600) and miR-433 (HR = 2.100) and high expression of miR-214 (HR = 2.400) were related to poor outcome in patients with GC.[14] The expression of miR-153 in tumor tissues was associated with worse overall survival (HR = 0.253) and disease-free survival (HR = 0.228) in patients with GC. Guo et al[131] found that high hsa-mir-3923 expression in patients with GC may be an independent prognosis element for poor survival. Chen et al developed a prognostic model that included six miRNAs (miRNA-549, miRNA-100, miRNA-653, miRNA-374a, miRNA-668, and miRNA-509-3). This miRNA signature differentiated between high- and low-risk patients (HR = 1.699) and forecast 3- and 5-year survival.[132] MiR-1236-3p expression in tumor tissues was significantly declined, which was an independent prognostic factor for overall survival in patients with GC.[133]
MiRNA expression in blood samples may also be used for prognostic prediction. Multivariate analysis showed that low miR-203 levels and high miR-25 levels in serum independently forecast metastasis and poor prognosis for patients with GC.[122,134] Hou et al[135] confirmed that low serum miRNA-206 expression may be used for diagnosis and to predict the recurrence of gastric carcinoma and patients’ prognosis. Liu et al[136] found that 7 miRNAs (miR-125b, let-7e, miR-222, miR-148a, miR-21, miR-26a, and miR-126) in plasma predicted the recurrence and prognosis of GC independently or in combination with a pathologic factors index (HR = 1.839 of seven miRNAs and HR = 3.379 of the combination in validation set). Further study showed that patients with stages II and III GC benefited from the prognostic prediction. Nishibeppu et al[137] reported that high levels of miR-1229-3p in plasma were an independent predictor of poor prognosis for recurrence-free survival in GC (HR = 3.710).
Previously published work provided information on specific miRNA expression, and a ton of data demonstrated that these miRNAs exhibited decent sensitivity and specificity for the diagnosis and prognosis prediction of GC. Although more independent extensive groups or cohorts are needed to verify the conclusions of previous studies, miRNAs may be a group of new, non-invasive biomarkers with ample accuracy for GC diagnostic and prognostic analyses.
Cancer treatment
The treatment for GC includes operative and non-operative treatments. Although surgery is conventional for solid tumors, over half of the patients experience a relapse or general metastasis.[10] Therefore, non-operative treatments are indispensable. The clinical use of miRNAs as a treatment for GC remains in the experimental phase in helping to enhance chemo-sensitivity or chemotherapy response. An ongoing clinical trial focused on the discovery and validation of miRNAs in predicting the effect of chemotherapy response in patients with GC. These researchers tried to combine biologic biomarkers and clinical factors to establish a prediction model for promoting the response of chemotherapy in GC (data from https://clinicaltrials.gov).
Except for being chemo-sensitive or chemo-resistant, miRNAs may also be used to enhance therapeutic efficacy in cancers. MiRNA-based drugs may have bright application prospects. Some miRNAs are being tested as an miRNA-based cancer therapy in clinical trials. A clinical trial of MRX34 (Mirna Therapeutics, Austin, Texas, USA) constructed a miR-34 mimetic to re-express miR-34 in cancer cells. The experiment was performed in patients with primary liver cancer or liver metastases in a phase I clinical trial protocol.[138,139] In a pre-clinical study for non-small cell lung cancer, MRX34 treatment had marked effects in decreasing the expression of the checkpoint signal PD-L1 and enhancing the expression of CD8(+) cells in cancer tissues.[140] MRX34 showed some curative effect in this experiment, but this clinical trial was stopped in 2016 because of the various and severe immune side effects in patients.[141]
It is obvious that the safety and efficiency of miRNA-based drugs remain a problem. More studies are required to focus on the use of miRNAs in laboratory-to-clinical transformation.
Prospect
As one of the most common malignant tumors in China, GC causes a high mortality rate annually. Recent research of miRNAs and cancers has drawn great attention from scientists, and many researchers examined the relationship between miRNAs and GC from laboratory to clinical applications. Numerous miRNAs were found and their functions, properties and signaling pathways were studied. However, it is important to study the interaction networks between miRNAs and investigate whether these molecules work synergistically or counteract each other. Problems still exist. The results of these studies are not consistent, and various miRNAs exhibited conflicting results of expression in different cohorts.[11] The expressions of miRNAs are the characteristic of heterogeneity in different studies, especially circulating miRNAs.[142,143] Even if specific miRNAs correlate with tumorigenesis and have diagnostic or prognostic values, most of the studies were case control studies with small sample sizes. Further investigation with large-scale validation is needed. Problems occurred during clinical transformation, and abundant evidence is needed before miRNAs may be adopted to treat people with cancer. Additionally, there are few large-scale investigations on the safety and efficacy of miRNA-based drugs.
The development of cancer is a multi-gene and multi-step continuous dynamic process, and we still have a long way to go. Fortunately, recent studies showed the broad prospects of miRNAs. The relatively stable property of miRNAs in the circulation makes them suitable for use as circulating biomarkers in cancer diagnostic and prognostic analysis. Furthermore, the acquisition of a miRNA signature compared to a single miRNA is more likely to provide useful information for clinical use on the individual-based management of diseases. Elucidation of the functions and relationships between these miRNAs and their target genes will increase the prospect for the effective therapeutic method for patients with GC. To achieve this goal, the optimization of laboratory techniques and experiment flow is necessary. Up until the present moment, the effect of miRNA application in the early diagnosis and risk stratification of precancerous lesions of GC is not clear. The application of miRNAs from laboratory to clinical transformation in therapeutics remains a big problem, but these molecules may be a promising research direction.
