Abstract
COVID-19 is caused by a novel beta coronavirus (SARS-CoV-2) strain that was first discovered in 2019 in the Wuhan city of China. Based on virus genome sequencing studies, the bat is suspected as the natural host of virus, and infection might be transmitted from bats via unknown intermediate hosts like reptiles and snakes etc., to infect humans. COVID-19 is transmitted from person to person contact, primarily via droplet infection within the incubation period or after clinical manifestations of fever, cough, sneezing, sputum, dyspnea, and pneumonia and through contaminated fomites. COVID-19 enters the respiratory tract through the ACE2 receptor on alveoli through binding of s-protein of the virus and causes injuries though the cytopathic effect, as well as cytokines and other mediators, released after developing sepsis. ACE 2 is almost 100-fold higher in kidneys than lung, and the virus can also involve the kidney in the same manner. Kidney involvement manifests in the form of proteinuria, hematuria, and an acute rise in serum creatinine. Kidney involvement is an independent risk factor for mortality. Diagnosis is primarly made by detecting viral RNA by reverse transcriptase polymerase chain reaction (rtPCR) in nasopharyngeal swab samples. Role of antibodies, both IgM and IgG are still evolving and at best restricted for epidemiological purpose. Though a large number of treatments, including hydroxychloroquine, anti-viral, convalescent plasma etc., are being tried, as of now treatment is symptomatic only.
Keywords: Acute kidney injury, COVID-19, coronavirus, epidemiology, mechanisms, sepsis, severe acute respiratory syndrome
Introduction
Coronaviruses (CoV) are amongst the newly emerging zoonotic virus which is transmitted between animals and human beings.[1] In the past, it has caused illness ranging from the common cold to more severe diseases such as Middle East Respiratory Syndrome (MERS-CoV) and Severe Acute Respiratory Syndrome (SARS-CoV).[2] The SARS-CoV was transmitted from civet cats to humans and MERS from dromedary camels to humans. Many other coronaviruses are still circulating in animals that are not found in humans to date.[3,4,5]
Corona Virus Disease-19 (COVID-19) is a novel CoV induced disease that was first discovered in December 2019, which was not previously reported in humans.[6] This CoV was renamed several times after discovery, first of all, as a newly identified β-coronavirus in Wuhan. On 12th January 2020, the World Health Organization (WHO) renamed it as the 2019-novel coronavirus (2019-nCoV), and subsequently on 11th February 2020, the Coronavirus Study Group of the International Committee on Taxonomy of viruses of WHO proposed the name SARS-CoV-2 for this virus, and the disease caused by the virus was called COVID-19.[7,8,9]
Global Situations of COVID-19
At the end of 2019, SARS-CoV-2 caused a cluster of pneumonia cases in Wuhan, a Chinese city, in the Hubei province of China. On 30th January, the outbreak was declared as Public Health Emergency of International Concern (PHEIC) and later a declared global pandemic by WHO.[10] The disease gradually gripped the entire world. The epicenter of pandemic later changed from Wuhan city in China to Europe and the USA.[11,12]
As on 21st March 2020, WHO reports, there were 266,073 confirmed cases, and 11,184 confirmed deaths in 183 countries. On 22nd April, WHO reported region-wise situation of COVID cases and death, globally 2,471,136 confirmed and 169,006 deaths. And region wise; the European Region 1219,486 cases and 109,952 deaths; Americas 925,291 cases and 44,775 deaths; the Eastern Mediterranean Region 139,349 cases and 6326 deaths; Western Pacific Region 136,271 cases and 5793 deaths; the South-East Asia Region 33,912 cases and 1427 deaths; and African Region 16,115 cases and 720 deaths.[13] WHO reported a very high level of risk assessment. The global map released by CDC as per the notification by WHO indicates that the entire world is affected by this COVID pandemic [Figure 1].[14]
Figure 1.
