Abstract
Gastric cancer (GC) is a common malignancy and is the third leading cause of cancer-related death. At present, there is no simple and effective screening method for early-stage GC, and the treatment results and prognosis are poor. With the continuous improvement of molecular biology techniques, research on circular RNA (circRNA) has gradually expanded over time. Much data supports the role of circRNA in tumorigenesis. Moreover, due to its structural specificity and biological stability, circRNA is anticipated to be a potential biomarker for tumor diagnosis. Studies have confirmed that circRNA can participate in the proliferation, invasion, metastasis, and apoptosis of GC. These findings will lead to novel directions for the diagnosis and treatment of GC. This article reviews the structure and function of circRNA, summarizes the current studies on circRNA, and discusses the potential diagnostic value of circRNA in GC.
Keywords: Circular RNA, Gastric cancer, Biomarker, Diagnosis
Introduction
Gastric cancer (GC) originates from the gastric mucosal epithelium. It is a common malignant tumor. According to the report from the World Health Organization's International Cancer Research Center, in 2018[1] GC ranked fifth in global cancer incidence and is the third leading cause of death amongst cancer patients. More than one million new cases were reported in 2018. The death toll is estimated at 783,000 per annum (equivalent to one per 12 deaths worldwide). Therefore, prevention and early diagnosis of GC are essential. At present, there is still no simple and efficient means for early screening of GC. Additionally, the sensitivity and specificity of existing biomarkers for clinical use need to be improved.
The continual advancements in the field of molecular biology and extensive use of technologies such as high-throughput sequencing have led to the rapid discovery of non-coding RNAs (ncRNAs). Amongst them, circular RNAs (circRNAs) have been determined to be an important class of ncRNAs. Research on circRNAs is gradually expanding. Several studies have shown that circRNA can play an important role in a variety of cancers by acting as an oncogene or tumor suppressor gene. Moreover, studies have confirmed that circRNA can interact with genes involved in GC, thus, participating in GC proliferation, invasion, apoptosis, and metastasis. For example, Zhang et al[2] found that the circLARP4 inhibits biological behaviors of GC cells by sponging miR-424. There is a binding site between miR-424 and large tumor suppressor kinase 1, a core part of the Hippo signaling pathway that functions as a tumor suppressor in GC. Thus, circRNA may play an important role in GC development. Due to its structural specificity and biological stability, circRNA may become a potential target for disease diagnosis and treatment. This article reviews the origin, characteristics, and effects of circRNA, and describes its role in GC development.
Definition and Characteristics of CircRNA
Definition of circRNA
NcRNA refers to RNA which is widely present in various living organisms, but cannot encode proteins, which play an important role in regulating life activities. In the human genome, genes encoding proteins account for only 1% to 2% of all genes.[3] Therefore, most genes produce ncRNA.
CircRNA is a precursor mRNA (pre-mRNA) derived from the transcription of RNA transcriptase II.[4–6] It is a special type of endogenous ncRNA. It does not contain the linear structure of the 5′ cap structure and the 3′ poly(A) tail of other RNAs. Instead, it forms a closed-loop structure, which is more stable than traditional linear RNA by covalent bond closure.[7–10]
CircRNA was discovered over 40 years ago. As early as 1976, Sanger et al[11] utilized virus-like research and found single-stranded circRNAs in higher plants. In 1979, Hsu et al[12] determined that the content of circRNA in Hela cells accounted for at least 1% to 2% of total RNA content using electron microscopy. However, due to the limited technical means at the time, circRNA was only considered to be a by-product of abnormal splicing of transcripts. Therefore, it did not attract much scientific attention. With the continuous development of molecular purification and high-throughput sequencing technologies, bioinformatic analysis technology is gradually progressing. In recent years, more and more circRNAs have been discovered and further researched. Intensive research has revealed that circRNA is widely present in various tissues, including nerve and liver, and so on.[13–15] and that the differences in circRNA contained in various tissues are also significant. As a result of recent studies, as many as 140,000 circRNAs have been discovered.[9,16] This suggests that circRNA may become the largest transcript in human cells.
Sources of circRNA
The circRNA of eukaryotic cells is a single-stranded circular structure lacking a free 5′ end cap and a 3′ poly(A) tail. The majority of circRNA is composed of exons; however, some may also contain one or two introns. Depending on the source of the exon and intron in the genome and its constituent sequences, eukaryotic circRNAs can generally be classified into the following three categories: (1) exon-derived circRNA molecules (exonic circRNA [ecRNA]), which contains only exons. These exons are connected by a 5′ end-3′ end,[8,17] (2) circular intronic RNA (ciRNA), which contains only introns and is looped through the 5′ end-2′ end,[18] and (3) exon and intron common source circRNA molecules (exon-intron circRNA [EIciRNA]) containing both exons and introns.[19] Studies have found that the types of circRNA increase with the degree of evolution of a species.[20] Therefore, the types of circRNAs can be very diverse.
Characteristics of circRNA
The structure of circRNA is different from that of traditional linear RNA, which is a closed RNA formed by non-covalent linkage of exons and/or introns. Studies have found that circRNA has the following characteristics: (1) circRNA is widely present in all eukaryotic organisms and is high in content; (2) The expression levels of circRNA in different tissues and different developmental stages of the same organism are different. Additionally, the content of circRNA is also very different in different tumor tissues; therefore, it has spatial and temporal specificity. (3) As circRNA does not have a 5′ end cap and a 3′ end poly(A) structure, it could not be easily cleaved by exonuclease giving its structural stability compared with linear RNA[21]; (4) circRNA is rich in microRNA (miRNA) binding sites, which can bind to corresponding miRNAs and act as a sponge[22]; (5) ciRNA has a highly conserved sequence, most of which does not mutate during biological evolution; (6) circRNA can be widely found in human exosomes (small vesicles containing complex RNAs and proteins). At present, there are more than 1000 types of circRNAs in human serum exosomes. The expression of these circRNAs in some tumor tissues is highly specific. Therefore, circRNA can be used as potential tumor markers.[23] Other features of circRNA remain to be elucidated.