Conclusions
Abundant research supports that miRNAs provide a novel understanding in the occurrence and development of GC. MiRNAs may function to target mRNAs for regulation and interaction with other non-coding RNAs. The characteristics of miRNAs, like presence in accessible body fluids, make miRNAs promising biomarkers and potential targets for therapeutic interventions. Although converting research of basic medicine to clinical practice remains a huge challenge, experimental results offer hope for further research.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81873554) and the Shaanxi Foundation for Innovation Team of Science and Technology (No. 2018TD-003).
Conflicts of interest
None.
Footnotes
How to cite this article: Wu SR, Wu Q, Shi YQ. Recent advances of miRNAs in the development and clinical application of gastric cancer. Chin Med J 2020;133:1856–1867. doi: 10.1097/CM9.0000000000000921
Si-Ran Wu and Qiong Wu contributed equally to this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Soerjomataram I, Lortet-Tieulent J, Parkin DM, Ferlay J, Mathers C, Forman D, et al. Global burden of cancer in 2008: a systematic analysis of disability-adjusted life-years in 12 world regions. Lancet 2012; 380:1840–1850. doi: 10.1016/S0140-6736(12)60919-2. [DOI] [PubMed] [Google Scholar]
- 3.Link A, Kupcinskas J. MicroRNAs as non-invasive diagnostic biomarkers for gastric cancer: current insights and future perspectives. World J Gastroenterol 2018; 24:3313–3329. doi: 10.3748/wjg.v24.i30.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet 2016; 388:2654–2664. doi: 10.1016/s0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 5.Hundahl SA, Phillips JL, Menck HR. The National Cancer Data Base Report on poor survival of U.S. gastric carcinoma patients treated with gastrectomy: Fifth Edition American Joint Committee on Cancer staging, proximal disease, and the “different disease” hypothesis. Cancer 2000; 88:921–932. [PubMed] [Google Scholar]
- 6.Zhang X, Li M, Chen S, Hu J, Guo Q, Liu R, et al. Endoscopic screening in Asian countries is associated with reduced gastric cancer mortality: a meta-analysis and systematic review. Gastroenterology 2018; 155:347–354.e9. doi: 10.1053/j.gastro.2018.04.026. [DOI] [PubMed] [Google Scholar]
- 7.Zong L, Abe M, Seto Y, Ji J. The challenge of screening for early gastric cancer in China. Lancet 2016; 388:2606.doi: 10.1016/s0140-6736(16)32226-7. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature 2004; 431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol 2014; 20:10432–10439. doi: 10.3748/wjg.v20.i30.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao NB, He YF, Li XQ, Wang K, Wang RL. The role of miRNA and lncRNA in gastric cancer. Oncotarget 2017; 8:81572–81582. doi: 10.18632/oncotarget.19197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 13.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006; 103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol 2010; 11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jinawath N, Furukawa Y, Hasegawa S, Li M, Tsunoda T, Satoh S, et al. Comparison of gene-expression profiles between diffuse- and intestinal-type gastric cancers using a genome-wide cDNA microarray. Oncogene 2004; 23:6830–6844. doi: 10.1038/sj.onc.1207886. [DOI] [PubMed] [Google Scholar]
- 16.Stock M, Otto F. Gene deregulation in gastric cancer. Gene 2005; 360:1–19. doi: 10.1016/j.gene.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 17.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006; 6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 18.Oliveira KCD, Araujo TMT, Albuquerque CI, Barata GA, Gigek CO, Leal MF, et al. Role of miRNAs and their potential to be useful as diagnostic and prognostic biomarkers in gastric cancer. World J Gastroentero 2016; 22:7951–7962. doi: 10.3748/wjg.v22.i35.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, et al. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest 2008; 88:1358–1366. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- 20.Motoyama K, Inoue H, Nakamura Y, Uetake H, Sugihara K, Mori M. Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin Cancer Res 2008; 14:2334–2340. doi: 10.1158/1078-0432.CCR-07-4667. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi Y, Tsujii M, Wang J, Kondo J, Akasaka T, Jin Y, et al. CagA mediates epigenetic regulation to attenuate let-7 expression in Helicobacter pylori-related carcinogenesis. Gut 2013; 62:1536–1546. doi: 10.1136/gutjnl-2011-301625. [DOI] [PubMed] [Google Scholar]
