Abstract
Background:
Non-coding RNAs have attracted considerable attention for their vital role in cancer. The purpose of this study was to determine the effects of non-coding RNAs on hepatocellular carcinoma (HCC) and reveal their regulatory mechanism in the pathophysiological process.
Methods:
We measured the expression of mucin 1 (MUC1) and miR-485-5p in tissues from 15 HCC patients and in liver cancer cell lines by quantitative real-time polymerase chain reaction and Western blot, screened for aberrantly expressed microRNAs (miRNAs) by miRNA microarrays. Bioinformatics tools were used to find the miRNA and circular RNA that regulated MUC1, which were validated by RNA immunoprecipitation assay and luciferase reporter assay. Cell counting kit-8, Transwell assays, and flow cytometry were used to conduct functional experiments. Proteins were examined by western blot and immunohistochemical staining assays. Significant differences between groups were estimated using the one-way analysis of variance. A P < 0.05 was considered statistically significant.
Results:
MUC1 was overexpressed in HCC tissues compared with that in paratumor tissues (normal vs. tumor, 1.007 ± 0.215 vs. 75.213 ± 18.403, t = 18.401, P < 0.001) while miR-485-5p was down-regulated (normal vs. tumor, 4.894 ± 0.684 vs. 1.586 ± 0.398, t = 16.191, P < 0.001). Inhibition of miR-485-5p promoted cell proliferation (73.33% ± 5.13% vs. 41.33% ± 3.51%, t = 8.913, P < 0.001), migration (102 ± 8 cells vs. 46 ± 8 cells, t = 8.681, P < 0.001), invasion (59 ± 7 cells vs. 28 ± 2 cells, t = 8.034, P < 0.01), and suppressed apoptosis (22.64% ± 6.97% vs. 36.33% ± 3.96%, t = 2.958, P < 0.05) of HepG2 cells with which MUC1 is knocked down. Mechanically, miR-485-5p binds to MUC1, while circHECTD1 binds to miR-485-5p, resulting in the indirect up-regulation of the MUC1 level.
Conclusions:
Our findings reveal that circHECTD1 facilitates HCC progression by sponging miR-485-5p to up-regulate MUC1.
Keywords: Mucin 1, CircHECTD1, MicroRNA-485-5p, Hepatocellular carcinoma, Competing endogenous RNA
Introduction
Liver cancer is a prevalent malignant tumor, is emerging as the fourth leading cause of tumor-associated mortality globally, and is severely threatening human health.[1] The primary liver cancer with the highest incidence is hepatocellular carcinoma (HCC), accounting for about 75% to 85% of cases. Currently, the main methods for treating HCC are surgery, chemotherapy, radiotherapy, and liver transplantation. However, because it is highly invasive, HCC is extremely prone to metastasis, and because patients are often already in the advanced stage when they are diagnosed, this leads to poor prognosis[2]; the survival rate of patients at 5 years is <18%.[3] Consequently, investigations into new targets for the identification and treatment of HCC are urgently needed.
Mucin 1 (MUC1) is a core member of the MUC family: it is composed of three parts, the extracellular domain (MUC1-N), the transmembrane region (MUC1-TM), and the cytoplasmic tail segment (MUC1-CT).[4] The tyrosine phosphorylation site of MUC1-CT interacts with various protein kinases and is phosphorylated by the protein kinases, which in turn participate in a variety of signal transduction pathways,[5] such as those involving Bcl-2-associated X-protein (BAX),[6] c-Jun N-terminal kinase/transforming growth factor beta (TGF-β),[7] p53,[8] nuclear factor-κB,[9] and mitogen-activated protein kinase-extracellular-signal-regulated kinase (ERK)/ERK (MEK-ERK),[10] and so on. In addition to being a transmembrane glycoprotein, it is also an oncogene. In normal tissues, MUC1 is expressed on the surface of epithelial cells with polar distribution, whereas in tumor tissues of various epithelial origins, it is overexpressed, abnormally glycosylated, and shows loss of polarity. Multiple works in the literature have reported that, in HCC cell lines and tissues, MUC1 is overexpressed[7,11,12] and promotes cell migration and invasion by activating the TGF-β/activator protein AP-1 pathway.[13] Targeting MUC1 by RNA interference and inhibitors can inhibit the progression of HCC.[7] Knockdown of MUC1 has been shown to significantly suppress tumor cell proliferation.[14] All of the above suggest that MUC1 acts as a key factor in HCC tumorigenesis; therefore, an in-depth study of the regulatory mechanism of MUC1 may be the key to a targeted molecular therapy for HCC.
CircRNA is a kind of single-stranded and covalently closed circular RNA with sequence conservation, and is formed by back-splicing from the precursor mRNA. It regulates gene or mRNA expression by sponging miRNA, interfering with splicing mechanisms, or sequestering other gene expression regulators.[15,16] In recent years, studies have found that circRNA, as a “molecular sponge,” can inhibit the function of miRNA.[17] Zhu et al[18] reported that the circ-0067934 circRNA regulated cancer-related miR-1324 and participated in the corresponding pathways, which suggested that circRNAs may be important regulators of cancer.[19] MiRNAs, a kind of single-stranded RNA with 21 to 23 nt length, bind to the corresponding mRNA molecules through specific miRNA binding sites to competitively inhibit the binding of mRNAs to targets, thereby regulating target gene expressions. Research has shown that miRNAs regulate cell proliferation, invasion, and migration in tumors.[20,21] Lin et al[22] analyzed the circRNA-miRNA-mRNA regulatory network in HCC using integrated microarray, which revealed that the mechanism of non-coding RNAs participated in the progression of HCC. In addition, Xu et al[23] reported that circSETD3 inhibited HCC proliferation by modulating mitogen-activated protein kinase 14 expression by sponging miR-421. Cao et al[24] found that hsa_circ_101280 promoted HCC proliferation and inhibited apoptosis by regulating miR-375/JAK2. Therefore, we speculated that the regulatory mechanism of MUC1, which acts as an oncogene, may also be involved in circRNA and miRNA.
In this study, we investigated the function of circHECTD1/miR-485-5p/MUC1 regulatory network in the HCC pathophysiological process and hoped to provide a potential therapeutic target for HCC.
Methods
Ethical approval
After approval from the ethics committees of the three hospitals (Tongde Hospital of Zhejiang Province, Hangzhou, China; Hangzhou First People's Hospital, Hangzhou, China; and Zhejiang Cancer Hospital, Hangzhou, China), the study was carried out according to the ethics guidelines of the Declaration of Helsinki. All selected patients signed the informed consent before the study commenced.
Patients and tissue samples
A total of 15 HCC patients participated in the study, of which ten were positive for hepatitis B and five were non-infectious (confirmed by pathologists). None of the patients received chemotherapy or radiotherapy before operation. Fresh HCC tissues (n = 15) and the corresponding adjacent normal tissues (n = 15) were obtained from these surgically treated patients in the Tongde Hospital of Zhejiang Province, Hangzhou, China; Hangzhou First People's Hospital, Hangzhou, China; and Zhejiang Cancer Hospital, Hangzhou, China. The tissue samples were immediately frozen in liquid nitrogen for transportation and stored at −80°C or fixed in formaldehyde (10%).
Cell lines
A normal liver cell THLE-3 and liver cancer cell lines, including HCCLM, MHCC97L, SMMC7721-1640, Hep3B, and HepG2, were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in media, such as bronchial epithelial growth medium (BEGM) (for THLE-3; Lonza/Clonetics Corporation, Walkersville, MD, USA; we discarded the gentamycin/amphotericin and epinephrine and added extra 5 ng/mL epidermal growth factor, 70 ng/mL phosphoethanolamine), Dulbecco's modification of Eagle's medium (for HCCLM, MHCC97L, and HepG2; Gibco, Thermo Fisher Scientific, Waltham, MA, USA), Roswell Park Memorial Institute (RPMI) 1640 medium (for SMMC7721; Gibco, Thermo Fisher Scientific), and minimum essential medium (MEM) (for Hep3B; Gibco, Thermo Fisher Scientific), mixed with 10% fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific) in an environment at 37°C with 5% CO2.