CircRNA Synthesis and Degradation Mechanism
CircRNA synthesis
The structural genes of most eukaryotes are split genes. The pre-mRNA directly generated by DNA transcription generally needs to be excised by splicing. The remaining exon sequences are then spliced to form a mature linear mRNA,[24] a split gene. Most circRNAs are also derived from pre-mRNA. However, the difference is in the splicing reaction. In circRNA, the upstream 5′ cleavage site and the downstream 3′ cleavage site are linked by a covalent bond to form a circular transcript. This splicing method is called reverse-splicing.[8,25] The synthesis of circRNA is shown in Figure 1. Studies have found that reverse-splicing of pre-mRNA can form three different circRNAs: ecRNA, ciRNA, and EIciRNA. The formation mechanisms of the three circRNAs are as follows:
Figure 1.
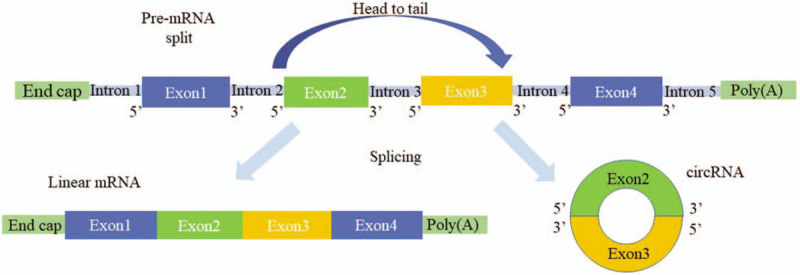
Circular RNA synthesis. Most pre-mRNA remove introns and splice the remaining exon sequences to form mature linear mRNA. In circRNA, the upstream 5′ cleavage site and the downstream 3′ cleavage site are linked by a covalent bond to form a circular transcript. circRNA: Circular RNA; pre-mRNA: Precursor mRNA.
Formation mechanism of ecRNA
The exon-derived circRNA molecule is mainly located in the cytoplasm and is produced by the exon cyclization pathway. Jeck et al[26] proposed a cyclization mechanism for two exons, namely lariat driven circularization and intron pairing driven circularization. The lariat driven circularization is also known as exon skipping. It refers to the exon skipping of pre-mRNA during post-transcriptional processing during the lariat driven circularization, which brings the distance of the non-adjacent exons closer. Then, by reverse splicing, the 3′ end of the downstream exon splice donor is covalently bound to the 5′ end of the splice acceptor of the upstream exon to form a lasso intermediate. The intron is then removed to form an ecRNA. In the intron pairing driven cyclization process, the introns flanking the circRNA exons are mainly mediated by reverse complementation, which promotes the exon to form a cyclized structure. The intron in the loop is then cleaved and ligated to form an ecRNA. The most common flanking complement is the complementary pairing of Alu and B1, which plays an important role in the process of exon cyclization. The perfectly matched complementary sequence can promote the expression of circRNA.[26,27]
Formation mechanism of ciRNA
Intron-derived circRNA molecules are produced by the intron cyclization pathway and are mainly distributed in the nucleus.[18] The formation of ciRNA needs to be driven by several conserved sequences, which mainly include a GU-rich sequence near the 5′-terminal cleavage site (the length of which is seven nucleotides [nt]) and a C-base-rich sequence near the 3′-end branch site (the length of which is 11 nt). Most introns form a lasso structure during the splicing process; however, these structures are degraded by debranching enzymes soon after debranching.[28] The specific sequences determined by these lengths are resistant to debranching enzyme degradation. Additionally, the lasso structure formed by the introns is prevented from being degraded. Thereby, a structurally stable circular structure ciRNA is formed, increasing the formation efficiency of ciRNA.[18,29] In the process of ciRNA formation, this specific sequence in pre-mRNA is required for the normalization of cyclization. So this specific sequence is a prerequisite for the synthesis of ciRNA by eukaryotes.[18]
Formation mechanism of EIciRNA
EIciRNA molecules are mainly present in the nucleus. In the cyclization process driven by intron pairing, if the intron sequence contained in the RNA molecule is not completely excised, but a part of it is stably retained, an EIciRNA in which the intron and the exon sequence coexist is formed. For example, in the nucleus of cervical cancer HeLa cells, reverse splicing forms the EIciRNA containing the intron.[19,30]
CircRNA degradation
Since circRNA does not have a 5′ end cap and a 3′ poly(A) tail, it is resistant to degradation by ribonuclease R (RNase R, a 3′-5′ exonuclease derived from the Escherichia coli superfamily). Therefore, it is more stable than linear RNA.[31] It may be degraded by other means.
At present, there are few reports on the mechanism of circRNA degradation. However, studies have found that miR-671 can rely on Argonaute protein (AGO) to directly degrade circRNA sponge for miR-7 (ciRS-7).[17] Experiments have also shown that higher concentrations of circRNAs than corresponding linear RNAs have been found in extracellular vesicles (EVs), such as exosomes and microvesicles recovered from cell culture fluids.[32] This suggests that circRNA can be excreted outside the cell by EVs which may be the cellular clearance mechanism of circRNA.