- 22.Li W, Li J, Mu H, Guo M, Deng H. MiR-503 suppresses cell proliferation and invasion of gastric cancer by targeting HMGA2 and inactivating WNT signaling pathway. Cancer Cell Int 2019; 19:164.doi: 10.1186/s12935-019-0875-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang T, Kang W, Zhang B, Wu F, Dong Y, Tong JH, et al. miR-508-3p concordantly silences NFKB1 and RELA to inactivate canonical NF-kappaB signaling in gastric carcinogenesis. Mol Cancer 2016; 15:9.doi: 10.1186/s12943-016-0493-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bai TL, Liu YB, Li BH. MiR-411 inhibits gastric cancer proliferation and migration through targeting SETD6. Eur Rev Med Pharmacol Sci 2019; 23:3344–3350. doi: 10.26355/eurrev_201904_17697. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z, Sun F, Hong Y, Liu Y, Fen M, Yin K, et al. MEG2 is regulated by miR-181a-5p and functions as a tumor suppressor gene to suppress the proliferation and migration of gastric cancer cells. Mol Cancer 2017; 16:133.doi: 10.1186/s12943-017-0695-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan T, Wang Y, Zhao ZJ, Gu H. Protein-tyrosine phosphatase PTPN9 negatively regulates ErbB2 and epidermal growth factor receptor signaling in breast cancer cells. J Biol Chem 2010; 285:14861–14870. doi: 10.1074/jbc.M109.099879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Q, Luo GH, Yang ZP, Zhu F, An YX, Shi YQ, et al. miR-17-5p promotes proliferation by targeting SOCS6 in gastric cancer cells. Febs Lett 2014; 588:2055–2062. doi: 10.1016/j.febslet.2014.04.036. [DOI] [PubMed] [Google Scholar]
- 28.Ni QF, Zhang Y, Yu JW, Hua RH, Wang QH, Zhu JW. miR-92b promotes gastric cancer growth by activating the DAB2IP-mediated PI3K/AKT signalling pathway. Cell Prolif 2020; 53:e12630.doi: 10.1111/cpr.12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Li B, Zhang L, Li Q, He Z, Zhang X, et al. miR-664a-3p functions as an oncogene by targeting Hippo pathway in the development of gastric cancer. Cell Prolif 2019; 52:e12567.doi: 10.1111/cpr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Deng S, Zhang X, Qin Y, Chen W, Fan H, Feng X, et al. miRNA-192 and -215 activate Wnt/beta-catenin signaling pathway in gastric cancer via APC. J Cell Physiol 2020; [Epub ahead of print]. doi: 10.1002/jcp.29550. [DOI] [PubMed] [Google Scholar]
- 31.Guo H, Ji F, Zhao X, Yang X, He J, Huang L, et al. MicroRNA-371a-3p promotes progression of gastric cancer by targeting TOB1. Cancer Lett 2019; 443:179–188. doi: 10.1016/j.canlet.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 32.Ding L, Li Q, Chakrabarti J, Munoz A, Faure-Kumar E, Ocadiz-Ruiz R, et al. MiR130b from Schlafen4(+) MDSCs stimulates epithelial proliferation and correlates with preneoplastic changes prior to gastric cancer. Gut 2020; [Epub ahead of print] doi: 10.1136/gutjnl-2019-318817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuo M, Nakada C, Tsukamoto Y, Noguchi T, Uchida T, Hijiya N, et al. MiR-29c is downregulated in gastric carcinomas and regulates cell proliferation by targeting RCC2. Mol Cancer 2013; 12:15.doi: 10.1186/1476-4598-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han TS, Hur K, Xu G, Choi B, Okugawa Y, Toiyama Y, et al. MicroRNA-29c mediates initiation of gastric carcinogenesis by directly targeting ITGB1. Gut 2015; 64:203–214. doi: 10.1136/gutjnl-2013-306640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu SJ, Wang G, Zhang PF, Zhang R, Huang YX, Lu YM, et al. MicroRNA-203 suppresses gastric cancer growth by targeting PIBF1/Akt signaling. J Exp Clin Cancer Res 2016; 35:47.doi: 10.1186/s13046-016-0323-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Fang Z, Yin S, Sun RC, Zhang SX, Fu M, Wu YL, et al. miR-140-5p suppresses the proliferation, migration and invasion of gastric cancer by regulating YES1. Mol Cancer 2017; 16:139.doi: 10.1186/s12943-017-0708-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, et al. Beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell 2012; 151:1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin L, Xiao J, Shi L, Chen W, Ge Y, Jiang M, et al. STRA6 exerts oncogenic role in gastric tumorigenesis by acting as a crucial target of miR-873. J Exp Clin Cancer Res 2019; 38:452.doi: 10.1186/s13046-019-1450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li T, Guo HQ, Zhao XD, Jin J, Zhang LF, Li H, et al. Gastric cancer cell proliferation and survival is enabled by a cyclophilin B/STAT3/miR-520d-5p signaling feedback loop. Cancer Res 2017; 77:1227–1240. doi: 10.1158/0008-5472.Can-16-0357. [DOI] [PubMed] [Google Scholar]
- 40.Min J, Han TS, Sohn Y, Shimizu T, Choi B, Bae SW, et al. microRNA-30a arbitrates intestinal-type early gastric carcinogenesis by directly targeting ITGA2. Gastric Cancer 2020; [Epub ahead of print]. doi: 10.1007/s10120-020-01052-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang M, Shi L, Yang C, Ge Y, Lin L, Fan H, et al. miR-1254 inhibits cell proliferation, migration, and invasion by down-regulating Smurf1 in gastric cancer. Cell Death Dis 2019; 10:32.doi: 10.1038/s41419-018-1262-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao J, Lin L, Luo D, Shi L, Chen W, Fan H, et al. Long noncoding RNA TRPM2-AS acts as a microRNA sponge of miR-612 to promote gastric cancer progression and radioresistance. Oncogenesis 2020; 9:29.doi: 10.1038/s41389-020-0215-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng G, Mou T, He J, Chen D, Lv D, Liu H, et al. Circular RNA circRHOBTB3 acts as a sponge for miR-654-3p inhibiting gastric cancer growth. J Exp Clin Cancer Res 2020; 39:1.doi: 10.1186/s13046-019-1487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Duan J, Qu Y, Deng T, Liu R, Zhang L, et al. Onco-miR-24 regulates cell growth and apoptosis by targeting BCL2L11 in gastric cancer. Protein Cell 2016; 7:141–151. doi: 10.1007/s13238-015-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duan Y, Hu L, Liu B, Yu B, Li J, Yan M, et al. Tumor suppressor miR-24 restrains gastric cancer progression by downregulating RegIV. Mol Cancer 2014; 13:127.doi: 10.1186/1476-4598-13-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang M, Gu H, Qian H, Zhu W, Zhao C, Zhang X, et al. miR-17-5p/20a are important markers for gastric cancer and murine double minute 2 participates in their functional regulation. Eur J Cancer 2013; 49:2010–2021. doi: 10.1016/j.ejca.