Abstract
In comparing with the popular Ac4ManNAz applied as cell labels via Cu-free click chemistry, two novel azido mannosamine lipids with C6 and C12 esters on anomeric hydroxyl groups were prepared and encapsulated in a liposome delivery system, which enhanced chemical stabilities and showed good cell-metabolizable labeling efficiency on MDA-MB-231 cells with strong fluorescence after the treatment of DBCO-Cy5 by triazole formation via click chemistry.
Abstract Graphic
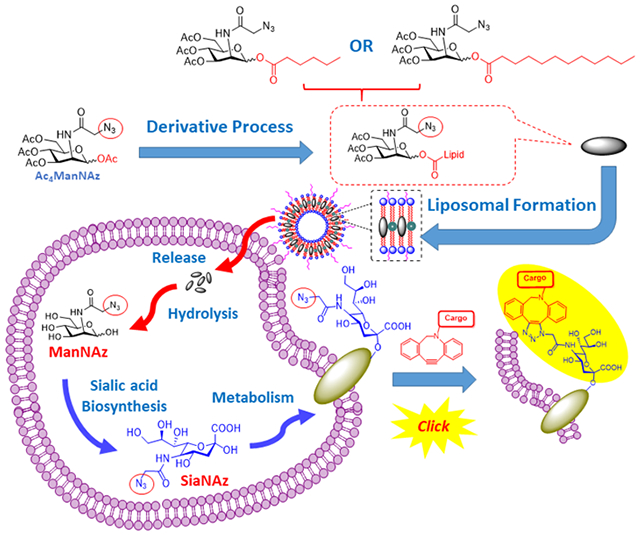
Metabolic oligosaccharide engineering of unnatural carbohydrates created by the Bertozzi group has become a powerful tool to introduce chemical receptors onto cell surfaces. 1–3 Based on the successful employment of modified N-azidoacetyl mannosamine (ManNAz) which can also be metabolized through the sialic acid biosynthetic pathway and incorporated onto cell surface sialoglycoconjugates as normal sialic acids, 4, 5 such biosynthetic precursors of the ManNAc derivative containing an azido group as a chemical probe can be used as metabolic tracers for labeling sialoglycans through different types of click chemistry on the azido group. 6, 7 Since then, the application of click chemistry in vitro and in vivo has been widely applied in the research of cell labeling due to the favorable features, especially on biocompatibility and high specificity with a good reaction rate. 8 This type of cell labeling strategy has been developed as an efficient method to incorporate selectively reactive chemical groups into cell-surface glycoproteins by novel molecular, 9, 10 nanoparticle 11–14 and liposomal strategies 15, 16 of delivery systems. Tetraacetyl-N-azidoacetylmannosamine (Ac4ManNAz, AAM) is one of the most extensively studied metabolic cell-labeling reagents, which can be typically metabolized in the cell through sialic acid biosynthesis to azido sialic acid on the cell membrane and then label the cell through click chemistry after treatment with dibenzocyclooctyne (DBCO)-bearing reagents. 3, 6
In order to improve the chemical stability and cell labeling efficiency of Ac4ManNAz (AAM), we prepared derivatives of AAM with different carbon chain length esters at the C-1-OH of Ac3ManNAzOH for hydrophobic lipids (Scheme 1). Herein, two novel liposomal azido mannosamine lipids loaded with hexanoic (C6) or dodecanoic (C12) ester tails instead of acetyl (C2) ester on the anomeric hydroxyl group of Ac3ManNAzOH were designed and prepared to test the cell labeling efficiency via Cu-free click chemistry. To overcome the low water solubility of the lipids with long ester tails (Scheme 2), we took advantage of the liposome system, and all the azido-lipids were encapsulated by the liposome system with good loading efficiencies.
Scheme 1.