Function of CircRNA
Regulation of gene expression
The primary biological role of ncRNA is to regulate gene expression. Studies have shown that, as a member of the ncRNA family, circRNA can also regulate the transcription process of genes.[33] CircRNA is widely distributed in the nucleus of eukaryotic cells and can promote gene transcription by binding to the RNA polymerase II complex or proteins involved in transcription, thereby regulating the expression of genes.[18] For example, Li et al[30] found that there are two nuclear EIciRNAs (circEIF3J and circPAIP2) which have sites for binding to small nuclear RNA (snRNA) in the U1 nucleus. These EIciRNAs can regulate gene expression by interacting with snRNA. EIciRNA first forms an EIciRNA-U1snRNP complex with U1 small nuclear ribonucleoprotein (snRNP), which interacts with the RNA polymerase II transcriptional complex in the promoter region of the parental gene to promote gene expression and regulate transcriptional genes.
miRNA sponge effect of circRNA
As a short-chain ncRNA molecule, miRNA can specifically bind to the 3′ untranslated region (UTR) of mRNA. This inhibits the translational function of the target mRNA or induces degradation of the target mRNA, and regulates gene expression after transcription.[34] Because miRNA can regulate the level of RNA that binds to it, this is called the molecular sponge of miRNA.[35,36] Studies have shown that gene expression can also be regulated by the interaction of circRNA with miRNA.[37] Many circRNAs bind to their corresponding miRNAs and act as competing endogenous RNAs (ceRNAs), thereby relieving the negative regulation of miRNA expression on target genes to increase target gene expression levels. This effect is called miRNA sponge action of circRNA.
The first circRNA reported to have such an effect is ciRS-7, which was found by Hansen et al.[22] Since ciRS-7 contains approximately 70 miR-7 miRNA response elements, it can act as miR-7 sponge. By adsorbing and strongly inhibiting the function of miR-7, it can be negatively regulated.
In eukaryotes, more and more circRNAs have been found to contain miRNA binding sites. For example, circHIPK3 can be used as a molecular sponge for many miRNAs.[16] Wang et al[38] found that heart-related circRNA can be used as a sponge for miR-223. Its binding to miR-233 can inhibit the activity of miRNA, thereby greatly increasing the gene expression downstream of miR-223, ultimately inhibiting cardiac hypertrophy and heart failure and slowing the progression of the disease.
MiRNAs can participate in normal physiological activities and play an important role in the disease development of diseases. CircRNA can regulate the activity of miRNAs in vivo because of its miRNA molecular sponge. Therefore, circRNA is undoubtedly involved in the regulation of disease development. Due to the structural stability of circRNA, the miRNA's sponge action is more durable than other types of competitive endogenous RNA.
CircRNA can interact with RNA binding proteins (RBPs)
As demonstrated with in-depth studies of circRNA molecules, circRNA not only binds directly to proteins, but also indirectly binds to certain proteins via RNA. CircRNA can bind to RBP to form an RNA-protein complex. This complex regulates the interaction between RNA and RNA-binding proteins to regulate gene transcription and protein activity, thereby affecting the expression level of post-transcriptional genes.[15]
Certain ecRNA molecules can be used as target sequence elements. They use their complementary sequences to bind RBP, DNA, or RNA through the principle of base complementation, which serves as a bridge between RBP, DNA, and RNA. For example, cerebellar degeneration-related protein 1 transcript (CDR1as) can bind tightly to the miRNA's effector AGO,[15,22] which is cleaved and degraded by miR-671. Ashwal-Fluss et al[4] also found in both human and fruit fly studies that the splicing factor (muscle blind, MBL/MBNL1) binds to the second exon to form circRNA (circMBL). The resulting circMBL has a binding site for MBL on the flanking intron. When the intracellular MBL protein content is too high, it promotes the production of circMBL, thereby reducing the mRNA production of the protein.
The translation role of circRNA
Studies have shown that some ncRNAs also have translational functions to produce functional proteins or peptides in vivo.[39–41] Researchers previously thought that circRNA is one of ncRNAs. There is no suitable internal ribosome entry site (IRES) due to its lack of 5′ end cap and 3′ poly(A) tail structure, so it is impossible to translate into protein. However, it has finally been discovered that there are many genes from which circRNAs are derived. Moreover, exon sequences can also be included in the structure, which may prove that circRNA has a translation function with a coding protein. If these circRNAs contain an IRES, the eukaryotic ribosomal 40S small subunit can enter the IRES of these circRNAs through this site, thereby guiding protein synthesis.[42]
The first circRNA discovered to have a protein-encoding function is the RNA of hepatitis D virus (HDV) found by Kos et al[43] HDV is a satellite virus of hepatitis B virus, a single-stranded circRNA molecule with a protein-encoding function. Its circRNA can express pathogenic viral proteins.[13] Later, AbouHaidar et al[44] also found that there is a highly stable circRNA molecule of about 220 nt long on the rice yellow spot virus. It is a covalently closed circRNA (RCCC RNA). This type of circRNA has an internal ribosome binding site that binds to the corresponding ribosome and translates a protein with a relative molecular mass of 16,000. Other studies have further confirmed that after the endogenous circRNA begins to be translated, its special structure may make it easier for ribosomes to circulate and participate in the elongation of the polypeptide chain.[45]
The functions of circRNA are illustrated in Figure 2.
Figure 2.