Global map showing entire world is affected with COVID pandemic. (source https://www.cdc.gov/coronavirus/2019-nc ov/cases-updates/world-map.html)
India-specific situation
India noticed the first case of the COVID pandemic on 30th January 2020. On March 23, the Indian Council of Medical Research (ICMR) data revealed a result of a total of 18,383 samples tested from 17,493 individuals; 415 were positive, and there were 7 deaths due to COVID-19.[15] The Center for Disease Dynamics, Economics and Policy (CDDEP), applying mathematical models used in the USA, pointed out that possibly 300 million (30 crore) cases will occur in India, out of them 10 crores will face severe COVID infection.[16] Looking at the incidence of 5.1% of AKI in severe cases,[17] there would be 5.1 million AKI patients because of SARS-CoV-2, and presumably many of them may require renal replacement therapy (RRT). It is estimated that with community spread COVID-19 in India with limited resources and health infrastructure, it could be challenging to combat the situation of patients with multiorgan failure and kidney failure, if the disease spreads fast within 2-3 months.
Lockdown effect in India
To curb the transmission of the virus in the community, India observed a 14-hour voluntary public curfew on 22 March 2020, and subsequently, on 24th March, a nationwide lockdown for 21 days was implemented, affecting the entire 1.3 billion population of India. On 14th April, the lockdown was extended till 3 May[18] and futher till 17th May, with the lifting of restriction as per the zones defined on number of new cases seen in the different areas.
The lockdown has reduced the rate of positivity from 16% on 14th April 2020 to 6% on 28th April 2020; and the death rate also declined from 9% to 6% during the same period. Till date, India successfully prevented widespread community spread. As of 28th April 2020, the Ministry of Health and Family Welfare has confirmed a total of 29,974 cases, 7,027 recoveries (including 1 migration), and 937 deaths in the country.[19] The COVID-19 affected almost all states of India, which has been highlighted in Figure 2. Experts suggest that the number of infections could be much higher as India's testing rates are among the lowest in the world. The infection rate of COVID-19 in India is reported to be 1.7, significantly lower than in the worst affected countries.[18] Despite lower testing rates, the case fatality rate in India and other south Asian countries appears low because of several reasons, few being the early implementation of lockdown[20,21]; and the universal BCG vaccination in childhood, which induces non-specific protection mediated via the induction of innate immune memory.[22] The hot and humid environment may be another factor as high COVID pandemic related fatality observed in latitude band with average temperature below 17degree Celsius and a Chinese study showed both one unit increase of temperature and absolute humidity were associated with the decreased COVID-19 death.[23] The individual host immunity and virulence factor may also affect the fatality.[24] Possibly the lower virulence of the COVID strain and proportionately lower elderly population than the western world in India and other parts of South Asia may be a plausible explanation of low fatality. However, these factors need to be confirmed in a scientific study.[25]
Figure 2.
Map of the 2019-nCoV outbreak in India as of 28 April 2020, 17:00 Hrs. 5000 + confirmed cases 1000–4999 confirmed cases 500–999 confirmed cases 100–499 confirmed cases 50–99 confirmed cases 1–49 confirmed cases shifted to another state (Source https://en.wikipedia.org/wiki/2020_corona virus_pandemic_in_India accessed on 28th April 2020)
Epidemiology
The impact of a pandemic depends on the number of infected persons, transmissibility, and the clinical severity of the infection. The world needs to expand public health activities, social and economic planning in reference to the SARS-CoV-2, and reduced the impact on public health, social and economic well-being of the people. Till March end, the epidemiology of COVID-19 was not very clear.[26] Now, with increasing information on the epidemiology of COVID-19, the newly affected parts, including India, evolved with the concept of lockdown and reduced the transmission as an effective way to curb the transmission.
Mode of transmission
Epidemiological studies in Wuhan at the beginning of the outbreak identified an initial association with a seafood market that sold live animals for food purposes, where most of the early patients had worked or at least visited there. The market was traced as the source, and subsequently, the market was closed for disinfection.[27] However, as the outbreak progressed, person-to-person spread became the primary mode of transmission.