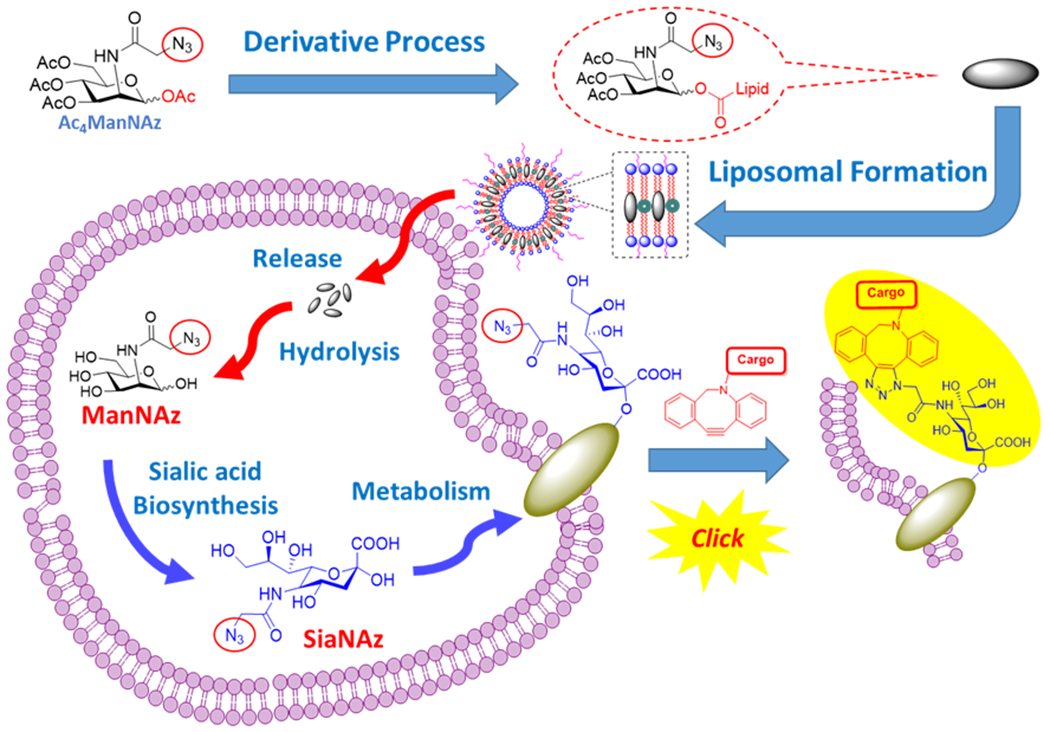
Design of Liposomal Azido Mannosamine Lipids on Metabolic Cell Labeling and Imaging
Scheme 2.
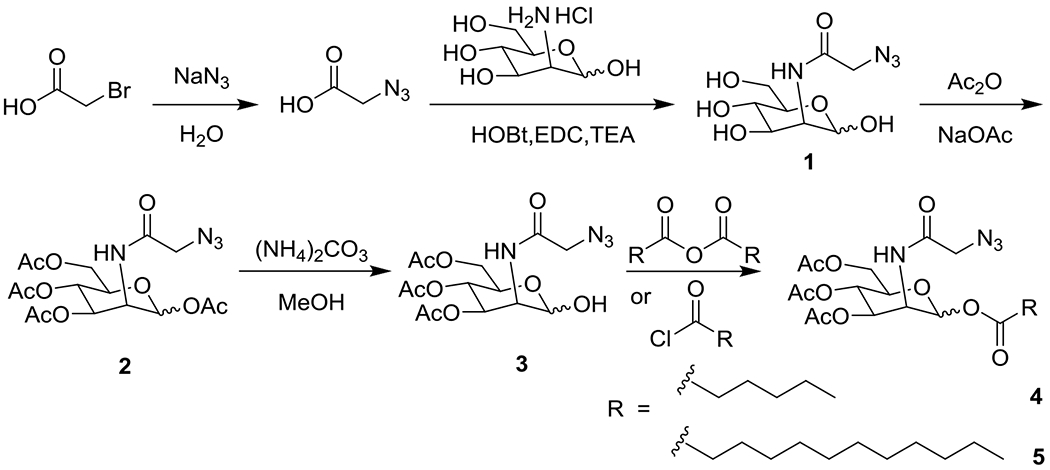
Synthesis of Ac4ManAz and Its Derivative Lipids
Results and Discussion
Starting with AAM from the previous work, 15, 17, 18 triacetyl-N-azidoacetylmannosamine lipid (Ac3ManNAzOH-lipid) was successfully synthesized through selective exposure of the hydroxyl group on the anomeric carbon 19 and acylation 20 with varying carbon chain length lipid tails of saturated C6 or C12 fatty acids in concise routes (Scheme 2). In the assessment of the hydrolytic degradation by HPLC, C6 and C12 lipids (Figure 1A) showed much more chemical stability than AAM in 37°C incubation in PBS (Figure S2 in the Supporting Information). Ac3ManNAzOH lipid-loaded liposomes were continually prepared by the highly efficient process 15, 21 by extrusion of the lipid mixture with 1,2-distearoyl-sn-glycero-3- phosphocholine (DSPC), cholesterol, and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N- [amino (polyethylene glycol)-2000] (DSPE-PEG2000, Figure 1B). The prepared Ac3ManNAzOH lipid-loaded liposomes had effective diameters of around 200 nm as determined by dynamic light scattering (DLS) and sugar loading efficiency of 72% (Figure S3 in the Supporting Information) on the C2 lipid (AAM), 41% (Figure S4 in the Supporting Information) on C6 lipid and 50% (Figure S5 in the Supporting Information) on the C12 lipid (Figure 1C). The C2 lipid (AAM) had the highest loading efficiency, which might be affiliate with its higher water solubility in the four types of lipids (Figure S1 in the supporting information), so AAM might not only distribute into the liposome membrane as other hydrophilic lipids but also partially dissolve in the aqueous fluid part of liposome.