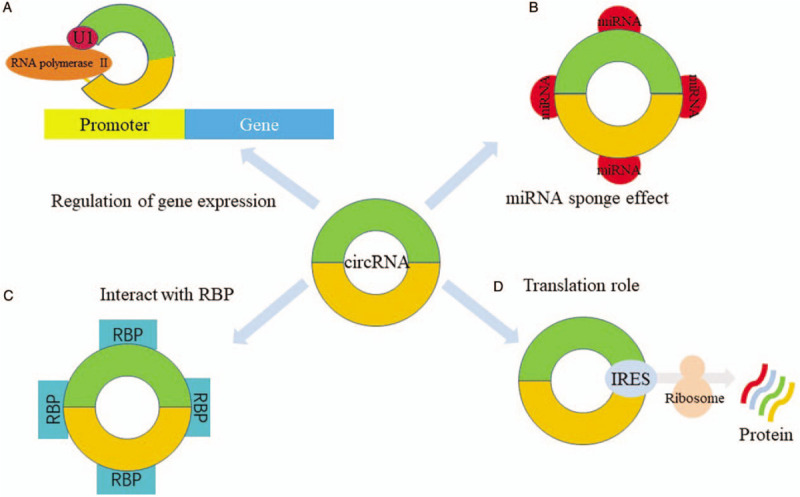
Function of circular RNA. (A) Regulation of gene expression: circRNA binds to snRNA in the U1 nucleus, and then binds to the promoter region of the parent gene together with RNA polymerase II (Pol II) to regulate its own expression; (B) miRNA sponge effect of circRNA: circRNAs have many miRNA binding sites, which can bind to their corresponding miRNAs and act as ceRNAs; (C) interact with RNA binding proteins: circRNA can bind to RBP to form an RNA-protein complex; (D) translation role of circRNA: circRNAs contain an IRES, the eukaryotic ribosomal 40S small subunit can enter the internal ribosome entry site of these circRNAs through this site, thereby guiding protein synthesis. ceRNA: Competing endogenous RNA; circRNA: Circular RNA; IRES: Internal ribosome entry site; miRNA: MicroRNA; RBP: RNA binding protein; snRNA: Small nuclear RNA.
Research Method of CircRNA
Expression analysis techniques
Including RNA sequencing, real-time quantitative polymerase chain reaction (RT-PCR), Northern blotting, two-dimensional polyacrylamide gel electrophoresis (2D-PAGE), RNase R and RNase H chip technology, RNA-fluorescence in situ hybridization, and biotin-coupled circRNA capture technology are used in circRNA research.
RNA sequencing is a transcriptomics research method based on second-generation gene sequencing technology. Early RNA sequencing was performed using Ploy(A) tail enrichment to construct a library for sequencing. However, circRNA lacks the Poly(A) tail due to the closed loop structure. Therefore, circRNA has been largely eliminated when constructing the library. Presently, RNA sequencing has begun to construct complementary DNA (cDNA) libraries using ribosomal ribonucleic acid removal and random primer methods combined with a series of algorithmic formulas. This has led to the discovery of abundant circRNA expression in a variety of eukaryotic organisms.[46] RT-PCR is the easiest and fastest way to identify circRNA. Primers designed for circRNA reverse splice site sequences allow amplification and quantification of specific circRNAs.
RNase R is an exonuclease, which is capable of degrading RNA containing 5′ and 3′ ends. It can be combined with RT-PCR to identify circRNA and can also be used for sequencing circRNA purification and sequencing results.[47] Starke et al[6] utilized a combination of Northern blotting and RNase H. verified that the exon circRNA has a closed loop structure. Northern blotting recognized the circularized exon sequence by the designed probe. RNase H is an endoribonuclease that cleaves RNA when DNA and RNA hybridize. This method effectively distinguishes between circRNA and trans-splicing products.
In 2D-PAGE, linear RNAs move diagonally according to their size, while circRNA moves in an arc. Therefore, 2D-PAGE can be used to enrich and quantify circRNA for sequencing and to quantify and characterize it.[48] In other methods, chip technology is mainly used to screen circRNA. The biotin-coupled circRNA capture technology is a circRNA northern blotting technique based on multi-biotin signal amplification with high sensitivity.
Functional research
It mainly includes circRNA overexpression by constructing related vectors, designing siRNA, and small hairpin RNA knockdown (knockout) circRNA. For example, by transfecting the pcDNA3-ciRS-7 recombinant plasmid into zebrafish embryos, ciRS-7 overexpression can be achieved. It has been found to affect brain development. Zhang et al[49] used siRNA technology to study the role of circRNA in the pathogenesis of colorectal cancer and found that hsa_circ_0007534 can inhibit the proliferation of colorectal cells and induce apoptosis.
CircRNA and miRNA interaction studies
At present, there are two main methods for studying the interaction between circRNA and miRNA, namely dual-luciferase reporter assay and RNA immunoprecipitation (RIP).[2] The dual-luciferase reporter assay first constructs a luciferase reporter vector containing the circRNA fragment of interest and then co-transfects the miRNA with the reporter gene into the cell. The binding of miRNA to circRNA is verified by detecting the activity of firefly and Renilla luciferase.
The interaction between circRNA and miRNA can also be achieved by RIP of AGO protein in combination with quantitative real-time PCR (qRT-PCR) analysis. Since circRNA, miRNA, and AGO proteins can co-localize in cells, AGO proteins can be enriched by antibody-specific binding purification to explore the number, changes, and interactions of circRNA and miRNA. This method can be used to verify the “sponge effect” of circRNA adsorption on miRNAs.
Functions of CircRNAs in GC
It has been reported that circRNA is differentially expressed in many cancers and can affect cancer occurrence, development, and metastasis. Therefore, circRNA plays an important role in GC.