Total RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
The RNAiso Plus kit (TaKaRa, Dalian, China) was used to isolate the total RNA from tissues and cells, and the RNA content was detected using a NanoDrop1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). The RevertAid RT Reverse Transcription Kit (Thermo Fisher Scientific) was used to obtain the complementary DNA from the RNA, the qRT-PCR reactions were performed on the ABI 7500 PCR Detection System (Applied Biosystems, Foster City, CA, USA), and the fluorescent signal was SYBR Green I (Roche, Mannheim, Germany). All operations were performed according to the manufacturer's recommendations. The following are the primer sequences: cir-forward: 5′-ACTCCGTCACCTCGATTAGC-3′; cir-reverse: 5′-ATCATCCCATGTTCTCCGGC-3′; MUC1-forward: 5′-CGCCGAAAGAACTACGGGCAGCTG-3′; MUC1-reverse: 5′-CAAGTTGGCAGAAGTGGCTGCCAC-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-forward: 5′-AATCCCATCACCATCTTCC-3′; GAPDH-reverse: 5′-CATCACGCCACAGTTTCC-3′; miR-forward: 5′-GGCTGGCCGTGATGAATTC-3′; miR-reverse: 5′-GCGAGCACAGAATTAATACGAC-3′; U6-forward: 5′-CTCGCTTCGGCAGCACA-3′; U6-reverse: 5′-AACGCTTCACGAATTTGCGT-3′. Sangon Biotech (Shanghai, China) was responsible for primer synthesis. The expression of the RNAs was evaluated by a relatively quantitative (2−ΔΔCt) method.
Microarray and data analysis
The miRNA expression profiles in HCC tissues and normal tissues were detected using Agilent Human Microarray (Release 19.0, Agilent, Santa Clara, USA) according to the manufacturer's recommendations. The acquired array images were analyzed with Agilent Feature Extraction software (version 11.0.1.1, Agilent Technologies). Quantile normalization and subsequent data processing were performed with the Gene Spring GX v11.5.1 software package (Agilent Technologies). Student's t test was used to judge the differences between the two groups and P < 0.05 was considered statistically significant.
Immunohistochemical staining
All samples (n = 30) were immersed in neutral-buffered formalin (10%; Sangon Biotech) for fixation, then paraffin-embedded and sliced. After dehydration using gradient ethanol and inactivation of the endogenous enzyme by H2O2, the slices were incubated at room temperature for 30 min with normal goat serum. Then the primary antibody MUC1 (1:100 dilution; Abcam, Shanghai, China) was added and incubated overnight at 4°C. The primary antibody in the negative control was replaced by phosphate-buffered saline (PBS). After washing three times with PBS, the slices were incubated at room temperature for 1 h with the secondary antibody, viz., horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G (IgG) (1:500 dilution; Boster, Wuhan, China). After washing again with PBS, a diaminobenzidine mixture was added (Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd, Beijing, China), and the reaction continued for 5 min in the dark at room temperature. Next, the slices were washed, counterstained, dehydrated, transparentized, and sealed. Finally, they were observed using a microscope (IX71; Olympus, Tokyo, Japan).
Western blot
The supernatant after cell lysis was collected with radioimmunoprecipitation assay lysis buffer (Beyotime, Shanghai, China). The concentration of proteins was measured by the bicinchoninic acid Protein Assay Kit (Sangon Biotech), and the same amount of protein was denatured with 5 × sodium dodecyl sulfate (SDS; Sangon Biotech) loading buffer in boiling water at 100°C for 5 min. To separate the proteins, SDS-polyacrylamide gel electrophoresis was used; then the separated proteins were transferred to a polyvinylidene fluoride (PVDF) membrane, and the PVDF membrane was incubated with 5% non-fat milk at room temperature for 1 h. Next, the anti-MUC1 antibody (1:1000; Abcam) and GAPDH (1:1000; Abcam) were added and incubated at 4°C overnight. Then the membrane was washed with Tris-buffered saline with Tween-20 three times; subsequently, the secondary antibody (HRP-labeled) was added and incubated at 20°C for 1 h. Finally, the protein was tested using an enhanced chemiluminescence kit (Millipore, Billerica, MA, USA).
Cell transfection
The synthesis of small interfering RNA specific to the HECTD1 gene (siHECTD1) (5′-ACCTATTGGGGAGCAGATTCT-3′), siMUC1 (5′-AAGGTACCATCAATGTCCACG-3′), and siControl (5′-CGCT TACCGATTCAGAATGG-3′) was carried out by Sangon Biotech.
Six-well plates were seeded with HepG2 cells (3 × 105 per well) or 96-well plates were planted with HepG2 cells (1 × 104 per well) and incubated overnight. Based on experimental needs, the miRNA or plasmid and Lipofectamine 3000 mixtures (Thermo Fisher Scientific) were prepared according to the manufacturer's instructions and transfected into the HepG2 cells. After 6 h, the medium was changed, and another 48 h later, RNA and protein could be extracted from the cells.
Luciferase reporter assay
The 3′-untranslated region (UTR) of MT-MUC1 (mutant-type), WT-MUC1 (wild-type), MT-HECTD1, and WT-HECTD1 genes were amplified by PCR respectively and cloned into the XbaI site of pGL3-control vector (Promega, Madison, WI, USA), downstream from the Firefly luciferase 3′-UTR. Six-well plates were coated with HepG2 cells and then co-transfected as indicated: (1) miR-485-5p mimics or the negative control (NC-miRNA) with pGL3-WT-MUC1 or pGL3-MT-MUC1; and (2) miR-485-5p mimics or NC-miRNA with pGL3-WT-HECTD1 or pGL3-MT-HECTD1. Luciferase activity was detected 48 h later and presented as relative activity.
Cell proliferation assay
The cells were collected when they were in the logarithmic growth phase and diluted to a cell suspension concentration of 1 × 105/mL; then 100 μL was added per well in a 96-well plate. The cells were incubated overnight and then transfected as described above. The cell proliferation ability was measured using the cell counting kit-8 (CCK-8) assay. Each well was supplemented with CCK-8 reagent (Sangon Biotech) (10 μL per well). Then the 96-well plate was placed in a cell incubator for 1 h, and finally, the optical density values were recorded at 450 nm using a microplate reader (Molecular Devices, USA).
Cell migration and invasion assays
The transfected cells were collected and tested in Transwell chambers. Cells with the medium (without serum) were planted into the upper compartment of the 24-well Transwell chambers (8 μm pore size; Corning, NY, USA), while in the lower compartment, complete medium (10% FBS) was filled in as a chemoattractant. After 48 h of incubation, the cells in the lower chamber were fixed with formaldehyde, stained with 0.2% crystal violet, and then observed under a microscope. The invasion assays were similar to the migration assay, with the difference being that the membrane was pretreated using Matrigel (BD Biosciences, San Diego, CA, USA) in the former.
Apoptosis detection
The cells transfected as described above were collected and stained using a PharMingen apoptosis detection kit (BD Biosciences) according to the manufacturer's recommendations, and then the apoptotic cells were examined using a flow cytometer (BD Biosciences).
RNA immunoprecipitation (RIP) assay
RIP assay was performed using the RNA binding protein immunoprecipitation kit (Millipore). First, mixtures of the HepG2 cell lysates (lysed with RIP lysis buffer) and magnetic beads (conjugated to human anti-Argonaute2 (Ago2) antibody [Millipore] or IgG antibody) were incubated. Next, the immunoprecipitated RNAs were isolated using proteinase K, and finally, qRT-PCR was used to detect the purified RNAs.
Statistical analysis
GraphPad Prism 6.0 statistical analysis software (La Jolla, CA, USA) was used to compare data among groups. The data are presented as the mean ± standard deviation of three independent experiments. The Kaplan-Meier method was used for the survival analysis and the Pearson correlation coefficient was used for correlation analysis. Significant differences between groups were estimated using a one-way analysis of variance. A P < 0.05 was considered statistically significant.