Figure 1.
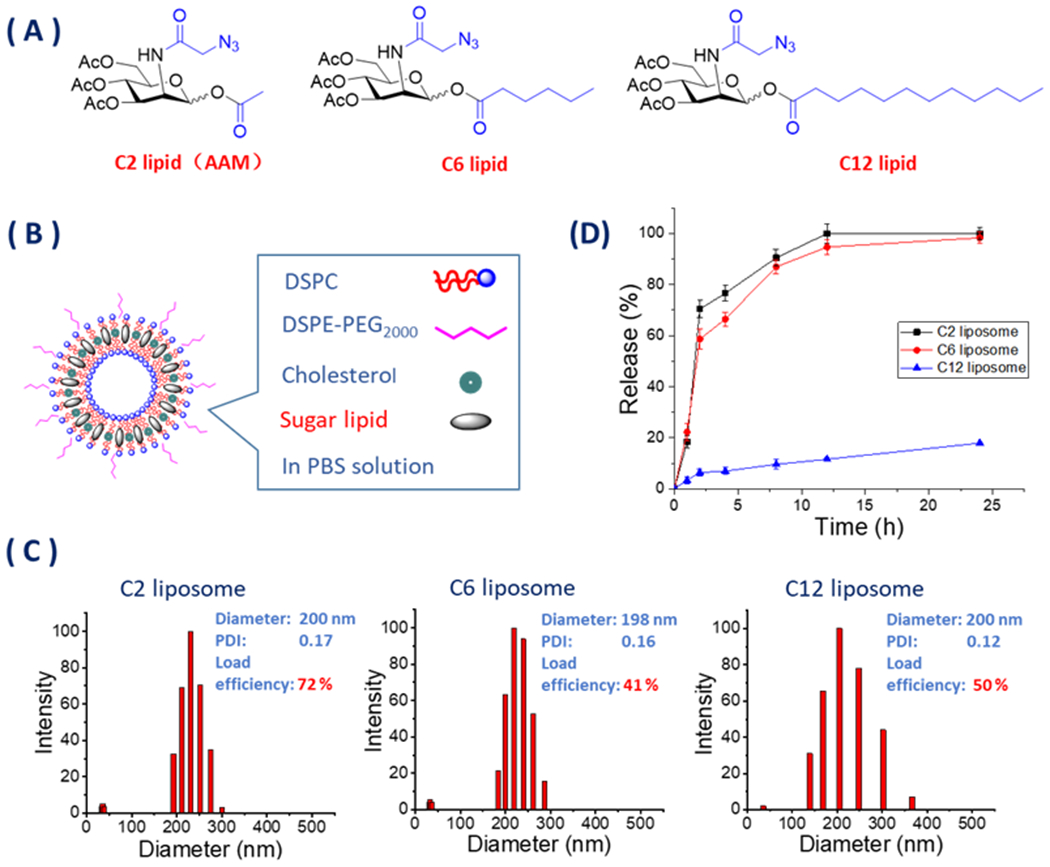
(A) Structure of Ac3ManNAzOH lipids with C2, C6, or C12 ester tail on anomeric hydroxyl group. (B) Preparation of Ac3ManNAzOH lipid-loaded liposomes. (C) Diameter and diametric distribution of Ac3ManNAzOH lipid-loaded liposomes. (D) Azido carbohydrate release of Ac3ManNAzOH lipid-loaded liposomes by reverse-titration method of DBCO-Cy3.
Sugar release test of Ac3ManNAzOH lipid-loaded liposomes by the reverse-titration method of DBCO-Cy3 (Figure S6 in the Supporting Information) indicated that the C2 and C6 lipid-loaded liposomes had the faster release rates, while C12 lipid-loaded liposomes had significantly lower release rates under the same physiological conditions (Figure 1D).