Differentially expressed circRNAs in GC
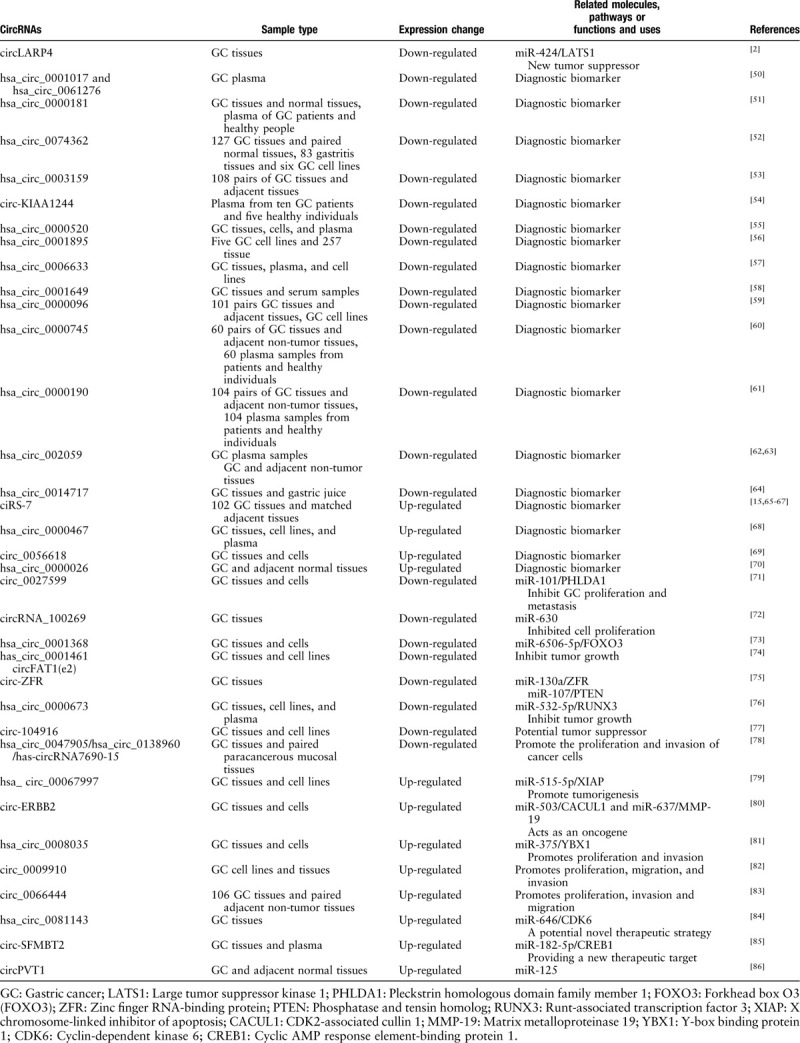
Since the discovery of circRNA, its relationship with tumors has been a hot topic of research. Researchers hope to further elucidate the pathogenesis of tumors through the study of circRNA, and find novel methods of diagnosis and treatment. A large number of studies have shown that there are several differentially expressed circRNAs in GC. Some expression levels were significantly elevated, while others were decreased. These differentially expressed circRNAs are summarized in Table 1. For example, in a study by Xie et al[52], hsa_circ_0074362 was significantly down-regulated in GC tissue when compared with adjacent tissues. Similarly, Li et al[50] also found that hsa_circ_0001017 and hsa_circ_0061276 were significantly decreased in GC patients’ plasma. In another example, Lu et al[70] found that hsa_circ_0000467 was up-regulated in GC tissues, plasma and cell lines. The discovery of these differentially expressed circRNAs indicates that circRNA is involved in GC development and progression, providing new clues for revealing the mechanism of GC and providing a new direction for its diagnosis and treatment.
Table 1.
Differential expression and functions of circRNAs in gastric cancer.
CircRNA can be used as a diagnostic marker for GC
Zhao et al[51] found that the level of hsa_circ_0000181 in GC patients’ tissues and plasma was significantly lower than that of non-tumor tissues or healthy human plasma. The expression of hsa_circ_0000181 in GC tissues was significantly correlated with tumor diameter, lymphatic metastasis, distant metastasis, and carbohydrate antigen 19-9 (CA19-9) (P < 0.05). The decrease of hsa_circ_0000181 in the plasma of patients was significantly correlated with differentiation and carcinoembryonic antigen (P < 0.05). Therefore, detection of hsa_circ_0000181 in tissues and plasma may be a biomarker for GC diagnosis. Tian et al[53] also found that the expression level of hsa_circ_0003159 was significantly negatively correlated with gender, distant metastasis, and tumor-lymph node metastasis. Therefore, hsa_circ_0003159 may be a potential tumor biomarker for GC.
Chen et al[61] found that hsa_circ_0000190 was down-regulated in GC patients’ tissues and plasma (P < 0.001) and its expression level was significantly correlated with tumor diameter, lymph node metastasis, distant metastasis, tumor, node, and metastasis (TNM) stage and CA19-9 level (P < 0.05). The combined determination of tissue and plasma circRNA can improve the accuracy of diagnosis, indicating that hsa_circ_0000190 may be a novel non-invasive biomarker for the diagnosis of GC.
CircRNA inhibits the occurrence, development, and metastasis of GC
Wang et al[71] studied the mechanism of action of circRNA in the GC suppressor gene pleckstrin homologous domain family member 1 (PHLDA1). The circRNA expression profile of the circRNA gene chip in GC tissues showed that hsa_circ_0027599 (circ_0027599) was significantly down-regulated in GC tissues when compared to the control. Overexpression of circ_0027599 inhibits proliferation and metastasis of GC cells. Using bioinformatics tools and luciferase reporter assays, it was confirmed that circ_0027599 is a sponge of miR-101-3p.1 (miR-101), which inhibits the survival and metastasis of cancer cells. It was also confirmed that PHLDA1 is regulated by circ_0027599 in GC cells. Therefore, the study found that PHLDA1 is regulated by circ_0027599/miR-101, which can inhibit the survival and metastasis of GC.
Lu et al[73] found that hsa_circ_0001368 was widely down-regulated in GC tissues and cells, which was associated with poor prognosis in patients with GC. Functional experiments have shown that knocking down hsa_circ_0001368 can promote cell proliferation and invasion. In addition, the knockdown of hsa_circ_0001368 also leads to accelerated growth of tumors in vivo. Subsequently, it was confirmed that hsa_circ_0001368 was used as a ceRNA of the sponge miR-6506-5p. Although forkhead box O3 (FOXO3) can serve as a functional target of miR-6506-5p, knocking out hsa_circ_0001368 reduced the expression of the tumor suppressor gene FOXO3. Therefore, research shows that in GC, hsa_circ_0001368 can exert tumor-suppressive effects through the miR-6506-5p/FOXO3 axis.