Results
MUC1 is highly expressed in HCC, which is linked to poor survival of patients
To verify the MUC1 expression in HCC tissues and paratumor tissues, qRT-PCR was used. The results were in line with expectations [Figure 1A]: in cancerous tissues an obviously higher level of MUC1 was observed (normal vs. tumor, 1.007 ± 0.215 vs. 75.213 ± 18.403, t = 18.401, P = 0.0008). This was further confirmed by immunohistochemical staining assays, with positive staining for MUC1 being observed in HCC tissue samples (representative images are shown in Figure 1B). Analysis using Kaplan-Meier survival curves showed that patients with high levels of MUC1 had a significantly shortened overall survival (high vs. low, 58.71 ± 27.77 vs. 75.22 ± 18.43 months, t = 2.138, P = 0.0412) [Figure 1C], which is supported by the The Cancer Genome Atlas (TCGA) database analysis [Figure 1D]. In addition, we tested the expressions of MUC1 in different hepatoma cell lines (HCCLM, MHCC97L, SMMC7721, Hep3B, and HepG2) by western blot, with normal human hepatocyte THLE-3 as a control. The results are shown in Figure 1E. MUC1 levels in liver cancer cells, especially in HepG2 cells, were obviously higher than in the control. All data indicate that overexpression of MUC1 is related to the development of HCC.
Figure 1.
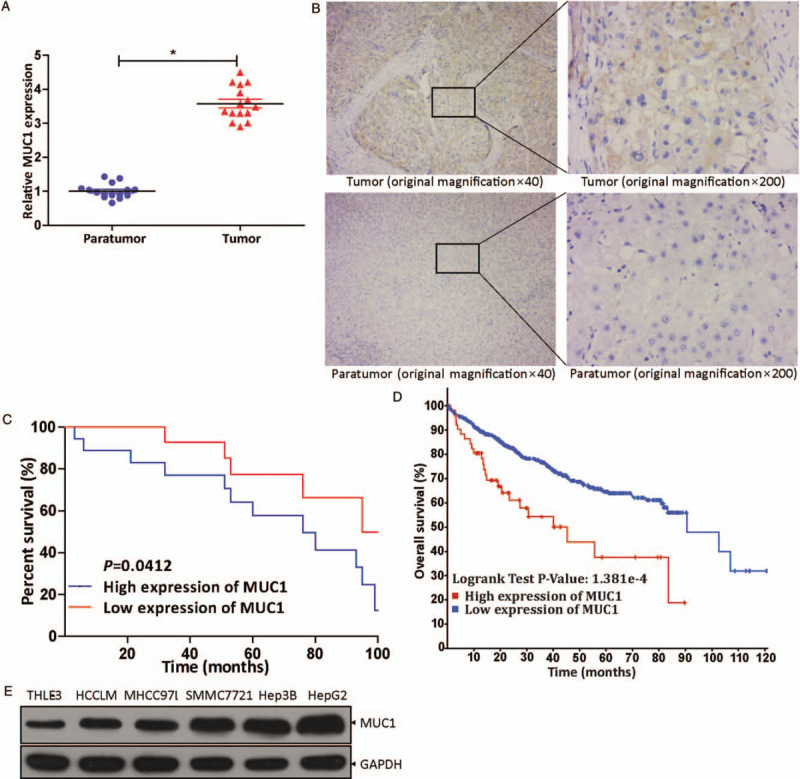
The expression of MUC1 in vivo and in vitro and its impact on overall patient survival. (A) qRT-PCR revealed MUC1 levels in HCC tissues (n = 15) and corresponding paratumor tissues (n = 15). ∗P < 0.001 compared with paratumor tissues. (B) MUC1 levels were examined in tissue samples (n = 30) by immunohistochemical staining assay (original magnification ×40 or ×200). (C) Analysis by Kaplan-Meier survival curves for the relationship between overall survival rate of patients and MUC1 expression level (P = 0.0412). (D) Analysis for the correlation between MUC1 expression level in HCC and overall survival rate of patients in the TCGA database. (E) The expression level of MUC1 in HCCLM, MHCC97L, SMMC7721, Hep3B, and HepG2 was examined by Western blot, with THLE-3 cells as control. HCC: Hepatocellular carcinoma; MUC1: Mucin 1; qRT-PCR: Quantitative real-time polymerase chain reaction.
MUC1 serves as a downstream gene of miR-485-5p
To determine the miRNAs that regulate MUC1, we first screened for aberrantly expressed miRNAs in HCC by miRNA microarrays and found ten miRNAs that were abnormally down-regulated, including miR-485-5p, which is considered to be an anti-tumor gene in HCC [Figure 2A]. Then, in HCC tissues (n = 15) and corresponding paratumor tissues (n = 15), qRT-PCR was conducted to detect miR-485-5p expression (normal vs. tumor, 4.894 ± 0.684 vs. 1.586 ± 0.398, t = 16.191, P = 0.0005). The negative correlation of MUC1 and miR-485-5p can be observed from Pearson analysis curve (P < 0.01) [Figure 2B] and the result was consistent with that in Figure 2A. By using bioinformatics tool TargetScan (www.targetscan.org), we discovered that there were binding regions between MUC1 and miR-485-5p, which indicated that miR-485-5p targeted MUC1 directly [Figure 2C]. Among the liver cancer cell lines mentioned above, MUC1 levels were highest in HepG2 cell line [Figure 1E]. Therefore, we selected the HepG2 cell line for subsequent research. The luciferase activity in the HepG2 cells was significantly depressed (NC-miRNA vs. miR-485-5p, 1.000 ± 0.120 vs. 0.417 ± 0.129, t = 5.735, P = 0.0046) when their WT-MUC1 was co-transfected with miR-485-5p mimics, while the luciferase activity was unchanged when miR-485-5p mimics were co-transfected with MT-MUC1 [Figure 2D]. This result suggests that miR-485-5p could combine with MUC1. When miR-485-5p levels were altered by miR-485-5p mimics (miR-485-5p mimics vs. NC-miRNA, 3.662 ± 0.416 vs. 1.000 ± 0.056, t = 10.681, P = 0.0004) or inhibitors (miR-485-5p inhibitor vs. NC-miRNA, 0.342 ± 0.071 vs. 1.000 ± 0.115, t = 8.474, P = 0.0011) in HepG2 cells, the expressions of MUC1 changed accordingly (miR-485-5p mimics vs. NC-miRNA, 0.373 ± 0.086 vs. 1.000 ± 0.017, t = 12.341, P = 0.0002; miR-485-5p inhibitor vs. NC-miRNA, 3.867 ± 0.669 vs. 1.000 ± 0.111, t = 7.321, P = 0.0019), and the levels of MUC1 and miR-485-5p were negatively correlated [Figure 2E and 2F], which proved the correlation between them once again. All data indicate that miR-485-5p targeted MUC1 directly.
Figure 2.