We next tested the cell-labeling efficiency of Ac3ManNAzOH lipids on MDA-MB-231 breast cancer cells. The cells were incubated with 10 μM Ac3ManNAzOH lipid-loaded liposomes for 2 or 12 h, and then treated with DBCO-Cy5 independently for another 1 h to detect the expression of the azido groups via Cu-free click chemistry. The cells without liposome treatment were used as a negative control in PBS. Flow cytometry measurements of the Cy5 fluorescence intensity (Figure 2A) showed that all the Ac3ManNAzOH lipid-loaded liposomes could label MDA-MB-231 cells efficiently in the contrast of PBS as the negative control after 12 h incubation. In contrast with the results of 12 h incubation group, fluorescence intensities after 2 h incubation showed no significant differences between the cells treated with Ac3ManNAzOH lipid-loaded liposomes and those treated with PBS. There were also no significant differences in fluorescence intensities between the cells treated with free AAM as the positive control compared with the AAM liposome (Figure 2C), which indicated that the difference between the 2 and 12 h incubation groups was probably due to the time needed for the biosynthesis and metabolic pathway.
Figure 2.
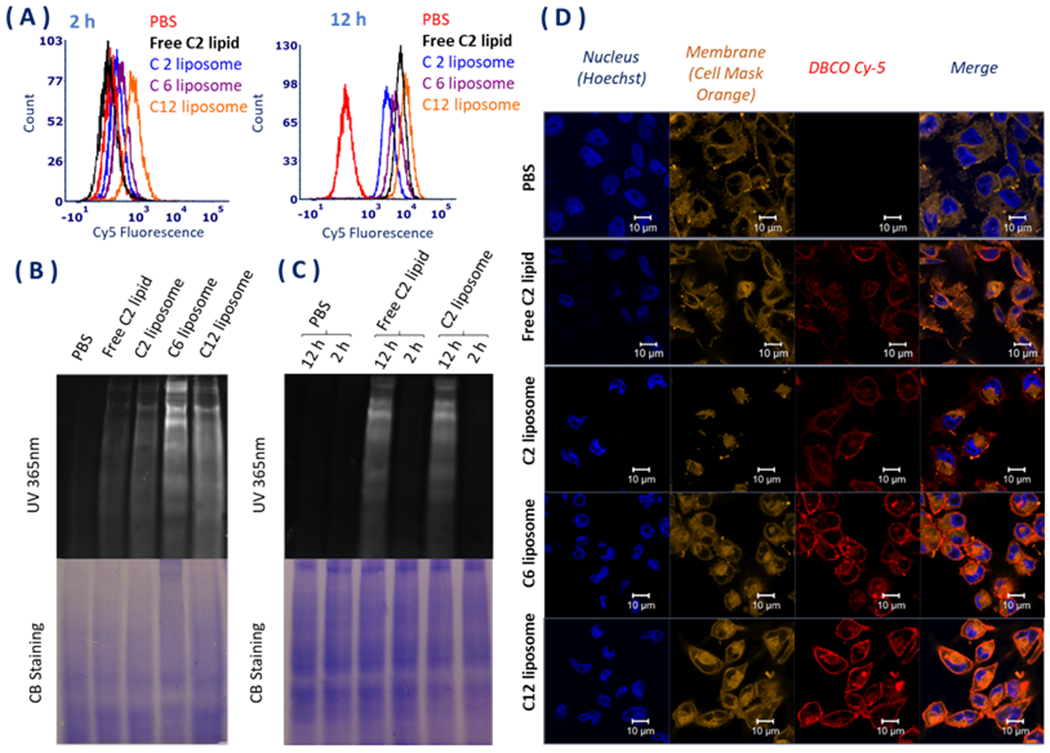
(A) Flow cytometry analysis of the cellular internalization of the liposomes after 2 and 12 h incubation with the liposome and free C2 lipid (AAM). (B) SDS-PAGE analysis of azide groups in glycoproteins of MDA-MB-231 cells in vitro after 2 and 12 h incubation with the liposomes compared to free C2 lipid (AAM). The gel was detected under 365 nm UV directly after DBCO-Cy5 incubation for another 1 h. The Coomassie stain showed the total amount of protein. (C) SDS-PAGE analysis of azide groups in glycoprotein of MDA-MB-231 cells in vitro after 2 and 12 h incubation with free C2 lipid and C2 liposome. The gel was detected under 365 nm UV directly after DBCO-Cy5 incubation for another 1 h. The Coomassie stain showed the total amount of protein. (D) Confocal microscopy images of MDA-MB-231 cells incubated with 10 μM of Ac3ManNAzOH-lipid-loaded liposomes and free AAM for 12 h at 37°C, followed by labeling with DBCO-Cy5 for another 1 h. Cells treated with PBS and labeled with DBCO-Cy5 were used as the control. The cell nucleus was stained with Hoechst and the membrane was stained with cell mask orange.