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 47.Xu Z, Li Z, Wang W, Xia Y, He Z, Li B, et al. MIR-1265 regulates cellular proliferation and apoptosis by targeting calcium binding protein 39 in gastric cancer and, thereby, impairing oncogenic autophagy. Cancer Lett 2019; 449:226–236. doi: 10.1016/j.canlet.2019.02.026. [DOI] [PubMed] [Google Scholar]
- 48.Zhu Z, Yang Q, Zhang B, Wu W, Yuan F, Zhu Z. miR-106b promotes metastasis of early gastric cancer by targeting ALEX1 in vitro and in vivo. Cell Physiol Biochem 2019; 52:606–616. doi: 10.33594/000000043. [DOI] [PubMed] [Google Scholar]
- 49.Zhou J, Chen Q. Poor expression of microRNA-135b results in the inhibition of cisplatin resistance and proliferation and induces the apoptosis of gastric cancer cells through MST1-mediated MAPK signaling pathway. FASEB J 2019; 33:3420–3436. doi: 10.1096/fj.201800618RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiao F, Gong P, Song Y, Shen X, Su X, Li Y, et al. Downregulated PITX1 modulated by MiR-19a-3p promotes cell malignancy and predicts a poor prognosis of gastric cancer by affecting transcriptionally activated PDCD5. Cell Physiol Biochem 2018; 46:2215–2231. doi: 10.1159/000489590. [DOI] [PubMed] [Google Scholar]
- 51.Ashrafizadeh M, Rafiei H, Mohammadinejad R, Farkhondeh T, Samarghandian S. Wnt-regulating microRNAs role in gastric cancer malignancy. Life Sci 2020; 250:117547.doi: 10.1016/j.lfs.2020.117547. [DOI] [PubMed] [Google Scholar]
- 52.Peng Y, Zhang XJ, Ma Q, Yan RB, Qin Y, Zhao YQ, et al. MiRNA-194 activates the Wnt/beta-catenin signaling pathway in gastric cancer by targeting the negative Wnt regulator, SUFU. Cancer Lett 2017; 385:117–127. doi: 10.1016/j.canlet.2016.10.035. [DOI] [PubMed] [Google Scholar]
- 53.Wang L, Yu T, Li W, Li M, Zuo Q, Zou Q, et al. The miR-29c-KIAA1199 axis regulates gastric cancer migration by binding with WBP11 and PTP4A3. Oncogene 2019; 38:3134–3150. doi: 10.1038/s41388-018-0642-0. [DOI] [PubMed] [Google Scholar]
- 54.Kang W, Tong JH, Lung RW, Dong Y, Zhao J, Liang Q, et al. Targeting of YAP1 by microRNA-15a and microRNA-16-1 exerts tumor suppressor function in gastric adenocarcinoma. Mol Cancer 2015; 14:52.doi: 10.1186/s12943-015-0323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang W, Tong JH, Chan AW, Lee TL, Lung RW, Leung PP, et al. Yes-associated protein 1 exhibits oncogenic property in gastric cancer and its nuclear accumulation associates with poor prognosis. Clin Cancer Res 2011; 17:2130–2139. doi: 10.1158/1078-0432.CCR-10-2467. [DOI] [PubMed] [Google Scholar]
- 56.Wang SM, Tie J, Wang WL, Hu SJ, Yin JP, Yi XF, et al. POU2F2-oriented network promotes human gastric cancer metastasis. Gut 2016; 65:1427–1430. doi: 10.1136/gutjnl-2014-308932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding L, Zhang S, Xu M, Zhang R, Sui P, Yang Q. MicroRNA-27a contributes to the malignant behavior of gastric cancer cells by directly targeting PH domain and leucine-rich repeat protein phosphatase 2. J Exp Clin Cancer Res 2017; 36:45.doi: 10.1186/s13046-017-0516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang JX, Xu Y, Gao Y, Chen C, Zheng ZS, Yun M, et al. Decreased expression of miR-939 contributes to chemoresistance and metastasis of gastric cancer via dysregulation of SLC34A2 and Raf/MEK/ERK pathway. Mol Cancer 2017; 16:18.doi: 10.1186/s12943-017-0586-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Wu Q, Yang Z, An Y, Hu H, Yin J, Zhang P, et al. MiR-19a/b modulate the metastasis of gastric cancer cells by targeting the tumor suppressor MXD1. Cell Death Dis 2014; 5:e1144.doi: 10.1038/cddis.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang F, Li K, Pan M, Li W, Wu J, Li M, et al. miR-589 promotes gastric cancer aggressiveness by a LIFR-PI3K/AKT-c-Jun regulatory feedback loop. J Exp Clin Cancer Res 2018; 37:152.doi: 10.1186/s13046-018-0821-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye G, Huang K, Yu J, Zhao L, Zhu X, Yang Q, et al. MicroRNA-647 targets SRF-MYH9 axis to suppress invasion and metastasis of gastric cancer. Theranostics 2017; 7:3338–3353. doi: 10.7150/thno.20512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan J, Yang B, Lin S, Xing R, Lu Y. Downregulation of miR-142-5p promotes tumor metastasis through directly regulating CYR61 expression in gastric cancer. Gastric Cancer 2019; 22:302–313. doi: 10.1007/s10120-018-0872-4. [DOI] [PubMed] [Google Scholar]