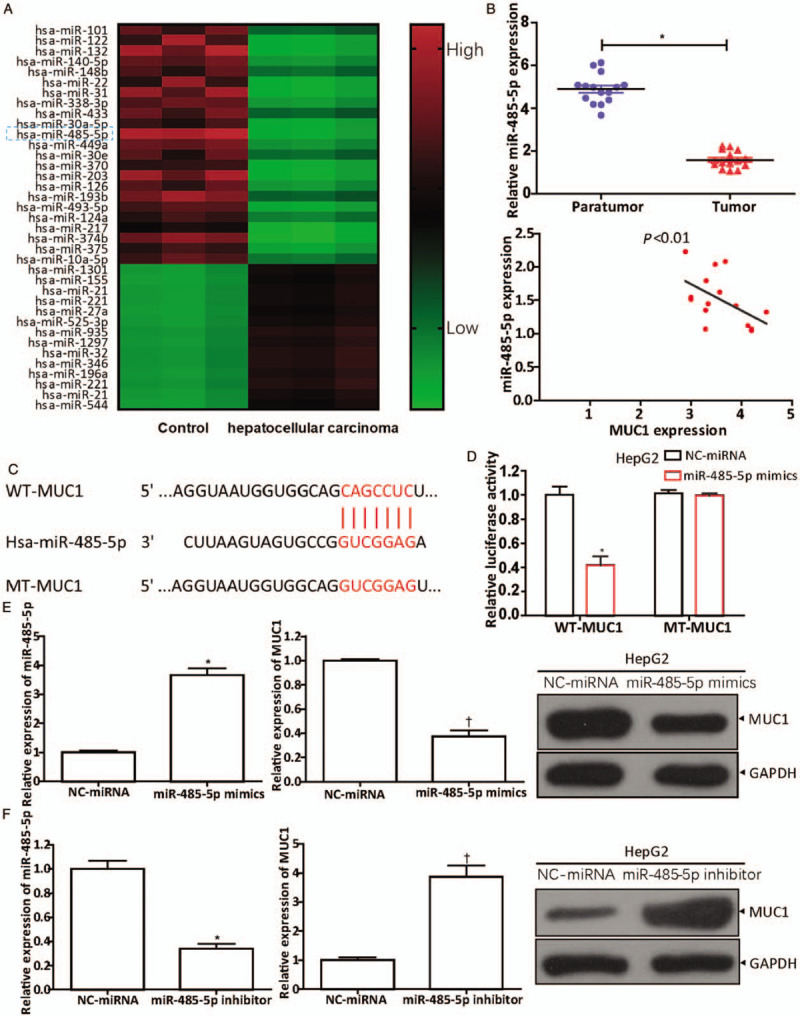
MUC1 is a downstream gene of miR-485-5p. (A) Screening for differential miRNAs in HCC tissues by miRNA microarray. (B) The miR-485-5p expression in HCC tissues (n = 15) and corresponding paratumor tissues (n = 15) measured by qRT-PCR. ∗P < 0.001 compared with paratumor tissues. The correlation of MUC1 and miR-485-5p analyzed by Pearson analysis (n = 15, P < 0.01). (C) The complementary regions of MUC1 and miR-485-5p investigated using the bioinformatics tool TargetScan. (D) Luciferase activity was detected after WT-MUC1 or MT-MUC1 cotransfection with NC-miRNA or miR-485-5p mimics for 48 h in HepG2 cells, according to the Lipofectamine 3000 manufacturer's instructions. ∗P < 0.01 compared with NC-miRNA. Western blot and qRT-PCR showed the expressions of MUC1 and miR-485-5p after transfection of (E) miR-485-5p mimics (miR-485-5p: ∗P < 0.001 compared with NC-miRNA group; MUC1: †P < 0.001 compared with NC-miRNA group) or (F) miR-485-5p inhibitor for 48 h in HepG2 cells, according to the Lipofectamine 3000 manufacturer's instructions (miR-485-5p: ∗P < 0.01 compared with NC-miRNA group; MUC1: †P < 0.01 compared with NC-miRNA group). The data in (D–F) are expressed as mean ± standard deviation (n = 3). HCC: Hepatocellular carcinoma; MiR: MicroRNA; MUC1: Mucin 1; MT-MUC1: Mutant-type MUC1; qRT-PCR: Quantitative real-time polymerase chain reaction; WT-MUC1: Wild-type MUC1.
Effects of miR-485-5p on HepG2 cells through regulation of the expression of MUC1
To confirm the function of MUC1 in HCC, the HepG2 cells were transfected with siMUC1 or its negative control. The results represented in Figure 3A showed that the knockdown of MUC1 blocks the proliferation of HepG2 cells (siMUC1 vs. siControl, 42.67% ± 4.04% vs. 100.00% ± 11.53%, t = 8.126, P = 0.0012). Further, siMUC1 and miR-485-5p inhibitors were co-transfected to verify whether miR-485-5p had any effect on hepatoma cells by regulating MUC1, and NC-miRNA was used as the control. The results showed that miR-485-5p inhibitors reversed the inhibition of siMUC1 on the proliferation of HepG2 cells ([siMUC1 + miR-485-5p inhibitor] vs. [siMUC1 + NC-miRNA], 73.33% ± 5.13% vs. 41.33% ± 3.51%, t = 8.913, P = 0.0009) [Figure 3A].
Figure 3.
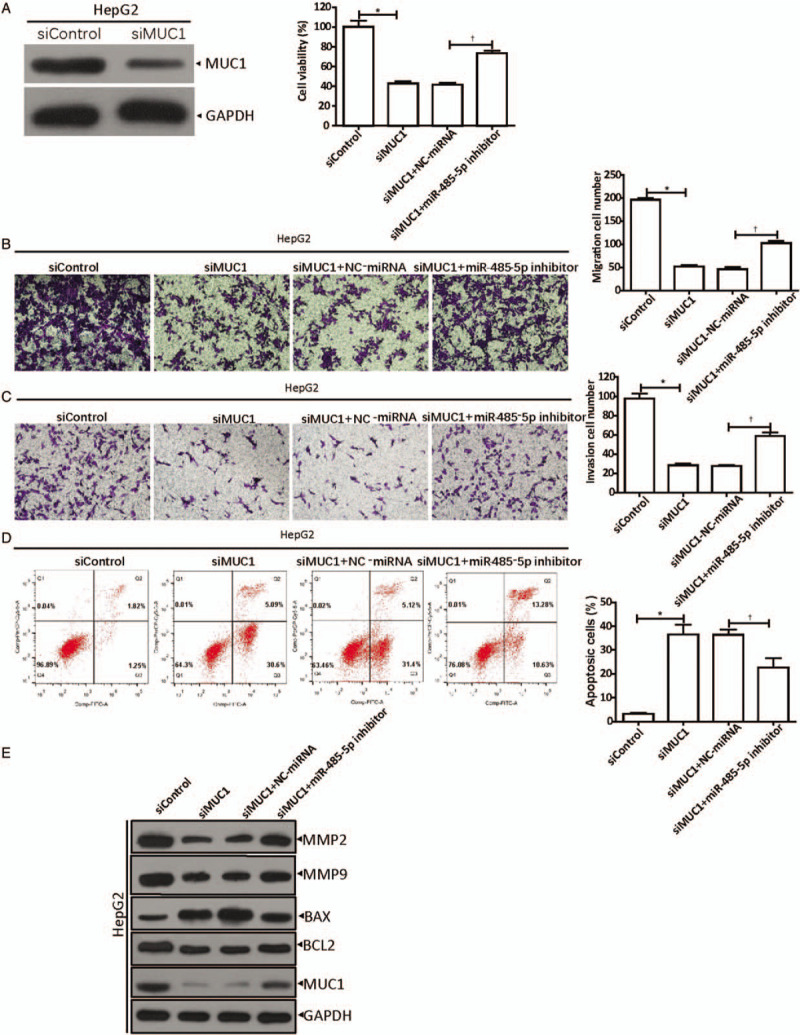
MiR-485-5p regulates MUC1 expression to facilitate the development of hepatocellular carcinoma. (A) Inhibition of miR-485-5p on HepG2 siMUC1 cell viability was detected by CCK-8. ∗P < 0.001 compared with siControl; †P < 0.01 compared with control group (siMUC1 + NC-miRNA). (B) Inhibition of miR-485-5p on the migration ability of HepG2 siMUC1 cells was analyzed by Transwell assays. ∗P < 0.001 compared with siControl; †P < 0.001 compared with control group (siMUC1 + NC-miRNA). (C) Inhibition of miR-485-5p on the invasion ability of HepG2 siMUC1 cells was analyzed by Transwell assays. ∗P < 0.001 compared with siControl; †P < 0.01 compared with control group (siMUC1 + NC-miRNA). (D) Promotion of miR-485-5p on HepG2 siMUC1 cell apoptosis was analyzed by flow cytometry. ∗P < 0.01 compared with siControl; †P < 0.05 compared with control group (siMUC1 + NC-miRNA). (E) Western blot showed the expressions of MMP-2, MMP-9, BAX, BCL2, and MUC1 in MUC1-silenced HepG2 cells with down-regulated miR-485-5p. The data in (A–D) are shown as mean ± standard deviation (n = 3). BAX: Bcl-2-associated X-protein; BCL2: B-cell lymphoma-2; CCK-8: Cell counting kit-8; MiR: MicroRNA; MMP: Matrix metalloproteinase; MUC1: Mucin 1.