Furthermore, the cell labeling results indicated that all the Ac3ManNAzOH lipids could be metabolized onto the cell membrane glycoprotein through sialic acid biosynthesis by SDS-PAGE compared with Coomassie blue staining (Figure 2B). Fluorescence intensity could be observed directly on the SDS-PAGE gel under 365 nm UV because of the highly effective formation of a stable triazole from azido-glycoprotein binding with the fluorescent dye DBCO-Cy5 through click chemistry.
Results of the confocal microscopy (Figure 2D) showed that all the cells treated with Ac3ManNAzOH lipid-loaded liposomes could generate fluorescence on the cell membrane after treatment with DBCO-Cy5 compared with PBS as the negative control and the free AAM as the positive control. Cell labeling efficiencies were significantly improved by the long ester tail lipid loaded liposome. The C6 and C12 lipid loaded liposomes showed better fluorescence intensities than C2 lipid-loaded liposomes, which indicated that long ester tails on C-1-OH could not only improve chemical stability of the sugar part but also enhance the labeling efficiency.
Summary and Conclusion
In summary, we developed an enhanced metabolic cell labeling strategy of a liposomal azido carbohydrate carrier by Ac3ManNAzOH lipids with C6 and C12 ester tails on the anomeric hydroxyl group. All the lipids with ester tails of different lengths could be delivered in liposome systems with good loading efficiency, and the metabolic cell labeling efficiencies maintained on MDA-MB-231 cells as AAM. The labeling efficiency seemed better after extending the length of the ester on anomeric carbon. This work may provide a promising cell labeling system based on metabolic oligosaccharide engineering via Cu-free click chemistry.
Experimental Section
General Experimental Details
All other reagents were used as obtained from commercial sources. Room temperature refers to ambient temperature. Temperatures of 0°C were maintained using an ice–water bath. Thin-layer chromatography was performed using silica gel 60 with F254 indicator on glass plates (Merck). Flash chromatography was performed using Merck 40–63 μm silica gel. Solvent ratios for the purification of compounds by flash chromatography are reported as percent volume (v/v). Reactions were performed using oven-dried glassware apparatus under an atmosphere of nitrogen or argon with anhydrous solvents. Anhydrous dichloromethane (DCM), hexane, and ethyl acetate were purified by passing them through alumina columns and kept anhydrous in molecular sieves. NMR spectroscopy was performed in deuterated solvents at 20°C. Chemical shifts (δ) are given in parts per million (ppm) relative to TMS, and the solvent residual signal is used as a reference. Abbreviations for peak multiplicities are s (singlet), d (doublet), t (triplet), dt (doublet of triplets), m (multiplet), and br (broad). Mass spectra were measured using an electrospray ionization (ESI) mass spectrometer. Details for the spectrometers and other methods are given in the Supporting Information.
Synthesis of Ac4ManNAz (2, AAM). Azidoacetic acid was prepared by adding sodium azide (NaN3, 2.6 g, 40.0 mmol) to bromoacetic acid (2.78 g, 20.0 mmol) in deionized water (30 mL) and stirring at room temperature for 48 h. Thin-layer chromatography (TLC) analysis using detection by bromocresol green stain indicated that the reaction was completed. The solution was extracted by ethyl acetate and washed with 1 M HCl aq.; the organic layer was washed with saturated NaCl aq., and dried over Na2SO4. The solvent was removed to obtain yellow oil (1.4 g, 69%). The crude product could be used directly for the next step. Compound 1 was synthesized as follows: D-Mannosamine hydrochloride (1.5 g, 7.0 mmol) was added to azidoacetic acid (0.98 g, 9.7 mmol) from the previous step. Triethylamine (TEA, 2.5 mL 17.0 mmol) was added and the reaction mixture was stirred for 5 min at room temperature. Then the solution was cooled to 0°C and N-hydroxybenzotriazole (HOBt, 0.95 g, 7.1 mmol) was added, followed by addition