The respiratory droplets released during cough, sneezing, talks, and mucus secretion are the dominant medium of transmission. The infection occurs if a person inhales such droplets or touches the contaminated surface with droplets and, subsequently, their own eyes, nose, and mouth.[28,29] The droplets do not travel more than 6 feet and do not linger in the air also. The aerosol spread is uncommon; however, the aerosol-generating procedures like intubation, suction, nebulization, etc., may transmit the disease. However, a report revealed that SARS-CoV-2 might remain viable in aerosols under experimental conditions for at least three hours.[28] The possibility of transmission is higher in the early phase as soon as symptoms appear as the viral RNA peaks during that period. However, it may transmit during the incubation period as well.[30] The incubation period is typically within two weeks of exposure, with the majority occurring within 4-5 days of exposure.
According to a joint WHO-China report, the rate of secondary COVID-19 ranged from 1-5% among tens of thousands of close contacts of confirmed patients in China.[31] In the United States, the symptomatic secondary attack rate was 0.45% among 445 close contacts of 10 established patients.[32] SARS-CoV-2 has been detected in non-respiratory specimens, including stool, blood, and ocular secretions, but the role of these sites in the transmission is uncertain.[33,34,35,36,37] Live viruses had also been cultured from stool; however, the fecal-oral transmission did not appear to be a significant factor in the spread of infection.[31]
Period of infectivity
The period of infectivity is uncertain in COVID-19. The detection of viral RNA from the respiratory tract does not always indicate the presence of an infectious virus. The patient might be more infectious during the earlier stage of infections. The viral RNA from the upper respiratory tract specimen after the onset of symptoms is usually higher than the later phase of the disease.[29,38,39] Additionally, in a study of nine patients with mild COVID-19, infectious virus was isolated from nasal/oropharyngeal and sputum specimens during the first week of illness, but not after this interval, despite continued viral RNA levels at these sites.[39] A modeling study from China showed that infectiousness started 2.3 days before symptom onset, peaked 0.7 days before symptom onset and declined within seven days, particularly after isolation of these patients.[29]
However, an asymptomatic individuals may also transmit the disease during the incubation period. A Singapore study in an analysis of 157 locally acquired COVID-19 showed the transmission rate of 6.4% during the incubation period. The exposures occurred one to three days before symptoms in these patients.[40] The exact estimation of asymptomatic infection will be possible with serologic testing, which is still in the phase of testing and validation.[40,41,42]
The duration of viral shedding varies with degree of severity. With milder illness, 90% showed negative test by ten days, while severe illness shows a more prolonged period of positivity. However, another study has shown more prolonged shedding of the virus of a median duration of 24 days. However, detection of viral RNA in sample does not correlate with infectivity. In the study of nine patients with mild COVID-19, the infectious virus was not detected from respiratory specimens when the viral RNA level was <106 copies/mL.[39]
Environmental contamination
Environmental contamination and fomite transmission from a contaminated surface to mucous membranes of the nose, eye, mouth etc., is possible in heavy viral contamination settings. A Singapore study showed the presence of viral RNA on handles, light switches, bed and handrails, interior doors and windows, toilet bowl and also the sink basin in the airborne infection isolation room of a patient with symptomatic mild COVID-19.[28] However, routine cleaning with sodium dichloroisocyanurate cleared the virus from these surfaces. SARS-CoV-2 may persist for differing time interval on different surfaces. The various disinfectants including ethanol at concentrations between 62 and 71% inactivate SARS-CoV-2 within one minute.[43] The duration of viral persistence on surfaces depends on type of surface, the ambient temperature, relative humidity, and the size of the initial inoculum.[44,45]
Risk of animal contact
Despite the thought of initial transmission from animals to human beings, there is no evidence to suggest domestic animals are a major source of infection in humans for SARS-CoV-2. There have been no reports of domesticated animals transmitting SARS-CoV-2 infection to humans. Experimental studies showed the virus replicated in cats after intranasal inoculation, but not in dogs.[46] CDC recommends that pets should be kept away from other animals or people outside of the household. People with confirmed or suspected COVID-19 try to avoid close contact with household pets, in similar fashion as with other human household members for the duration of their quarantine period.[47]
Protective Immunity and risk of reinfection
The definitive evidence of the development of protective antibody in infected COVID patients is still emerging.[38,39] The presence of neutralizing activity in convalescent plasma has been reported.[48] In a study on 23 recovered COVID-19 patients revealed, antibodies to the receptor-binding domain of the spike protein and the nucleocapsid protein appears by day 14 following onset of symptoms. The study also showed that antibody titers by enzyme-linked immunosorbent assay (ELISA) correlated with neutralizing activity.[39] Having neutralising activities, these antibodies are likely to be protective. The US-FDA has granted emergency use authorization for tests that qualitatively identify antibodies against SARS-CoV-2 in serum or plasma.[49] The serologic screening will be an important tool to understand population immunity and distinguishing peoples at lower risk for reinfection.