Then, Transwell experiments were performed with the transfected HepG2 cells to test the migration and invasion abilities. The cells were treated as described in cell proliferation assays. The results are shown in Figure 3B and 3C. When MUC1 was knocked down, compared to their controls, the amounts of migrated (siMUC1 vs. siControl, 52 ± 6 cells vs. 196 ± 7 cells, t = 29.390, P < 0.0001) and invading cells (siMUC1 vs. siControl, 28 ± 4 cells vs. 98 ± 9 cells, t = 12.752, P = 0.0002) were significantly reduced, while the inhibition effect was reversed by the miR-485-5p inhibitor (migration: [siMUC1 + miR-485-5p inhibitor] vs. [siMUC1 + NC-miRNA], 102 ± 8 cells vs. 46 ± 8 cells, t = 8.681, P = 0.0009; invasion: [siMUC1 + miR-485-5p inhibitor] vs. [siMUC1 + NC-miRNA], 59 ± 7 cells vs. 28 ± 2 cells, t = 8.034, P = 0.0013).
Next, HepG2 cells were transfected as described above, and cell apoptosis was examined by flow cytometry [Figure 3D]. The results indicated that in the siMUC1 group, there were more apoptotic cells than in the siControl group (siMUC1 vs. siControl, 36.49% ± 7.03% vs. 3.13% ± 0.73%, t = 8.173, P = 0.0012), whereas after co-transfection of siMUC1 with the miR-485-5p inhibitor, the apoptotic cells were reduced significantly ([siMUC1 + miR-485-5p inhibitor] vs. [siMUC1 + NC-miRNA], 22.64% ± 6.97% vs. 36.33% ± 3.96%, t = 2.958, P = 0.0416).
Furthermore, we examined the expression of the proteins involved in tumorigenesis, such as matrix metalloproteinase (MMP)-2, MMP-9, BAX, B-cell lymphoma-2 (BCL2), and MUC1 [Figure 3E], by Western blot. The results confirm our speculation that the knockdown of MUC1 in HepG2 cells inhibited the expressions of MMP-2, MMP-9, and BCL2, while it promoted the BAX expression level. Subsequently, when siMUC1 and miR-485-5p inhibitors were co-transfected, the inhibitory effect of miR-485-5p on MUC1 expression was removed, and the expression of MMP-2, MMP-9, and BCL2 were increased, whereas the BAX expression level decreased.
The results of these functional experiments revealed that the abilities of HepG2 cells in terms of proliferation, migration, and invasion were inhibited by miR-485-5p, while cell apoptosis was promoted.
CircHECTD1 binds to miR-485-5p
The circRNA that regulates miR-485-5p was found by analyzing the Starbase 2.0 database, and we found that there are complementary sequences between circHECTD1 and miR-485-5p [Figure 4A]. Next, qRT-PCR was used to examine the circHECTD1 expression in HCC tissues (n = 15) and matched paratumorta tissues (n = 15). In the HCC tissues, circHECTD1 was up-regulated dramatically (normal vs. tumor, 1.500 ± 0.398 vs. 4.494 ± 0.730, t = 19.361, P = 0.0005), and through Pearson analysis, we concluded that the miR-485-5p level was negatively related to the circHECTD1 level (P < 0.01) [Figure 4B], which suggested that circHECTD1 could target miR-485-5p. Then, luciferase assays were used to demonstrate this theory. The luciferase activity in HepG2 cells decreased significantly when WT-HECTD1 were transfected with miR-485-5p mimics (NC-miRNA vs. miR-485-5p mimics, 1.000 ± 0.076 vs. 0.490 ± 0.156, t = 5.092, P = 0.0070), whereas the luciferase activity was steadily maintained when MT-HECTD1 were co-transfected with miR-485-5p mimics [Figure 4C]. This result suggests that miR-485-5p could combine with circHECTD1. Subsequently, a RIP assay was used. In the Ago2 pellets, there was more circHECTD1 (Ago2 vs. IgG, 7.740 ± 0.454 vs. 1.000 ± 0.110, t = 24.991, P = 0.0000) and miR-485-5p (Ago2 vs. IgG, 6.893 ± 0.315 vs. 1.000 ± 0.150, P < 0.0001) [Figure 4D] than in the IgG pellets, which indicates that circHECTD1 may act as a sponge for miR-485-5p in HepG2 cells. Moreover, compared to their normal controls, when the circHECTD1 was silenced (siHECTD1 vs. siControl, 0.447 ± 0.049 vs. 1.000 ± 0.131, t = 6.841, P = 0.0024), miR-485-5p expression was obviously up-regulated (siHECTD1 vs. siControl, 4.240 ± 0.355 vs. 1.000 ± 0.200, t = 13.772, P = 0.0002) [Figure 4E], whereas up-regulated circHECTD1 (circHECTD1 vs. Control, 4.003 ± 0.115 vs. 1.000 ± 0.070, t = 38.631, P < 0.0001) produced the opposite result (circHECTD1 vs. Control, 0.463 ± 0.112 vs. 1.000 ± 0.089, t = 6.519, P = 0.0029) [Figure 4F]. Overall, the data shown in Figure 4 prove that circHECTD1 is a sponge for miR-485-5p.
Figure 4.