CircRNA can promote the occurrence of GC
Zhang et al[79] verified the expression of circ_0067997 in GC tissues and cell lines by qRT-PCR. High circ_0067997 expression was associated with low overall survival rate in GC patients. Knocking out circ_0067997 significantly reduced cell proliferation and invasion. In addition, X chromosome-linked inhibitor of apoptosis (XIAP) can be targeted and regulated by miR-515-5p, while circ_0067997 is directly identified as a sponge of miR-515-5p. Therefore, circ_0067997 can promote GC development by regulating the miR-515-5p/XIAP axis.
Huang et al[81] screened the up-regulated circRNA-hsa_circ_0008035 in microarray data. Functional experiments showed that siRNA-transfected hsa_circ_0008035 reduced the proliferation and invasion of GC cells. Hsa_circ_0008035 acts as a sponge for miR-375 and reduces its expression, while miR-375 can target Y-box binding protein 1 (YBX1) 3′-UTR. Therefore, hsa_circ_0008035 promotes proliferation and invasion of GC cells by modulating the miR-375/YBX1 axis.
CircRNA can be used to analyze the prognosis of GC patients
Chen et al[86] found a significant up-regulation of circPVT1 levels in a series of differentially expressed circRNA studies in GC. After predictive analysis and experiments, circPVT1 can be used as a “sponge” of miR-125, which competes with the miR-125 family and let-7 to regulate the expression of these miRNA target genes. This promotes the proliferation of GC cells. CircPVT1 is closely related to GC patient prognosis. High circPVT1 levels have been correlated with higher survival rates, which may be related to miR-125 with an anti-cancer effect. Additionally, circPVT1 can be used as a prognostic indicator of overall survival and disease-free survival independent of clinicopathological parameters such as tumor size and TNM staging. Therefore, circPVT1 can be used as an independent parameter for analyzing the prognosis of GC patients and is a new proliferative factor and prognostic indicator for GC.
CircRNA can be a potential new therapeutic target for the treatment of GC
Xue et al[84] found that hsa_circ_0081143 was up-regulated in GC tissues. Correlation analysis showed that the high expression of hsa_circ_0081143 was associated with poor TNM staging, lymph node metastasis, and overall survival. Then, through functional experiments, it was found that hsa_circ_0081143 inhibition not only decreases the viability of GC cells, reduces their invasive ability, but also induces GC cell sensitivity to cisplatin (DDP). Next, the interaction between hsa_circ_0081143 and miR-646 was determined. Hsa_circ_0081143 was demonstrated to act as an endogenous sponge by directly binding to miR-646. The down-regulation of miR-646 effectively reversed the inhibitory effect of hsa_circ_0081143 knockdown on cyclin-dependent kinase 6 (CDK6). In addition, hsa_circ_0081143 silencing inhibited tumorigenesis in vivo and significantly enhanced the inhibitory effect of DDP on GC cells. Therefore, hsa_circ_0081143 can be a potential new therapeutic strategy for GC therapy through the role of the hsa_circ_0081143/miR-646/CDK6 axis.
CircRNA regulates GC metabolism
Pan et al[67] found that ciRS-7 can interfere with the inhibitory effect of miR-7 on the phosphatase and tensin homolog (PTEN)/phosphatidylinositol 3-kinase (PI3K)/protein kinase B (PKB/AKT) pathway, thereby promoting GC cell proliferation. Previous studies have shown that the PTEN/PI3K/AKT pathway stimulates aerobic glycolysis. Thus, ciRS-7 may regulate the metabolism of GC cells through this pathway. It can be seen that the circRNA/miRNA regulatory axis may also play an important role in regulating the metabolism of gastrointestinal tumors.
CircRNA regulates the tumor microenvironment of GC
Studies have shown that the tumor cell's own circRNA is closely related to the gastrointestinal tumor microenvironment. The occurrence of epithelial-mesenchymal transition (EMT) in tumor cells is associated with a decrease in E-cadherin and an increase in vimentin in the microenvironment. Li et al[77] demonstrated that the expression of circ-104916 was down-regulated in GC tissues and cell lines. Overexpression of circ-104916 can down-regulate the content of vimentin in the microenvironment of GC and up-regulate the content of E-cadherin, ultimately inhibiting the EMT process of GC cells. Therefore, the use of circRNA to regulate the tumor microenvironment of GC will have potential application value.
Conclusions
More and more studies show that circRNAs may have potential for early diagnosis and treatment targets.[87] However, there is still a lack of multi-center clinical research data and their specific functions in the pathophysiology of GC development and molecular biological behavior are still not very clear. This topic requires further in-depth investigation. In the near future, they are likely to be useful and effective targets for the screening, diagnosis, and treatment of GC.
Funding
This work was supported by grants from the Scientific Research Foundation of Shanxi Province Healthy Commission (No. 2017068), the Natural Science Foundation of Shanxi Province (No. 201801D221259), and the Leading Clinical Department Sponsored by “136” Healthcare Improvement Project of Shanxi Province.
Conflicts of interest
None.