of 1-[3-(dimethylamino) propyl]-3-ethylcarbodiimide hydrochloride (EDC, 2.68 g, 14.0 mmol). The reaction was allowed to warm to room temperature overnight, at which point TLC analysis with cerric ammonium molybdate (CAM) stain indicates that the reaction is complete. The solution was concentrated to obtain crude 1 and then purified by silica gel chromatography with DCM–methanol (9:1, v/v) to colorless semisolid (3.1 g, 84%) which was acetylated directly. To a suspension of 1 (0.9 g, 3.4 mmol) in acetic anhydride (15 mL) was added anhydrous NaOAc (0.31 g, 3.8 mmol) and the mixture stirred at 140 °C for 10 h. The solution was resuspended in DCM and washed with saturated NaHCO3 aq. saturated NaCl aq. The organic phase was dried over Na2SO4, filtered, and concentrated. The crude material was then purified by silica gel chromatography, eluting with hexane–ethyl acetate (2:1, v/v) to obtain a pain white solid (0.6 g, 41.0%, 1/1 α/β isomers). 1H NMR (500 MHz, CDCl3) δ 6.67 (d, J = 9.0 Hz, 1H), 6.60 (d, J = 9.3 Hz, 1H), 6.06 (s, 1H), 5.91 (s, 1H), 5.35 (dd, J = 10.2, 4.3 Hz, 1H), 5.23 (d, J = 10.1 Hz, 1H), 5.18 (t, J = 9.8 Hz, 1H), 5.07 (dd, J = 9.9, 3.9 Hz, 1H), 4.75 (ddd, J = 9.1, 4.0, 1.7 Hz, 1H), 4.63 (ddd, J = 9.4, 4.3, 2.0 Hz, 1H), 4.26 (ddd, J = 12.0, 7.4, 4.4 Hz, 2H), 4.16 (dd, J = 12.5, 2.4 Hz, 1H), 4.13 (d, J = 2.5 Hz, 2H), 4.10–4.09 (m, 2H), 4.06 (d, J = 16.7 Hz, 2H), 3.84 (ddd, J = 9.7, 4.6, 2.5 Hz, 1H), 2.20 (s, 3H), 2.13 (d, J = 2.1 Hz, 9H), 2.08 (s, 6H), 2.02 (d, J = 2.3 Hz, 6H).13C NMR (126 MHz, CDCl3) δ 170.48, 170.12, 170.07, 169.55, 169.53, 168.32, 168.08, 167.29, 166.71, 91.31, 90.26, 73.38, 71.42, 70.26, 68.85, 65.13, 64.96, 61.78, 61.69, 52.60, 52.42, 49.72, 49.27, 20.85, 20.77, 20.75, 20.70, 20.66, 20.63, 20.59. ESI-HRMS (m/z) C16H22N4O10: calculated 430.1336; found: 453.1224 [M+Na]+.
Synthesis of Ac3ManNAzOH (3). Ac4ManNAz (2, 0.3 g, 0.7 mmol) and ammonium carbonate (0.074 g, 0.77 mmol) were dissolved in methanol (10 mL) and stirred at room temperature for 8 h. TLC analysis using detection by CAM indicated that the reaction was finished, the solvent was removed, and the crude product was purified by silica gel column chromatography with DCM–ethyl acetate (2:1, v/v) to yield a white solid (0.2 g, 73.6%, 5/1 α/β isomers). 1H NMR (500 MHz, CDCl3) δ 6.60 (d, J = 9.1 Hz, 1H), 5.39 (dd, J = 10.1, 4.2 Hz, 1H), 5.20–5.14 (m, 2H), 4.58 (ddd, J = 9.2, 4.3, 1.9 Hz, 1H), 4.47 (s, 1H), 4.26–4.23 (m, 1H), 4.19 (d, J = 4.2 Hz, 1H), 4.16 (d, J = 2.6 Hz, 1H), 4.06 (d, J = 3.6 Hz, 1H), 4.03 (s, 1H), 2.11 (s, 3H), 2.05 (s, 3H), 1.98 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 170.82, 170.35, 169.90, 167.16, 93.06, 69.16, 68.03, 65.78, 62.31, 60.48, 52.40, 50.86, 20.84, 20.71, 20.68.
Synthesis of C6 Ester of Ac3ManNAzOH (4) Ac3ManNAzOH (43 mg, 0.11 mmol) and hexanoic anhydride (0.1 mL, 0.42 mmol) were dissolved in anhydrous acetone (5 mL) at 0°C. 4-Dimethylaminopyridine (DMAP, 5 mg, 0.41 μmol) was added. The mixture was stirred overnight at room temperature and then concentrated. The residue was dissolved in DCM and then washed with saturated NaHCO3 aq. and saturated NaCl aq. The organic phase was dried over Na2SO4, filtered, and concentrated. The crude material was then purified by silica gel chromatography, eluting with hexane–ethyl acetate (1:1, v/v) to obtain a colorless semisolid (45 mg, 84%, 5/1 α/β isomers). 1H NMR (500 MHz, CDCl3) δ 6.56 (d, J = 9.3 Hz, 1H), 6.05 (s, 1H), 5.33 (dd, J = 10.1, 4.1 Hz, 1H), 5.21 (t, J = 10.1 Hz, 1H), 4.61 (dd, J = 9.2, 4.1 Hz, 1H), 4.22 (dd, J = 12.2, 4.3 Hz, 1H), 4.13–4.10 (m, 1H), 4.09 (s, 1H), 4.06 (s, 1H), 4.02 (d, J = 6.7 Hz, 1H), 2.42 (t, J = 7.4 Hz, 2H), 2.11 (s, 3H), 2.06 (s, 3H), 2.00 (s, 3H), 1.68–1.65 (m, 2H), 1.33 (dt, J = 6.9, 3.6 Hz, 4H), 0.90 (t, J = 6.6 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ 170.90, 170.47, 170.05, 169.53, 166.66, 91.11, 77.29, 77.04, 76.78, 70.28, 68.92, 65.15, 61.80, 52.44, 49.31, 33.99, 31.12, 24.34, 22.25, 20.75, 20.64, 20.58, 13.86. ESI-HRMS (m/z) C20H30N4O10: calculated 486.1962; found: 509.1862 [M+Na]+.