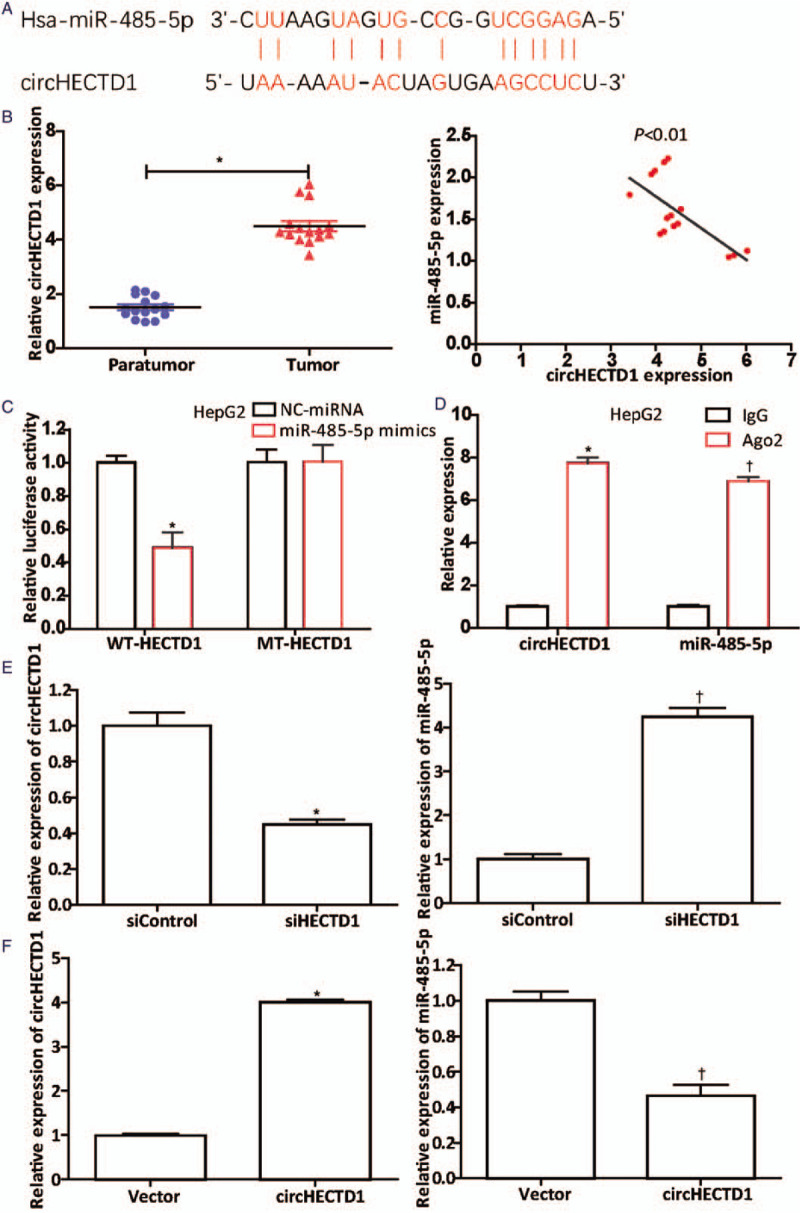
CircHECTD1 acts as a miR-485-5p sponge. (A) Complementary sequences between circHECTD1 and miR-485-5p discovered from the Starbase 2.0 database analysis. (B) qRT-PCR showed that circHECTD1 in HCC tissues (n = 15) was dramatically up-regulated. ∗P < 0.001 compared with the level in paratumor tissues. The correlation between their expression levels was negative (P < 0.01). (C) Luciferase activity was detected after cotransfection of WT-HECTD1- or MT-HECTD1-containing luciferase reporter gene with miR-485-5p mimics or NC-miRNA into HepG2 cells, according to the Lipofectamine 3000 manufacturer's instructions. ∗P < 0.01 compared with NC-miRNA group. (D) The circHECTD1 and miR-485-5p content in Ago2 pellets was verified using the RIP assay. ∗P < 0.001 (circHECTD1) compared with that in IgG pellets; †P < 0.001 (miR-485-5p) compared with that in IgG pellets. Further, qRT-PCR was conducted to investigate the effects of (E) down-regulation (circHECTD1: ∗P < 0.01 compared with the siControl group; miR-485-5p: †P < 0.001 compared with the siControl group) or (F) up-regulation of circHECTD1 (circHECTD1: ∗P < 0.001 compared with the vector; miR-485-5p: †P < 0.01 compared with the vector) on the miR-485-5p level. The data in (C–F) are presented as mean ± standard deviation (n = 3). HCC: Hepatocellular carcinoma; MiR: MicroRNA; MT-HECTD1: Mutant-type HECTD1; qRT-PCR: Quantitative real-time polymerase chain reaction; RIP: RNA immunoprecipitation; WT-HECTD1: Wild-type HECTD1.
CircHECTD1 sponges miR-485-5p to positively regulate MUC1
To determine whether circHECTD1 promoted HCC progression through miR-485-5p, we conducted a series of verifications. First of all, we up-regulated [Figure 5A] or down-regulated [Figure 5B] circHECTD1 and observed the MUC1 expression by western blot and qRT-PCR (Figure 5A: circHECTD1 vs. Control, 4.253 ± 0.399 vs. 1.000 ± 0.089, t = 13.790, P = 0.0002; Figure 5B: circHECTD1 vs. Control, 0.367 ± 0.051 vs. 1.000 ± 0.200, t = 5.313, P = 0.0060). The results showed that they were positively correlated. Then, functional experiments revealed that the proliferation (siHECTD1 vs. siControl, 38.00% ± 5.57% vs. 100.30% ± 20.53%, t = 5.076, P = 0.0071) [Figure 5C], migration (siHECTD1 vs. siControl, 78 ± 10 cells vs. 214 ± 10 cells, t = 16.712, P = 0.0001) [Figure 5D], and invasion (siHECTD1 vs. siControl, 36 ± 4 cells vs. 104 ± 6 cells, t = 15.632, P = < 0.0001) [Figure 5E] of HepG2 cells that were inhibited by circHECTD1 knockdown, were reversed by inhibiting miR-485-5p (proliferation: [siHECTD1 + miR-485-5p inhibitor] vs. [siHECTD1 + NC-miRNA], 70.67% ± 3.79% vs. 36.67% ± 7.64%, t = 6.908, P = 0.0023; migration: [siHECTD1 + miR-485-5p inhibitor] vs. [siHECTD1 + NC-miRNA], 121 ± 3 vs. 77 ± 10, t = 7.212, P = 0.0020; invasion: 58 ± 5 vs. 35 ± 4, t = 6.528, P = 0.0028). Moreover, the enhancement of HepG2 cell apoptosis (siHECTD1 vs. siControl, 45.83% ± 13.01% vs. 3.39% ± 0.40%, t = 5.647, P = 0.0048) by circHECTD1 knockdown was also reversed by inhibiting miR-485-5 ([siHECTD1 + miR-485-5p inhibitor] vs. [siHECTD1 + NC-miRNA], 25.92% ± 2.63% vs. 46.29% ± 12.00%, t = 2.871, P = 0.0454) [Figure 5F]. In addition, protein (MMP-2, MMP-9, BAX, BCL2, and MUC1) expression levels were measured by western blot [Figure 5G]. From Figure 5G, we can see that in the circHECTD1-silenced cells, expressions of proteins, such as MMP-2, MMP-9, BCL2, and MUC1, were down-regulated, whereas the BAX level was up-regulated. The expression of these proteins, either down-regulation or up-regulation, brought about by circHECTD1 knockdown, was reversed by inhibiting miR-485-5p. Collectively, the data support the hypothesis that circHECTD1 sponges miR-485-5p to positively regulate MUC1.
Figure 5.
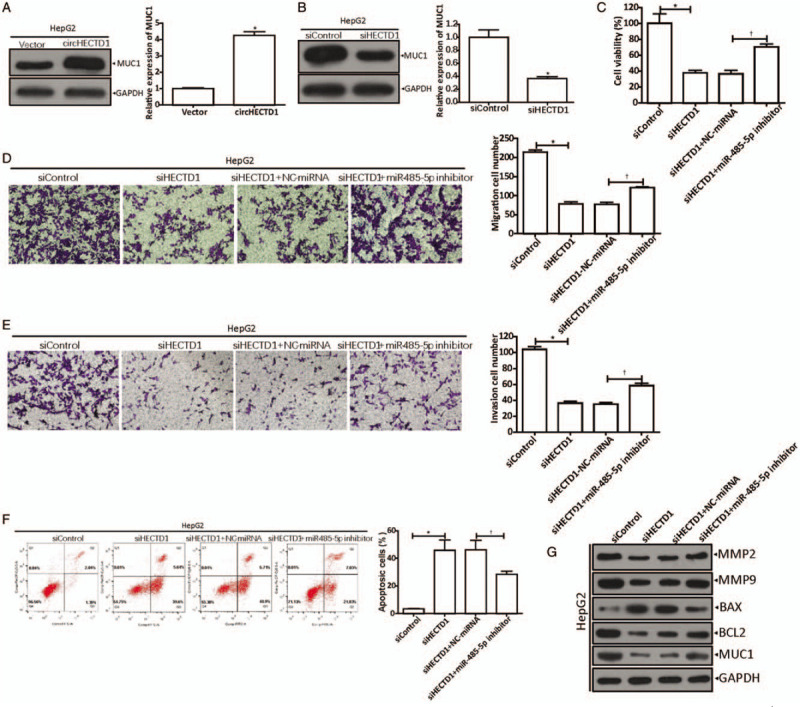
CircHECTD1 positively regulates MUC1 by sponging miR-485-5p. (A) MUC1 expressions measured by western blot and qRT-PCR after transfection with circHECTD1. ∗P < 0.001 compared with vector control. (B) MUC1 levels in circHECTD1-silenced HepG2 cells, detected by western blot and qRT-PCR. ∗P < 0.01 compared with siControl group. (C) Inhibition of miR-485-5p on HepG2 siHECTD1 cell viability was detected by CCK-8. ∗P < 0.01 compared with siControl, †P < 0.01 compared with control group (siHECTD1 + NC-miRNA). (D) Inhibition of miR-485-5p on HepG2 siHECTD1 cell migration capacity was analyzed by Transwell assays. ∗P < 0.001 compared with siControl; †P < 0.01 compared with control group (siHECTD1 + NC-miRNA). (E) Inhibition of miR-485-5p on HepG2 siHECTD1 cell invasion capacity was analyzed by Transwell assays. ∗P < 0.001 compared with siControl, †P < 0.01 compared with control group (siHECTD1 + NC-miRNA). (F) Promotion of miR-485-5p on HepG2 siHECTD1 cell apoptosis was tested by flow cytometry. ∗P < 0.01 compared with siControl; †P < 0.05 compared with control group (siHECTD1 + NC-miRNA). (G) Western blot showed the expressions of MMP2, MMP9, BAX, BCL2, and MUC1 in circHECTD1-silenced HepG2 cells with downregulated miR-485-5p. The data in (A–F) are shown as mean ± standard deviation (n = 3). BAX: Bcl-2-associated X-protein; BCL2: B-cell lymphoma-2; CCK-8: Cell counting kit-8; MiR: MicroRNA; MMP: Matrix metalloproteinase; MUC1: Mucin 1; qRT-PCR: Quantitative real-time polymerase chain reaction.