- 63.Zhou Y, Huang T, Siu HL, Wong CC, Dong Y, Wu F, et al. IGF2BP3 functions as a potential oncogene and is a crucial target of miR-34a in gastric carcinogenesis. Mol Cancer 2017; 16:77.doi: 10.1186/s12943-017-0647-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X, Li Z, Xuan Z, Xu P, Wang W, Chen Z, et al. Novel role of miR-133a-3p in repressing gastric cancer growth and metastasis via blocking autophagy-mediated glutaminolysis. J Exp Clin Cancer Res 2018; 37:320.doi: 10.1186/s13046-018-0993-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.He C, Wang L, Zhang J, Xu H. Hypoxia-inducible microRNA-224 promotes the cell growth, migration and invasion by directly targeting RASSF8 in gastric cancer. Mol Cancer 2017; 16:35.doi: 10.1186/s12943-017-0603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mousa H, Yuan M, Zhang X, Li X, Shopit A, Almoiliqy M, et al. MicroRNA-4316 inhibits gastric cancer proliferation and migration via directly targeting VEGF-A. Cancer Cell Int 2020; 20:62.doi: 10.1186/s12935-020-1132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deng M, Liu B, Song H, Yu R, Zou D, Chen Y, et al. beta-Elemene inhibits the metastasis of multidrug-resistant gastric cancer cells through miR-1323/Cbl-b/EGFR pathway. Phytomedicine 2020; 69:153184.doi: 10.1016/j.phymed.2020.153184. [DOI] [PubMed] [Google Scholar]
- 68.Ye TB, Yang MH, Huang DC, Wang X, Xue BQ, Tian N, et al. MicroRNA-7 as a potential therapeutic target for aberrant NF-kappa B-driven distant metastasis of gastric cancer. J Exp Clin Canc Res 2019; 38:55.doi: 10.1186/s13046-019-1074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang HY, Bai M, Deng T, Liu R, Wang X, Qu YJ, et al. Cell-derived microvesicles mediate the delivery of miR-29a/c to suppress angiogenesis in gastric carcinoma. Cancer Lett 2016; 375:331–339. doi: 10.1016/j.canlet.2016.03.026. [DOI] [PubMed] [Google Scholar]
- 70.Deng T, Zhang H, Yang H, Wang H, Bai M, Sun W, et al. Exosome miR-155 derived from gastric carcinoma promotes angiogenesis by targeting the c-MYB/VEGF axis of endothelial cells. Mol Ther Nucleic Acids 2020; 19:1449–1459. doi: 10.1016/j.omtn.2020.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Zhou Z, Zhang H, Deng T, Ning T, Liu R, Liu D, et al. Exosomes carrying microRNA-155 target forkhead box O3 of endothelial cells and promote angiogenesis in gastric cancer. Mol Ther Oncolytics 2019; 15:223–233. doi: 10.1016/j.omto.2019.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Du J, Liang Y, Li J, Zhao JM, Wang ZN, Lin XY. Gastric cancer cell-derived exosomal microRNA-23a promotes angiogenesis by targeting PTEN. Front Oncol 2020; 10:326.doi: 10.3389/fonc.2020.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X, Chen X, Tian Y, Jiang D, Song Y. Long noncoding RNA RGMB-AS1 acts as a microRNA-574 sponge thereby enhancing the aggressiveness of gastric cancer via HDAC4 upregulation. Onco Targets Ther 2020; 13:1691–1704. doi: 10.2147/OTT.S234144. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.Cao Y, Xiong JB, Zhang GY, Liu Y, Jie ZG, Li ZR. Long noncoding RNA UCA1 regulates PRL-3 expression by sponging microRNA-495 to promote the progression of gastric cancer. Mol Ther Nucleic Acids 2020; 19:853–864. doi: 10.1016/j.omtn.2019.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Wang S, Tang D, Wang W, Yang Y, Wu X, Wang L, et al. circLMTK2 acts as a sponge of miR-150-5p and promotes proliferation and metastasis in gastric cancer. Mol Cancer 2019; 18:162.doi: 10.1186/s12943-019-1081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y, Liu C, Luo M, Zhang Z, Gong J, Li J, et al. Chemotherapy-induced miRNA-29c/Catenin-delta signaling suppresses metastasis in gastric cancer. Cancer Res 2015; 75:1332–1344. doi: 10.1158/0008-5472.CAN-14-0787. [DOI] [PubMed] [Google Scholar]
- 77.Li BW, Wang WZ, Li Z, Chen Z, Zhi XF, Xu JH, et al. MicroRNA-148a-3p enhances cisplatin cytotoxicity in gastric cancer through mitochondrial fission induction and cyto-protective autophagy suppression. Cancer Lett 2017; 410:212–227. doi: 10.1016/j.canlet.2017.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen Z, Li Z, Soutto M, Wang W, Piazuelo MB, Zhu S, et al. Integrated analysis of mouse and human gastric neoplasms identifies conserved microRNA networks in gastric carcinogenesis. Gastroenterology 2019; 156:1127–1139.e8. doi: 10.1053/j.gastro.2018.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pang X, Zhou Z, Yu Z, Han L, Lin Z, Ao X, et al. FOXO3a-dependent miR-633 regulates chemotherapeutic sensitivity in gastric cancer by targeting Fas-associated death domain (vol 16, pg 233, 2019). RNA Biol 2019; 16:1074–1080. doi: 10.1080/15476286.2019.1565665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luo Y, Wu J, Wu Q, Li X, Wu J, Zhang J, et al. miR-577 regulates TGF-beta induced cancer progression through a SDPR-modulated positive-feedback loop with ERK-NF-kappaB in gastric cancer. Mol Ther 2019; 27:1166–1182. doi: 10.1016/j.ymthe.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shang Y, Zhang Z, Liu Z, Feng B, Ren G, Li K, et al. miR-508-5p regulates multidrug resistance of gastric cancer by targeting ABCB1 and ZNRD1. Oncogene 2014; 33:3267–3276. doi: 10.1038/onc.2013.297. [DOI] [PubMed] [Google Scholar]