Synthesis of C12 Ester of Ac3ManNAzOH (5). Ac3ManNAzOH (20 mg, 51.5 μmol) and TEA (10 μL, 70 μmol) were dissolved in anhydrous DCM (3 mL). Dodecanoic acid chloride (12.8 μL, 55.8 μmol) or octadecanoic acid chloride (23.7 mg, 78 μmol) was added at 0°C. The mixture was warmed to room temperature and stirred overnight. The reaction solution was washed with saturated NaHCO3 aq. and saturated NaCl aq. The organic phase was dried over Na2SO4, filtered, and concentrated. The crude material was then purified by silica gel chromatography, eluting with hexane–ethyl acetate (1:1, v/v) to obtain colorless semisolid (20.0 mg, 68%, 5/1 α/β isomers). 1H NMR (500 MHz, CDCl3) δ 6.56 (d, J = 9.2 Hz, 1H), 6.06 (s, 1H), 5.34 (dd, J = 10.2, 4.2 Hz, 1H), 5.22 (t, J = 10.1 Hz, 1H), 4.61 (dd, J = 7.6, 4.1 Hz, 1H), 4.24 (dd, J = 12.1, 3.9 Hz, 1H), 4.13 (d, J = 8.0 Hz, 1H), 4.10 (s, 1H), 4.07 (s, 1H), 4.03 (d, J = 10.1 Hz, 2H), 2.42 (t, J = 7.5 Hz, 2H), 2.12 (s, 3H), 2.07 (s, 3H), 2.00 (s, 3H), 1.68–1.63 (m, 3H), 1.25 (d, J = 3.6 Hz, 16H), 0.88 (s, 4H). 13C NMR (126 MHz, CDCl3) δ 170.91, 170.47, 170.06, 169.53, 166.63, 91.11, 77.27, 77.02, 76.76, 70.27, 68.93, 65.14, 61.77, 52.46, 49.31, 34.03, 31.91, 29.60, 24.66, 22.69, 20.59, 14.12. ESI-HRMS (m/z) C26H42N4O10: calculated 570.2901; found: 593.2782 [M+Na]+.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Health of US (1R01EB02565101 and partially 1R01CA207584) and National Natural Science Foundation of China (No. 81402845).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconjchem.9b00509.
General methods, including water solubility and chemical stability, liposome preparation, sugar release, biological evaluations, and NMR and MS spectra (PDF)
The authors declare no competing financial interest.