Discussion
With the development of molecular biology, researchers have discovered a variety of genes related to tumor progression[25-27] and regard them as targets for cancer gene therapy. Thus, the regulatory mechanisms of these targets have become new research hotspots.
MUC1 is abnormally expressed in tumor cells, mainly in epithelial cell-derived malignant tumors, leading to its acknowledgment as a potential tumor marker.[28-30] Multiple previous studies have reported that miRNA serves as a regulator in several human cancers,[31,32] including HCC.[33] In this investigation, we discovered that in HCC tissues and cell lines, the MUC1 level increased abnormally which correlated with a low survival rate of patients. MiRNA microarrays and bioinformatics assays suggested that MUC1 might be regulated by miR-485-5p; therefore, a series of tests was performed to verify this speculation. First, the hypothesis that miR-485-5p was directly linked to MUC1 was verified using the luciferase reporter assay; then inhibition of MUC1 expression by miR-485-5p was shown using qRT-PCR and western blot. Therefore, we conclude and report for the first time that MUC1 serves as a downstream gene of miR-485-5p.
To validate whether miR-485-5p had a regulatory effect on MUC1 in HCC, we silenced MUC1 in HepG2 cells by transfection with siMUC1. The results showed that down-regulation of MUC1 suppressed the cell viability, as well as migration and invasion abilities; however, it facilitated cell apoptosis, which could be reversed by inhibiting miR-485-5p expression. Furthermore, as we all know, certain proteins act as key factors in tumor development. Herein, western blot showed that the expressions of MMP-2 (related to tumor neovascularization and tumor metastasis), MMP-9 (involved in angiogenesis by releasing vascular endothelial growth factor), BCL2 (apoptotic protein) and BAX (proapoptotic protein) were related to MUC1 levels, which were modulated by miR-485-5p.
Non-coding RNAs are attracting increasing attention for their regulatory roles in cancer.[34,35] The first circRNA was discovered as early as in 1991[15]; however, until the emergence of next-generation sequencing, they were labeled as “products of splicing errors” and even considered to be “transcription noise.”[36-38] Later, with the progression of biotechnology, their crucial roles in biology, especially in cancer biology, were gradually discovered. Nowadays, mounting evidence demonstrates that circRNAs are involved in human malignancies,[19,39,40] mainly acting as miRNA molecular sponges to compete with other endogenous RNAs.[41,42] Recent research shows that circHECTD1 is associated with gastric cancer (GC) progression. In GC, circHECTD1 promotes ubiquitin carboxyl-terminal hydrolase 5 expression by targeting miR-1256.[43] In the present study, we found complementary sequences between circHECTD1 and miR-485-5p through Starbase 2.0 analysis, and subsequently, it was proved by RIP and luciferase reporter assays that circHECTD1 interacted with miR-485-5p directly. Knockdown of circHECTD1 resulted in up-regulation of miR-485-5p, which further confirmed that circHECTD1 sponged miR-485-5p.
Next, several experiments were conducted to clarify the circHECTD1/miR-485-mediated HCC procession from a mechanical perspective. MiR-485-5p binds to MUC1 directly, while circHECTD1 binds to miR-485-5p, resulting in the up-regulation of the MUC1 level indirectly. Then, functional experiments indicated that down-regulation of circHECTD1 suppressed cell viability, invasion ability, and migration ability, which could be reversed by inhibiting miR-485-5p expression. In addition, in circHECTD1-knockdown cells, expressions of proteins (MMP-2, MMP-9, BCL2, and BAX) showed the same trend as in MUC1-knockdown cells.
MMP-2 and MMP-9 belong to the MMP family, which can degrade multiple proteins in the extracellular matrix and facilitate the invasion and metastasis of tumor cells. MMP expression can be induced by extracellular matrix metalloproteinase inducer (EMMPRIN), which can be down-regulated by miR-485-5p.[33,44] This suggests that on one hand, circHECTD1 promoted MUC1 expression by sponging miR-485-5p, while MUC1 improved the expressions of both MMP-2 and MMP-9 to accelerate the progression of HCC; on the other hand, miR-485-5p down-regulated EMMPRIN, which, in turn, inhibited MMP-2 and MMP-9 expression levels, which restricted tumor invasion and migration.
Moreover, it has been reported that MUC1 is involved in apoptosis-related signaling pathways (5). In human hepatoma SMMC-7721 cells, silenced MUC1 inhibits cell proliferation through pathways that include the death receptor apoptotic ones mediated by caspase-8 and mitochondrial ones mediated by BAX.[6] In this study, the expressions of apoptosis-related BCL2 and BAX proteins were determined, and the results showed that down-regulation of MUC1 or circHECTD1 resulted in decreased BCL2 level, while the BAX level increased. However, the caspase proteins were not examined.
Collectively, these results further validated that circHECTD1 facilitates MUC1 expression by targeting miR-485-5p and then promotes HCC progression, the circHECTD1/miR-485-5p/MUC1 regulatory network might play a role in the HCC pathophysiological process, and circHECTD1 might be a potential therapeutic target for HCC in the future.
Funding
This work was supported by a grant from the Medical Health Science and Technology Program of Zhejiang Province (No. 2019PY003).
Conflicts of interest
None.