Footnotes
How to cite this article: Li XW, Yang WH, Xu J. Circular RNA in gastric cancer. Chin Med J 2020;133:1868–1877. doi: 10.1097/CM9.0000000000000908
Xue-Wei Li and Wen-Hui Yang contributed equally to this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer 2017; 16:151.doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis C, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014; 56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 5.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014; 28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, et al. Exon circularization requires canonical splice signals. Cell Rep 2015; 10:103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, et al. Scrambled exons. Cell 1991; 64:607–613. doi:10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 8.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J 1993; 7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 9.Pasman Z, Been MD, Garcia-Blanco MA. Exon circularization in mammalian nuclear extracts. RNA 1996; 2:603–610. [PMC free article] [PubMed] [Google Scholar]
- 10.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol 2015; 12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976; 73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979; 280:339–340. doi:10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 13.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014; 32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012; 7:e30733.doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 16.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016; 7:11215.doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 2011; 30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell 2013; 51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet 2013; 9:e1003777.doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong R, Ma XK, Chen LL, Yang L. Increased complexity of circRNA expression during species evolution. RNA Biol 2017; 14:1064–1074. doi: 10.1080/15476286.2016.1269999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Zheng QP, Bao CY, Li SY, Guo WJ, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015; 25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res 2015; 43:2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet 2016; 32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdelmohsen K, Panda AC, De S, Grammatikakis I, Kim J, Ding J, et al. Circular RNAs in monkey muscle: age-dependent changes. Aging (Albany NY) 2015; 7:903–910. doi: 10.18632/aging.100834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang XO, Wang HB, Zhang Y, Lu XH, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell 2014; 159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-Trelles F, Tarrio R, Ayala FJ. Origins and evolution of spliceosomal introns. Annu Rev Genet 2006; 40:47–76. doi: 10.1146/annurev.genet.40.110405.090625. [DOI] [PubMed] [Google Scholar]
- 29.Rearick D, Prakash A, McSweeny A, Shepard SS, Fedorova L, Fedorov A. Critical association of ncRNA with introns. Nucleic Acids Res 2011; 39:2357–2366. doi: 10.1093/nar/gkq1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015; 22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res 2006; 34:e63.doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lasda E, Parker R. Circular RNAs co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PLoS One 2016; 11:e0148407.doi: 10.1371/journal.pone.0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016; 17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 34.Pasquinelli AE, Hunter S, Bracht J. MicroRNAs: a developing story. Curr Opin Genet Dev 2005; 15:200–205. doi: 10.1016/j.gde.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 2007; 39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 36.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011; 147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA 2015; 6:563–579. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016; 37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 39.Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu FF, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016; 351:271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lauressergues D, Couzigou JM, Clemente HS, Martinez Y, Dunand C, Becard G, et al. Primary transcripts of microRNAs encode regulatory peptides. Nature 2015; 520:90–U205. doi: 10.1038/nature14346. [DOI] [PubMed] [Google Scholar]
- 41.Magny EG, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA, et al. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 2013; 341:1116–1120. doi: 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
- 42.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 1995; 268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 43.Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature 1986; 323:558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 44.AbouHaidar MG, Venkataraman S, Golshani A, Liu B, Ahmad T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci U S A 2014; 111:14542–14547. doi: 10.1073/pnas.1402814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perriman R, Ares M., Jr Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA 1998; 4:1047–1054. doi: 10.1017/s135583829898061x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Storr SJ, Safuan S, Ahmad N, El-Refaee M, Jackson AM, Martin SG. Macrophage-derived interleukin-1beta promotes human breast cancer cell migration and lymphatic adhesion in vitro. Cancer Immunol Immunother 2017; 66:1287–1294. doi: 10.1007/s00262-017-2020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet 2016; 17:679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development 2016; 143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang R, Xu J, Zhao J, Wang X. Silencing of hsa_circ_0007534 suppresses proliferation and induces apoptosis in colorectal cancer cells. Eur Rev Med Pharmacol Sci 2018; 22:118–126. doi: 10.26355/eurrev_201801_14108. [DOI] [PubMed] [Google Scholar]
- 50.Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med 2018; 96:85–96. doi: 10.1007/s00109-017-1600-y. [DOI] [PubMed] [Google Scholar]
- 51.Zhao Q, Chen S, Li T, Xiao B, Zhang X. Clinical values of circular RNA 0000181 in the screening of gastric cancer. J Clin Lab Anal 2018; 32:e22333.doi: 10.1002/jcla.22333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie Y, Shao Y, Sun W, Ye G, Zhang X, Xiao B, et al. Downregulated expression of hsa_circ_0074362 in gastric cancer and its potential diagnostic values. Biomark Med 2018; 12:11–20. doi: 10.2217/bmm-2017-0114. [DOI] [PubMed] [Google Scholar]
- 53.Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal 2018; 32:e22281.doi: 10.1002/jcla.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang W, Fu K, Sun H, Rong D, Wang H, Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer 2018; 17:137.doi: 10.1186/s12943-018-0888-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun H, Tang W, Rong D, Jin H, Fu K, Zhang W, et al. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark 2018; 21:299–306. doi: 10.3233/CBM-170379. [DOI] [PubMed] [Google Scholar]