- 82.Shao L, Chen Z, Soutto M, Zhu S, Lu H, Romero-Gallo J, et al. Helicobacter pylori-induced miR-135b-5p promotes cisplatin resistance in gastric cancer. FASEB J 2019; 33:264–274. doi: 10.1096/fj.201701456RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zheng H, Wang JJ, Yang XR, Yu YL. Upregulation of miR-34c after silencing E2F transcription factor 1 inhibits paclitaxel combined with cisplatin resistance in gastric cancer cells. World J Gastroenterol 2020; 26:499–513. doi: 10.3748/wjg.v26.i5.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu Q, Li RT, Qian HQ, Wei J, Xie L, Shen J, et al. Targeted delivery of miR-200c/DOC to inhibit cancer stem cells and cancer cells by the gelatinases-stimuli nanoparticles. Biomaterials 2013; 34:7191–7203. doi: 10.1016/j.biomaterials.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 85.Ji R, Zhang X, Gu H, Ma J, Wen X, Zhou J, et al. miR-374a-5p: a new target for diagnosis and drug resistance therapy in gastric cancer. Mol Ther Nucleic Acids 2019; 18:320–331. doi: 10.1016/j.omtn.2019.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang XY, Zhang HY, Bai M, Ning T, Ge SH, Deng T, et al. Exosomes serve as nanoparticles to deliver anti-miR-214 to reverse chemoresistance to cisplatin in gastric cancer. Mol Ther 2018; 26:774–783. doi: 10.1016/j.ymthe.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu X, Lu Y, Xu Y, Hou S, Huang J, Wang B, et al. Exosomal transfer of miR-501 confers doxorubicin resistance and tumorigenesis via targeting of BLID in gastric cancer. Cancer Lett 2019; 459:122–134. doi: 10.1016/j.canlet.2019.05.035. [DOI] [PubMed] [Google Scholar]
- 88.Li LQ, Pan D, Chen Q, Zhang SW, Xie DY, Zheng XL, et al. Sensitization of gastric cancer cells to 5-FU by microRNA-204 through targeting the TGFBR2-mediated epithelial to mesenchymal transition. Cell Physiol Biochem 2018; 47:1533–1545. doi: 10.1159/000490871. [DOI] [PubMed] [Google Scholar]
- 89.Zhang F, Li K, Yao X, Wang H, Li W, Wu J, et al. A miR-567-PIK3AP1-PI3K/AKT-c-Myc feedback loop regulates tumor growth and chemoresistance in gastric cancer. EBioMedicine 2019; 44:311–321. doi: 10.1016/j.ebiom.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu S, Li D, Li T, Qiao L, Li K, Guo L, et al. miR-494 sensitizes gastric cancer cells to TRAIL treatment through downregulation of survivin. Cell Physiol Biochem 2018; 51:2212–2223. doi: 10.1159/000495867. [DOI] [PubMed] [Google Scholar]
- 91.Wei B, Sun X, Geng Z, Shi M, Chen Z, Chen L, et al. Isoproterenol regulates CD44 expression in gastric cancer cells through STAT3/MicroRNA373 cascade. Biomaterials 2016; 105:89–101. doi: 10.1016/j.biomaterials.2016.07.040. [DOI] [PubMed] [Google Scholar]
- 92.Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv L, et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer 2020; 19:62.doi: 10.1186/s12943-020-01185-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ma L, Wang Z, Xie M, Quan Y, Zhu W, Yang F, et al. Silencing of circRACGAP1 sensitizes gastric cancer cells to apatinib via modulating autophagy by targeting miR-3657 and ATG7. Cell Death Dis 2020; 11:169.doi: 10.1038/s41419-020-2352-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang X, Li Z, Zhang Q, Wang W, Li B, Wang L, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer 2019; 18:71.doi: 10.1186/s12943-019-0969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang XX, Zhang Q, Hu H, Jin Y, Zeng AL, Xia YB, et al. A novel circular RNA circFN1 enhances cisplatin resistance in gastric cancer via sponging miR-182-5p. J Cell Biochem 2020; [Epub ahead of print]. doi: 10.1002/jcb.29641. [DOI] [PubMed] [Google Scholar]
- 96.Liu YY, Zhang LY, Du WZ. Circular RNA circ-PVT1 contributes to paclitaxel resistance of gastric cancer cells through the regulation of ZEB1 expression by sponging miR-124-3p. Biosci Rep 2019; 39: doi: 10.1042/BSR20193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yan YM, Zuo XS, Wei DY. Concise review: emerging role of CD44 in cancer stem cells: a promising biomarker and therapeutic target. Stem Cell Transl Med 2015; 4:1033–1043. doi: 10.5966/sctm.2015-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Misso G, Di Martino MT, De Rosa G, Farooqi AA, Lombardi A, Campani V, et al. Mir-34: a new weapon against cancer? Mol Ther Nucleic Acids 2014; 3:e194.doi: 10.1038/mtna.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li XJ, Ren ZJ, Tang JH. MicroRNA-34a: a potential therapeutic target in human cancer. Cell Death Dis 2014; 5:e1327.doi: 10.1038/cddis.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jang E, Kim E, Son HY, Lim EK, Lee H, Choi Y, et al. Nanovesicle-mediated systemic delivery of microRNA-34a for CD44 overexpressing gastric cancer stem cell therapy. Biomaterials 2016; 105:12–24. doi: 10.1016/j.biomaterials.2016.07.036. [DOI] [PubMed] [Google Scholar]