Risk factors
Older age, diabetes mellitus, chronic lung disease, cardiovascular disease, obesity, immunocompromised states, chronic kidney disease and liver diseases are the risk factor for the COVID-19. All ages and both sexes are are affected. Middle-aged adults and elderly above 60 years are most commonly affected with predominantly severe disease and increased mortality.[50,51,52] The US-CDC advocated that the people with the immunocompromised state, severe obesity, and liver disease are at a risk for severe illness.[53] Males had disproportionately high number of deaths in cohorts from China, Italy and the United States.[10,51,52] The pre-existing comorbidities are associated with a higher mortality.
Structure and viral genome of coronavirus in brief
The SARS-CoV-2 is a β-coronavirus, which is enveloped non-segmented positive-sense RNA virus of subfamily Orthocoronavirinae of the Coronaviridae family.[11,54] CoVs are divided into four genera called alpha (α), beta (β), gamma (γ) and delta (δ) CoV. α- and β-CoV can infect mammals, while γ- and δ-CoV tend to affect birds. Members of this large family of viruses can cause respiratory, enteric, hepatic, and neurological diseases in different animal species, including camels, cattle, cats, and bats.[54,55] Six CoVs have been discovered which can affect human, among which α-CoVs HCoV-229E and HCoV-NL63, and β-CoVs HCoV-HKU1 and HCoV-OC43 had low pathogenicity, and cause common cold like milder respiratory symptoms. The other two known β-CoVs, SARS-CoV, and MERS-CoV lead to severe and fatal respiratory tract infections.[2,54] SARS-CoV-2 is 29.9 kb,[55] While SARS-CoV and MERS-CoV have positive-sense RNA genomes of 27.9 kb and 30.1 kb, respectively.[56] It has been shown that the genome of CoVs contains a variable number (6-11) of open reading frames (ORFs).[57]
Figure 3 depicts the structure of the CoVs. CoVs are round or elliptic and often pleomorphic form, with a diameter of approximately 60–140 nm. The single-stranded RNA genome contains 29891 nucleotides, encoding for 9860 amino acids. Two-thirds of viral RNA, mainly located in the first open reading frame (ORF 1a/b), encodes 16 non-structure proteins (NSPs). The rest part of the virus genome encodes four essential structural proteins, including spike (S) glycoprotein, small envelope (E) protein, matrix (M) protein, and nucleocapsid (N) protein, and also several accessory proteins that interfere with host immune response.[55,58] The primary functions that direct coronavirus RNA synthesis and processing reside in nonstructural protein (nsp) 7 to 16, which are cleavage products of two large replicase polyproteins translated from the coronavirus genome.[59,60,61]
Figure 3.