- 56.Shao Y, Chen L, Lu R, Zhang X, Xiao B, Ye G, et al. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumour Biol 2017; 39:1010428317699125.doi: 10.1177/1010428317699125. [DOI] [PubMed] [Google Scholar]
- 57.Lu R, Shao Y, Ye G, Xiao B, Guo J. Low expression of hsa_circ_0006633 in human gastric cancer and its clinical significances. Tumour Biol 2017; 39:1010428317704175.doi: 10.1177/1010428317704175. [DOI] [PubMed] [Google Scholar]
- 58.Li WH, Song YC, Zhang H, Zhou ZJ, Xie X, Zeng QN, et al. Decreased expression of Hsa_circ_00001649 in gastric cancer and its clinical significance. Dis Markers 2017; 2017:4587698.doi: 10.1155/2017/4587698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li P, Chen H, Chen S, Mo X, Li T, Xiao B, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer 2017; 116:626–633. doi: 10.1038/bjc.2016.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol 2017; 23:6330–6338. doi: 10.3748/wjg.v23.i34.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta 2017; 466:167–171. doi: 10.1016/j.cca.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 62.Wang Y, Wang ZF. Efficient backsplicing produces translatable circular mRNAs. RNA 2015; 21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 2015; 444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 64.Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med 2017; 6:1173–1180. doi: 10.1002/cam4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okuda H, Xing F, Pandey PR, Sharma S, Watabe M, Pai SK, et al. miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res 2013; 73:1434–1444. doi: 10.1158/0008-5472.CAN-12-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem 2009; 284:5731–5741. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 67.Pan HY, Li T, Jiang YG, Pan CC, Ding YL, Huang ZG, et al. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem 2018; 119:440–446. doi: 10.1002/jcb.26201. [DOI] [PubMed] [Google Scholar]
- 68.Lu J, Zhang PY, Xie JW, Wang JB, Lin JX, Chen QY, et al. Hsa_circ_0000467 promotes cancer progression and serves as a diagnostic and prognostic biomarker for gastric cancer. J Clin Lab Anal 2018; 33:e22726.doi: 10.1002/jcla.22726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H, Yao G, Feng B, Lu X, Fan Y. Circ_0056618 and CXCR4 act as competing endogenous in gastric cancer by regulating miR-206. J Cell Biochem 2018; 119:9543–9551. doi: 10.1002/jcb.27271. [DOI] [PubMed] [Google Scholar]
- 70.Huang YS, Jie N, Zou KJ, Weng Y. Expression profile of circular RNAs in human gastric cancer tissues. Mol Med Rep 2017; 16:2469–2476. doi: 10.3892/mmr.2017.6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang L, Shen J, Jiang Y. Circ_0027599/PHDLA1 suppresses gastric cancer progression by sponging miR-101-3p.1. Cell Biosci 2018; 8:58.doi: 10.1186/s13578-018-0252-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y, Liu H, Li WD, Yu J, Li J, Shen ZY, et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging 2017; 9:1585–1594. doi: 10.18632/aging.101254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu J, Zhang PY, Li P, Xie JW, Wang JB, Lin JX, et al. Circular RNA hsa_circ_0001368 suppresses the progression of gastric cancer by regulating miR-6506-5p/FOXO3 axis. Biochem Biophys Res Commun 2019; 512:29–33. doi: 10.1016/j.bbrc.2019.02.111. [DOI] [PubMed] [Google Scholar]
- 74.Fang J, Hong H, Xue X, Zhu X, Jiang L, Qin M, et al. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett 2019; 442:222–232. doi: 10.1016/j.canlet.2018.10.040. [DOI] [PubMed] [Google Scholar]
- 75.Liu T, Liu S, Xu Y, Shu R, Wang F, Chen C, et al. Circular RNA-ZFR inhibited cell proliferation and promoted apoptosis in gastric cancer by sponging miR-130a/miR-107 and modulating PTEN. Cancer Res Treat 2018; 50:1396–1417. doi: 10.4143/crt.2017.537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Chang P, Wang F, Li Y. Hsa_circ_0000673 is down-regulated in gastric cancer and inhibits the proliferation and invasion of tumor cells by targetting miR-532-5p. Biosci Rep 2018; 38:BSR20180538.doi: 10.1042/BSR20180538. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 77.Li J, Zhen L, Zhang Y, Zhao L, Liu H, Cai D, et al. Circ-104916 is downregulated in gastric cancer and suppresses migration and invasion of gastric cancer cells. Onco Targets Ther 2017; 10:3521–3529. doi: 10.2147/OTT.S136347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lai Z, Yang Y, Yan Y, Li T, Li Y, Wang Z, et al. Analysis of co-expression networks for circular RNAs and mRNAs reveals that circular RNAs hsa_circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 are candidate oncogenes in gastric cancer. Cell Cycle 2017; 16:2301–2311. doi: 10.1080/15384101.2017.1380135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang H, Wang X, Huang H, Wang Y, Zhang F, Wang S. Hsa_circ_0067997 promotes the progression of gastric cancer by inhibition of miR-515-5p and activation of X chromosome-linked inhibitor of apoptosis (XIAP). Artif Cells Nanomed Biotechnol 2019; 47:308–318. doi: 10.1080/21691401.2018.1553787. [DOI] [PubMed] [Google Scholar]
- 80.Li X, He M, Guo J, Cao T. Upregulation of circular RNA circ-ERBB2 predicts unfavorable prognosis and facilitates the progression of gastric cancer via miR-503/CACUL1 and miR-637/MMP-19 signaling. Biochem Biophys Res Commun 2019; 511:926–930. doi: 10.1016/j.bbrc.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 81.Huang S, Zhang X, Guan B, Sun P, Hong CT, Peng J, et al. A novel circular RNA hsa_circ_0008035 contributes to gastric cancer tumorigenesis through targeting the miR-375/YBX1 axis. Am J Transl Res 2019; 11:2455–2462. [PMC free article] [PubMed] [Google Scholar]
- 82.Liu M, Liu KD, Zhang L, Cai J, Yao HW, Bai YK, et al. Circ_0009910 regulates growth and metastasis and is associated with poor prognosis in gastric cancer. Eur Rev Med Pharmacol Sci 2018; 22:8248–8256. doi: 10.26355/eurrev_201812_16519. [DOI] [PubMed] [Google Scholar]
- 83.Rong D, Dong C, Fu K, Wang H, Tang W, Cao H. Upregulation of circ_0066444 promotes the proliferation, invasion, and migration of gastric cancer cells. Onco Targets Ther 2018; 11:2753–2761. doi: 10.2147/OTT.S156516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xue M, Li G, Fang X, Wang L, Jin Y. Zhou Q. hsa_circ_0081143 promotes cisplatin resistance in gastric cancer by targeting miR-646/CDK6 pathway. Cancer Cell Int 2019; 19:25.doi: 10.1186/s12935-019-0737-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun H, Xi P, Sun Z, Wang Q, Zhu B, Zhou J, et al. Circ-SFMBT2 promotes the proliferation of gastric cancer cells through sponging miR-182-5p to enhance CREB1 expression. Cancer Manag Res 2018; 10:5725–5734. doi: 10.2147/CMAR.S172592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett 2017; 388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 87.Li D, Yang Y, Li ZQ, Li LC, Zhu XH. Circular RNAs: from biogenesis and function to diseases. Chin Med J 2019; 132:2457–2464. doi: 10.1097/CM9.0000000000000465. [DOI] [PMC free article] [PubMed] [Google Scholar]