- 101.Li P, Shan JX, Chen XH, Zhang D, Su LP, Huang XY, et al. Epigenetic silencing of microRNA-149 in cancer-associated fibroblasts mediates prostaglandin E2/interleukin-6 signaling in the tumor microenvironment. Cell Res 2015; 25:588–603. doi: 10.1038/cr.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang X, Wang C, Zhang X, Hua R, Gan L, Huang M, et al. Bmi-1 regulates stem cell-like properties of gastric cancer cells via modulating miRNAs. J Hematol Oncol 2016; 9:90.doi: 10.1186/s13045-016-0323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ni SJ, Zhao LQ, Wang XF, Wu ZH, Hua RX, Wan CH, et al. CBX7 regulates stem cell-like properties of gastric cancer cells via p16 and AKT-NF-kappaB-miR-21 pathways. J Hematol Oncol 2018; 11:17.doi: 10.1186/s13045-018-0562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Peng XD, Kang QJ, Wan R, Wang ZW. miR-26a/HOXC9 dysregulation promotes metastasis and stem cell-like phenotype of gastric cancer. Cell Physiol Biochem 2018; 49:1659–1676. doi: 10.1159/000493502. [DOI] [PubMed] [Google Scholar]
- 105.Fan D, Ren B, Yang X, Liu J, Zhang Z. Upregulation of miR-501-5p activates the wnt/beta-catenin signaling pathway and enhances stem cell-like phenotype in gastric cancer. J Exp Clin Cancer Res 2016; 35:177.doi: 10.1186/s13046-016-0432-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao M, Hou Y, Du YE, Yang L, Qin Y, Peng M, et al. Drosha-independent miR-6778-5p strengthens gastric cancer stem cell stemness via regulation of cytosolic one-carbon folate metabolism. Cancer Lett 2020; 478:8–21. doi: 10.1016/j.canlet.2020.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Han TS, Voon DC, Oshima H, Nakayama M, Echizen K, Sakai E, et al. Interleukin 1 up-regulates microRNA 135b to promote inflammation-associated gastric carcinogenesis in mice. Gastroenterology 2019; 156:1140–1155.e4. doi: 10.1053/j.gastro.2018.11.059. [DOI] [PubMed] [Google Scholar]
- 108.Wu Q, Yang ZP, Wang F, Hu SJ, Yang L, Shi YQ, et al. MiR-19b/20a/92a regulates the self-renewal and proliferation of gastric cancer stem cells. J Cell Sci 2013; 126:4220–4229. doi: 10.1242/jcs.127944. [DOI] [PubMed] [Google Scholar]
- 109.Li T, Guo H, Li H, Jiang Y, Zhuang K, Lei C, et al. MicroRNA-92a-1-5p increases CDX2 by targeting FOXD1 in bile acids-induced gastric intestinal metaplasia. Gut 2019; 68:1751–1763. doi: 10.1136/gutjnl-2017-315318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu H, Liu B, Chen Z, Li G, Zhang Z. MSC-induced lncRNA HCP5 drove fatty acid oxidation through miR-3619-5p/AMPK/PGC1alpha/CEBPB axis to promote stemness and chemo-resistance of gastric cancer. Cell Death Dis 2020; 11:233.doi: 10.1038/s41419-020-2426-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Prinz C, Weber D, Micro RNA. (miR) dysregulation during Helicobacter pylori-induced gastric inflammation and cancer development: critical importance of miR-155. Oncotarget 2020; 11:894–904. doi: 10.18632/oncotarget.27520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhao XD, Lu YY, Guo H, Xie HH, He LJ, Shen GF, et al. MicroRNA-7/NF-kappaB signaling regulatory feedback circuit regulates gastric carcinogenesis. J Cell Biol 2015; 210:613–627. doi: 10.1083/jcb.201501073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yang F, Xu Y, Liu C, Ma C, Zou S, Xu X, et al. NF-kappaB/miR-223-3p/ARID1A axis is involved in Helicobacter pylori CagA-induced gastric carcinogenesis and progression. Cell Death Dis 2018; 9:12.doi: 10.1038/s41419-017-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li N, Wang J, Yu W, Dong K, You F, Si B, et al. MicroRNA146a inhibits the inflammatory responses induced by interleukin17A during the infection of Helicobacter pylori. Mol Med Rep 2019; 19:1388–1395. doi: 10.3892/mmr.2018.9725. [DOI] [PubMed] [Google Scholar]
- 115.Pachathundikandi SK, Blaser N, Backert S. Mechanisms of inflammasome signaling, microRNA induction and resolution of inflammation by Helicobacter pylori. Curr Top Microbiol Immunol 2019; 421:267–302. doi: 10.1007/978-3-030-15138-6_11. [DOI] [PubMed] [Google Scholar]
- 116.Li S, Liang X, Ma L, Shen L, Li T, Zheng L, et al. MiR-22 sustains NLRP3 expression and attenuates H. pylori-induced gastric carcinogenesis. Oncogene 2018; 37:884–896. doi: 10.1038/onc.2017.381. [DOI] [PubMed] [Google Scholar]
- 117.Pachathundikandi SK, Backert S. Helicobacter pylori controls NLRP3 expression by regulating hsa-miR-223-3p and IL-10 in cultured and primary human immune cells. Innate Immun 2018; 24:11–23. doi: 10.1177/1753425917738043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zou M, Wang F, Jiang A, Xia A, Kong S, Gong C, et al. MicroRNA-3178 ameliorates inflammation and gastric carcinogenesis promoted by Helicobacter pylori new toxin, Tip-alpha, by targeting TRAF3. Helicobacter 2017; 22: doi: 10.1111/hel.12348. [DOI] [PubMed] [Google Scholar]
- 119.Blosse A, Levy M, Robe C, Staedel C, Copie-Bergman C, Lehours P. Deregulation of miRNA in Helicobacter pylori-induced gastric MALT lymphoma: from mice to human. J Clin Med 2019; 8: doi: 10.3390/jcm8060845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut 2010; 59:579–585. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 121.Yu L, Wu D, Gao H, Balic JJ, Tsykin A, Han TS, et al. Clinical utility of a STAT3-regulated miRNA-200 family signature with prognostic potential in early gastric cancer. Clin Cancer Res 2018; 24:1459–1472. doi: 10.1158/1078-0432.CCR-17-2485. [DOI] [PubMed] [Google Scholar]
- 122.Kong Y, Ning L, Qiu F, Yu Q, Cao B. Clinical significance of serum miR-25 as a diagnostic and prognostic biomarker in human gastric cancer. Cancer Biomark 2019; 24:477–483. doi: 10.3233/Cbm-182213. [DOI] [PubMed] [Google Scholar]