References
- (1).Mahal LK; Yarema KJ; Bertozzi CR Engineering Chemical Reactivity on Cell Surfaces Through Oligosaccharide Biosynthesis. Science 1997, 276, 1125–1128. [DOI] [PubMed] [Google Scholar]
- (2).Prescher JA; Dube DH; Bertozzi CR Chemical remodelling of cell surfaces in living animals. Nature 2004, 430, 873–877. [DOI] [PubMed] [Google Scholar]
- (3).Saxon E; Bertozzi CR Cell Surface Engineering by a Modified Staudinger Reaction. Science 2000, 287, 2007–2010. [DOI] [PubMed] [Google Scholar]
- (4).Keppler OT; Horstkorte R; Pawlita M; Schmidt C; Reutter W. Biochemical engineering of the N-acyl side chain of sialic acid: biological implications. Glycobiology 2001, 11, 11R–18R. [DOI] [PubMed] [Google Scholar]
- (5).Dube DH; Bertozzi CR Metabolic oligosaccharide engineering as a tool for glycobiology. Curr. Opin. Chem. Biol. 2003, 7, 616–625. [DOI] [PubMed] [Google Scholar]
- (6).Laughlin ST; Bertozzi CR Metabolic labeling of glycans with azido sugars and subsequent glycan-profiling and visualization via Staudinger ligation. Nat. Protoc. 2007, 2, 2930–2944. [DOI] [PubMed] [Google Scholar]
- (7).Chang PV; Prescher JA; Sletten EM; Baskin JM; Miller IA; Agard NJ; Lo A; Bertozzi CR Copper-free click chemistry in living animals. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 1821–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Codelli JA; Baskin JM; Agard NJ; Bertozzi CR, Second-Generation Difluorinated Cyclooctynes for Copper-Free Click Chemistry. J. Am. Chem. Soc. 2008, 130, 11486–11493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wang H; Wang R; Cai K; He H; Liu Y; Yen J; Wang Z; Xu M; Sun Y; Zhou X et al. Selective in vivo metabolic cell-labeling-mediated cancer targeting. Nat. Chem. Biol. 2017, 13, 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Shim MK; Yoon HY; Ryu JH; Koo H; Lee S; Park JH; Kim J-H; Lee S; Pomper MG; Kwon IC et al. Cathepsin B-Specific Metabolic Precursor for In Vivo Tumor-Specific Fluorescence Imaging. Angew. Chem., Int. Ed. 2016, 55, 14698–14703. [DOI] [PubMed] [Google Scholar]
- (11).Koo H; Lee S; Na JH; Kim SH; Hahn SK; Choi K; Kwon IC; Jeong SY; Kim K Bioorthogonal Copper-Free Click Chemistry In Vivo for Tumor-Targeted Delivery of Nanoparticles. Angew. Chem., Int. Ed. 2012, 51, 11836–11840. [DOI] [PubMed] [Google Scholar]
- (12).Kim JB; Park K; Ryu J; Lee JJ; Lee MW; Cho HS; Nam HS; Park OK; Song JW; Kim TS et al. Intravascular optical imaging of high-risk plaques in vivo by targeting macrophage mannose receptors. Sci. Rep. 2016, 6, 22608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Cai K; He X; Song Z; Yin Q; Zhang Y; Uckun FM; Jiang C; Cheng J Dimeric Drug Polymeric Nanoparticles with Exceptionally High Drug Loading and Quantitative Loading Efficiency. J. Am. Chem. Soc. 2015, 137, 3458–3461. [DOI] [PubMed] [Google Scholar]
- (14).Cai K; Wang AZ; Yin L; Cheng J Bio-nano interface: The impact of biological environment on nanomaterials and their delivery properties. J. Controlled Release 2017, 263, 211–222. [DOI] [PubMed] [Google Scholar]
- (15).Wang H; Gauthier M; Kelly JR; Miller RJ; Xu M; O’Brien. WD Jr.; Cheng J. Targeted Ultrasound-Assisted Cancer-Selective Chemical Labeling and Subsequent Cancer Imaging using Click Chemistry. Angew. Chem., Int. Ed. 2016, 55, 5452–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Xie R; Dong L; Du Y; Zhu Y; Hua R; Zhang C; Chen X In vivo metabolic labeling of sialoglycans in the mouse brain by using a liposome-assisted bioorthogonal reporter strategy. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 5173–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Luchansky SJ; Hang HC; Saxon E; Grunwell JR; Yu C; Dube DH; Bertozzi CR Constructing Azide-Labeled Cell Surfaces Using Polysaccharide Biosynthetic Pathways. Methods in Enzymology 2003, 362, 249–272. [DOI] [PubMed] [Google Scholar]
- (18).Laughlin ST; Agard NJ; Baskin JM; Carrico IS; Chang PV; Ganguli AS; Hangauer MJ; Lo A; Prescher JA; Bertozzi CR Metabolic Labeling of Glycans with Azido Sugars for Visualization and Glycoproteomics. Methods in Enzymology 2006, 415, 230–250 [DOI] [PubMed] [Google Scholar]
- (19).Fiandor J; García-López MT; De Las Heras FG; Méndez-Castrillón PP A Facile Regioselective 1-O-Deacylation of Peracylated Glycopyranoses. Synthesis 1985, 12, 1121–1123. [Google Scholar]
- (20).Elmouelhi N; Aich U; Paruchuri VDP; Meledeo MA; Campbell CT; Wang JJ; Srinivas R; Khanna HS; Yarema KJ Hexosamine Template. A Platform for Modulating Gene Expression and for Sugar-Based Drug Discovery. J. Med. Chem. 2009, 52, 2515–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Blume G; Cevc G Liposomes for the sustained drug release in vivo. Biochim. Biophys. Acta, Biomembr. 1990, 1029, 91–97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