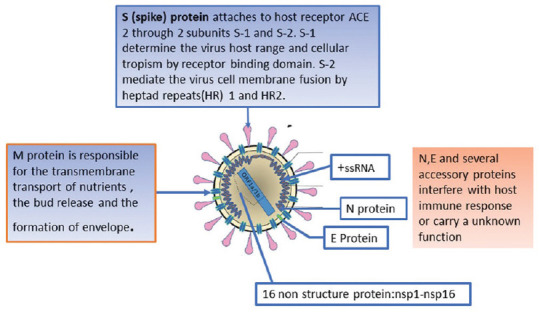
Depicting the structure of the Coronavirus and the role of different proteins
The sequencing studies of Wu et al.[60] revealed genomic and phylogenetic similarity of the SARS-CoV-2 with SARS-CoV, particularly in the S-protein gene and the receptor binding domain (RBD). This indicated the capability of direct human transmission like SARS-CoV. The whole-genome sequence studies showed that COVID-19 appears closer to the SARS-like bat CoVs as compared to the known SARS-CoV and MERS-CoV. Chan et al. have proven that the genome of the new HCoV, isolated from a cluster-patient with atypical pneumonia after visiting Wuhan, had 89% nucleotide identity with bat SARS-like-CoVZXC21 and 82% with that of human SARS-CoV.[4] For this reason, the new virus was called SARS-CoV-2.[59] The majority of genomically encoded proteins of SARS CoV-2 and SARS-CoVs were similar, except few differences in some amino acid substitutions in NSP2, NSP3, spike protein and receptor binding domains.[60,61]
Another recent research suggested[61] that the mutation in NSP2 and NSP3 play a role in infectious capability and differentiation mechanism of COVID-19. A study by Zhang et al.[62] revealed that SARS CoV-2 was mutating in different patients in China. Tang et al.[63] conducted a population genetic analysis of 103 COVID-19 genomes and classified out two prevalent types of COVID, L type (approximately 70%) and S type (approximately 30%). The strains in L type, derived from S type, are evolutionarily more aggressive and contagious. There is a need to keep an eye over this novel CoVs for their virulence and epidemic spread over the globe, at present.
It was also found that the genome sequence of SARS-CoV-2 is 96.2% identical to a bat CoV RaTG13, whereas it shares 79.5% identity to SARS-CoV. Based on virus genome sequencing results and evolutionary analysis, the bat is suspected as the natural host of virus origin, and COVID-19 might be transmitted from bats via unknown intermediate hosts like pangolins, other reptiles, and snakes etc., to infect humans.
Overall molecular mechanism of injury by COVID-19 (SARS-CoV-2)
S-protein of SARS-CoV-2 binds to host cell receptors, angiotensin-converting enzyme 2 (ACE2), which is a critical step for virus entry into cell.[64] ACE2 is cell receptor for COVID-19 and regulates the transmission across the species and between human beings as well.[65,66] S-protein contain two subunits, S1 and S2.[67,68,69] S1 determines the virus-host interaction and cellular tropism with the vital function domain-RBD, while S2 mediates virus-cell membrane fusion by two tandem domains, heptad repeats 1 (HR1)[68] and HR2.[66,67,68]
S-protein and ACE2 binding efficiency of COVID-19 is 10- to 20- fold higher than that of SARS-CoV.[70] For SARS-CoV, the cleavage of trimer S protein is triggered by the cell surface-associated transmembrane protease serine 2 (TMPRSS2)[71] and cathepsin,[72] however, the possible molecules facilitated membrane invagination for SARS-CoV-2 endocytosis are still under investigations.
After membrane fusion, the viral genome RNA is released into the cytoplasm, and the uncoated RNA translates two polyproteins, pp1a and pp1ab,[73] which encode non-structural proteins, and form replication-transcription complex (RTC) in double-membrane vesicle.[74] Continuously RTC replicates and synthesizes a nested set of subgenomic RNAs,[75] which encode accessory proteins and structural proteins. Mediating endoplasmic reticulum (ER) and Golgi bodies,[76] newly formed genomic RNA, nucleocapsid proteins, and envelope glycoproteins assemble and form viral particle buds. Lastly, the virion-containing vesicles fuse with the plasma membrane to release the virus leading to viremia.