Footnotes
How to cite this article: Jiang QL, Feng SJ, Yang ZY, Xu Q, Wang SZ. CircHECTD1 up-regulates mucin 1 expression to accelerate hepatocellular carcinoma development by targeting microRNA-485-5p via a competing endogenous RNA mechanism. Chin Med J 2020;133:1774–1785. doi: 10.1097/CM9.0000000000000917
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ge S, Huang D. Systemic therapies for hepatocellular carcinoma. Drug Discov Ther 2015;9:352–362. doi: 10.5582/ddt.2015.01047. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 4.Shimizu M, Yamauchi K. Isolation and characterization of mucin-like glycoprotein in human milk fat globule membrane. J Biochem 1982;91:515–524. doi: 10.1093/oxfordjournals.jbchem.a133724. [DOI] [PubMed] [Google Scholar]
- 5.Martínez-Sáez N, Peregrina JM, Corzana F. Principles of mucin structure: implications for the rational design of cancer vaccines derived from MUC1-glycopeptides. Chem Soc Rev 2017;46:7154–7175. doi: 10.1039/c6cs00858e. [DOI] [PubMed] [Google Scholar]
- 6.Yuan H, Wang J, Wang F, Zhang N, Li Q, Xie F, et al. Mucin 1 gene silencing inhibits the growth of SMMC-7721 human hepatoma cells through Bax-mediated mitochondrial and caspase-8-mediated death receptor apoptotic pathways. Mol Med Rep 2015;12:6782–6788. doi: 10.3892/mmr.2015.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Ni WH, Hu KB, Zhai XY, Xie F, Jie J, et al. Targeting MUC1 and JNK by RNA interference and inhibitor inhibit the development of hepatocellular carcinoma. Cancer Sci 2017;108:504–511. doi: 10.1111/cas.13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye J, Wei X, Shang Y, Pan Q, Yang M, Tian Y, et al. Core 3 mucin-type O-glycan restoration in colorectal cancer cells promotes MUC1/p53/miR-200c-dependent epithelial identity. Oncogene 2017;36:6391–6407. doi: 10.1038/onc.2017.241. [DOI] [PubMed] [Google Scholar]
- 9.Xiang S, Zou P, Wu J, Zheng F, Tang Q, Zhou J, et al. Crosstalk of NF-(B/P65 and LncRNA HOTAIR-mediated repression of MUC1 expression contribute to synergistic inhibition of castration-resistant prostate cancer by polyphyllin 1-enzalutamide combination treatment. Cell Physiol Biochem 2018;47:759–773. doi: 10.1159/000490028. [DOI] [PubMed] [Google Scholar]
- 10.Rajabi H, Hata T, Li W, Long MD, Hu Q, Liu S, et al. MUC1-C represses the RASSF1A tumor suppressor in human carcinoma cells. Oncogene 2019;38:7266–7277. doi: 10.1038/s41388-019-0940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamaki N, Kuno A, Matsuda A, Tsujikawa H, Yamazaki K, Yasui Y, et al. Serum wisteria floribunda agglutinin-positive sialylated mucin 1 as a marker of progenitor/biliary features in hepatocellular carcinoma. Sci Rep 2017;7:244 doi: 10.1038/s41598-017-00357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhai X, Zhu H, Wang W, Zhang S, Zhang Y, Mao G. Abnormal expression of EMT-related proteins, S100A4, vimentin and E-cadherin, is correlated with clinicopathological features and prognosis in HCC. Med Oncol 2014;31:970 doi: 10.1007/s12032-014-0970-z. [DOI] [PubMed] [Google Scholar]
- 13.Li Q, Liu G, Shao D, Wang J, Yuan H, Chen T, et al. Mucin1 mediates autocrine transforming growth factor beta signaling through activating the c-Jun N-terminal kinase/activator protein 1 pathway in human hepatocellular carcinoma cells. Int J Biochem Cell Biol 2015;59:116–125. doi: 10.1016/j.biocel.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 14.Li Q, Wang F, Liu G, Yuan H, Chen T, Wang J, et al. Impact of Mucin1 knockdown on the phenotypic characteristics of the human hepatocellular carcinoma cell line SMMC-7721. Oncol Rep 2014;31:2811–2819. doi: 10.3892/or.2014.3136. [DOI] [PubMed] [Google Scholar]
- 15.Che NLL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 16.Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Hansen TB, Jensen T, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles functions as efficient microRNA spongs. Nature 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 18.Zhu Q, Lu G, Luo Z, Gui F, Wu J, Zhang D, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR 1324/FZD5/Wnt/β-actenin axis. Biochem Biophys Res Commun 2018;497:626–632. doi: 10.1016/j.bbrc.2018.02.119. [DOI] [PubMed] [Google Scholar]
- 19.Arnaiz E, Sole C, Manterola L, Iparraguirre L, Otaegui D, Lawrie CH. CircRNAs and cancer: biomarkers and master regulators. Semin Cancer Biol 2019;58:90–99. doi: 10.1016/j.semcancer.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Ni J, Bucci J, Chang L, Malouf D, Graham P, Li Y. Targeting microRNAs in prostate cancer radiotherapy. Theranostics 2017;7:3243–3259. doi: 10.7150/thno.19934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest 2017;127:761–771. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin X, Chen Y. Identification of potentially functional circRNA-miRNA-mRNA regulatory network in hepatocellular carcinoma by integrated microarray analysis. Med Sci Monit Basic Res 2018;24:70–78. doi: 10.12659/MSMBR.909737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu L, Feng X, Hao X, Wang P, Zhang Y, Zheng X, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res 2019;38:98 doi: 10.1186/s13046-019-1041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao S, Wang G, Wang J, Li C, Zhang L. Hsa_circ_101280 promotes hepatocellular carcinoma by regulating miR-375/JAK2. Immunol Cell Biol 2019;97:218–228. doi: 10.1111/imcb.12213. [DOI] [PubMed] [Google Scholar]
- 25.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 26.Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med 2014;20:332–342. doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parrales A, Iwakuma T. Targeting oncogenic mutant p53 for cancer therapy. Front Oncol 2015;5:288 doi: 10.3389/fonc.2015.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miklavcic JJ, Schnabl KL, Mazurak VC, Thomson AB, Clandinin MT. Dietary ganglioside reduces proinflammatory signaling in the intestine. J Nutr Metab 2012;2012:280286 doi: 10.1155/2012/280286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miklavcic JJ, Hart TD, Lees GM, Shoemaker GK, Schnabl KL, Larsen BM, et al. Increased catabolism and decreased unsaturation of ganglioside in patients with inflammatory bowel disease. World J Gastroenterol 2015;21:10080–10090. doi: 10.3748/wjg.v21.i35.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohkawa Y, Miyazaki S, Miyata M, Hamamura K, Furukawa K, Furukawa K. Essential roles of integrin-mediated signaling for the enhancement of malignant properties of melanomas based on the expression of GD3. Biochem Biophys Res Commun 2008;373:14–19. doi: 10.1016/j.bbrc.2008.05.149. [DOI] [PubMed] [Google Scholar]
- 31.Jing LL, Mo XM. Reduced miR-485-5p expression predicts poor prognosis in patients with gastric cancer. Eur Rev Med Pharmacol Sci 2016;20:1516–1520. [PubMed] [Google Scholar]
- 32.Wang M, Cai WR, Meng R, Chi JR, Li YR, Chen AX. miR-485-5p suppresses breast cancer progression and chemosensitivity by targeting survivin. Biochem Biophys Res Commun 2018;501:48–54. doi: 10.1016/j.bbrc.2018.04.129. [DOI] [PubMed] [Google Scholar]
- 33.Sun X, Liu Y, Li M, Wang M, Wang Y. Involvement of miR-485-5p in hepatocellular carcinoma progression targeting EMMPRIN. Biomed Pharmacother 2015;72:58–65. doi: 10.1016/j.biopha.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 34.Wu J, Zhao W, Wang Z, Xiang X, Zhang S, Liu L. Long non-coding RNA SNHG20 promotes the tumorigenesis of oral squamous cell carcinoma via targeting miR-197/LIN28 axis. J Cell Mol Med 2019;23:680–688. doi: 10.1111/jcmm.13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao W, Ma X, Liu L, Chen Q, Liu Z, Zhang Z, et al. SNHG20: a vital lncRNA in multiple human cancers. J Cell Physiol 2019;234:14519–14525. doi: 10.1002/jcp.28143. [DOI] [PubMed] [Google Scholar]
- 36.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 38.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 39.Zhang M, Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J Hematol Oncol 2018;11:21 doi: 10.1186/s13045-018-0569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu J, Qi X, Liu L, Hu X, Liu J, Yang J, et al. Emerging epigenetic regulation of circular RNAs in human cancer. Mol Ther Nucleic Acids 2019;16:589–596. doi: 10.1016/j.omtn.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li D, Yang Y, Li ZQ, Li LC, Zhu XH. Circular RNAs: from biogenesis and function to diseases. Chin Med J 2019;132:2457–2464. doi: 10.1097/CM9.0000000000000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai J, Chen Z, Wang J, Wang J, Chen X, Liang L, et al. circHECTD1 facilitates glutaminolysis to promote gastric cancer progression by targeting miR-1256 and activating β-catenin/c-Myc signaling. Cell Death Dis 2019;10:576 doi: 10.1038/s41419-019-1814-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaushik DK, Hahn JN, Yong VW. EMMPRIN, an upstream regulator of MMPs, in CNS biology. Matrix Biol 2015;44-46:138–146. doi: 10.1016/j.matbio.2015.01.018. [DOI] [PubMed] [Google Scholar]