- 123.Zhu XL, Ren LF, Wang HP, Bai ZT, Zhang L, Meng WB, et al. Plasma microRNAs as potential new biomarkers for early detection of early gastric cancer. World J Gastroenterol 2019; 25:1580–1591. doi: 10.3748/wjg.v25.i13.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shimada H, Noie T, Ohashi M, Oba K, Takahashi Y. Clinical significance of serum tumor markers for gastric cancer: a systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer 2014; 17:26–33. doi: 10.1007/s10120-013-0259-5. [DOI] [PubMed] [Google Scholar]
- 125.Shin VY, Ng EK, Chan VW, Kwong A, Chu KM. A three-miRNA signature as promising non-invasive diagnostic marker for gastric cancer. Mol Cancer 2015; 14:202.doi: 10.1186/s12943-015-0473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huang ZB, Zhu DX, Wu LR, He MF, Zhou X, Zhang L, et al. Six serum-based mirnas as potential diagnostic biomarkers for gastric cancer. Cancer Epidem Biomar 2017; 26:188–196. doi: 10.1158/1055-9965.Epi-16-0607. [DOI] [PubMed] [Google Scholar]
- 127.Cui L, Zhang XJ, Ye GL, Zheng T, Song HJ, Deng HX, et al. Gastric juice microRNAs as potential biomarkers for the screening of gastric cancer. Cancer 2013; 119:1618–1626. doi: 10.1002/cncr.27903. [DOI] [PubMed] [Google Scholar]
- 128.Virgilio E, Giarnieri E, Giovagnoli MR, Montagnini M, Proietti A, D’Urso R, et al. Gastric juice microRNAs as potential biomarkers for screening gastric cancer: a systematic review. Anticancer Res 2018; 38:613–616. doi: 10.21873/anticanres.12265. [DOI] [PubMed] [Google Scholar]
- 129.Iwasaki H, Shimura T, Yamada T, Okuda Y, Natsume M, Kitagawa M, et al. A novel urinary microRNA biomarker panel for detecting gastric cancer. J Gastroenterol 2019; 54:1061–1069. doi: 10.1007/s00535-019-01601-w. [DOI] [PubMed] [Google Scholar]
- 130.Zhang ZL, Sun JL, Bai ZH, Li HJ, He SC, Chen R, et al. MicroRNA-153 acts as a prognostic marker in gastric cancer and its role in cell migration and invasion. Oncotargets Ther 2015; 8:357–364. doi: 10.2147/Ott.S78236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang X, Zhang Z, Zhang L, Zhou L. MicroRNA hsa-mir-3923 serves as a diagnostic and prognostic biomarker for gastric carcinoma. Sci Rep 2020; 10:4672.doi: 10.1038/s41598-020-61633-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen J, Hu B, Wang W, Qian XJ, Shan BJ, He YF. A six-microRNA signature to predict outcomes of patients with gastric cancer. Febs Open Bio 2019; 9:538–547. doi: 10.1002/2211-5463.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.An JX, Ma ZS, Ma MH, Shao S, Cao FL, Dai DQ. MiR-1236-3p serves as a new diagnostic and prognostic biomarker for gastric cancer. Cancer Biomark 2019; 25:127–132. doi: 10.3233/CBM-171026. [DOI] [PubMed] [Google Scholar]
- 134.Imaoka H, Toiyama Y, Okigami M, Yasuda H, Saigusa S, Ohi M, et al. Circulating microRNA-203 predicts metastases, early recurrence, and poor prognosis in human gastric cancer. Gastric Cancer 2016; 19:744–753. doi: 10.1007/s10120-015-0521-0. [DOI] [PubMed] [Google Scholar]
- 135.Hou CG, Luo XY, Li G. Diagnostic and prognostic value of serum microRNA-206 in patients with gastric cancer. Cell Physiol Biochem 2016; 39:1512–1520. doi: 10.1159/000447854. [DOI] [PubMed] [Google Scholar]
- 136.Liu XY, Zhang XW, Zhang Z, Chang JJ, Wang ZC, Wu Z, et al. Plasma microRNA-based signatures to predict 3-year postoperative recurrence risk for stage II and III gastric cancer. Int J Cancer 2017; 141:2093–2102. doi: 10.1002/ijc.30895. [DOI] [PubMed] [Google Scholar]
- 137.Nishibeppu K, Komatsu S, Imamura T, Kiuchi J, Kishimoto T, Arita T, et al. Plasma microRNA profiles: identification of miR-1229-3p as a novel chemoresistant and prognostic biomarker in gastric cancer. Sci Rep 2020; 10:3161.doi: 10.1038/s41598-020-59939-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pichler M, Calin GA. MicroRNAs in cancer: from developmental genes in worms to their clinical application in patients. Brit J Cancer 2015; 113:569–573. doi: 10.1038/bjc.2015.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Farooqi AA, Fayyaz S, Shatynska-Mytsyk I, Javed Z, Jabeen S, Yaylim I, et al. Is miR-34a a well-equipped Swordsman to conquer temple of molecular oncology? Chem Biol Drug Des 2016; 87:321–334. doi: 10.1111/cbdd.12634. [DOI] [PubMed] [Google Scholar]
- 140.Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, et al. PDL1 regulation by p53 via miR-34. J Natl Cancer Inst 2016; 108: doi: 10.1093/jnci/djv303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs 2017; 35:180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wei H, Pu K, Liu XG, Li BX, Zhang HS, Wang H, et al. The diagnostic value of circulating microRNAs as a biomarker for gastric cancer: a meta-analysis. Oncol Reports 2019; 41:87–102. doi: 10.3892/or.2018.6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tsujiura M, Komatsu S, Ichikawa D, Shiozaki A, Konishi H, Takeshita H, et al. Circulating miR-18a in plasma contributes to cancer detection and monitoring in patients with gastric cancer. Gastric Cancer 2015; 18:271–279. doi: 10.1007/s10120-014-0363-1. [DOI] [PubMed] [Google Scholar]