The S2 subunit of Covid-19 containing a fusion peptide, a transmembrane domain, and cytoplasmic domain is highly conserved, which could be a target for antiviral targeting against S-2 (anti-S2) compounds. The spike RBD presents only a 40% amino acid identity with other SARS-CoVs. The ORF3b has no homology with that of SARS-CoVs and a secreted protein (encoded by ORF8), which is structurally different from those of SARS-CoV, may be an area of interest and research in the future.[77]
Mechanisms of kidney and other organ injuries
Host susceptibility, particularly elderly and peoples with underlying diseases, hypertension, cardiac diseases, bronchial asthma, diabetes, chronic kidney disease etc., influence the risk of acquiring and progression of COVID-19. The mechanism of kidney injury by COVID-19 appears multifactorial and, although precisely, remains unknown.[77] The direct viral cytopathic effect on kidney tissue is a postulated mechanism, which is supported by the finding of viral nucleic acid material of CoV in blood and urine in SARS-CoV as well as COVID-19 patients.[17,78] The molecular study showed CoV uses ACE2 receptor for cell entry. ACE2 expression is 100-fold higher in kidney tissues than the lung.[50] It makes sense to postulate that ACE2 dependent pathway may be used by CoV to infect kidneys more severely than the lung. However, clinical observation is different from more lung involvement than the kidney.
The direct effector T cell-mediated injury and the immune complex-mediated glomerular injury with viral antigen and specific antibody could be another plausible mechanism. However, the present evidence of information with normal glomerular aspect on microscopy and absence of electron-dense deposit in SARS-CoV patients do not support this hypothesis.[79] Furthermore, there is report of collapsing glomerulonephritis in COVID-19 and proteinuria and hematuria in significant number of patients.
The other mechanism could be by inducing sepsis and the cytokine storm theory.[80] The cytokines and other mediators are released after CoV infection leading to sustained inflammatory response leading to hypotension, hypoxia, shock, and target organ injuries. The clinical pictures of patients with COVID-19 with sepsis support this hypothesis. The manifestations are particularly severe, with a wide range of signs and symptoms of multiorgan involvement. These signs and symptoms include respiratory events such as severe dyspnea and hypoxemia, renal impairment with reduced urine output, tachycardia, altered mental status, and functional alterations of organs expressed as laboratory data of hyperbilirubinemia, acidosis, high lactate, coagulopathy, and thrombocytopenia. However, these findings suggest the probable mechanism of AKI in many terminal cases. Wang et al. showed that 138 patients with COVID-19 disease, who were admitted in ICU, showed a tendency towards increased creatine kinase levels.[81] It contributes to AKI indirectly through the effects on renal tissues, because of hypotension, hypoxia, shock, and rhabdomyolysis.
Clinical manifestations
There are no specific clinical features that can yet reliably distinguish COVID-19 from other viral respiratory infections. Pneumonia appears to be the most severe and frequent manifestation of infection, characterized primarily by fever, cough, dyspnea and bilateral lung infiltrates on chest imaging.[50,51,82] In a study describing 138 patients with COVID-19 pneumonia in Wuhan, the most common clinical features at the onset of illness were fever (99%), fatigue (70%), dry cough (59%), anorexia (40%), myalgias (35%), dyspnea (31%) and sputum production (27%).[51,82]
However, different clinical manifestations were noticed from other parts of the world. A study report from NewYork city of USA, one of the worst affected city in the world showed the median age of 62 years, predominantly males (61%), and obese (36%). The clinical symptoms were cough (79%) more common than China, fever (77%) relatively less than China, and dyspnea (57%). Gastrointestinal symptoms (diarrhea, 24%; nausea and vomiting, 19%) were relatively common.[83] The US-CDC[84] added a few new symptoms in COVID checker list with chills, repeated shaking with chills, muscle pain, headache, sore throat and new loss of taste or smell.
The mild disease is usually characterized by fever, malaise, cough, upper respiratory symptoms, in the absence of dyspnea. Most of these patients do not need hospitalization. The severe disease is characterized by hypoxia (oxygen saturation ≤93 and on room air or PaO2/FiO2 < 300 mmHg), tachypnea (respiratory rate >30 breaths per minute) or respiratory distress, more than 50% involvement of the lung parenchyma on chest imaging and other organ involvement.[85]
Respiratory failure needing to ventilatory requirement also varied in different countries. Respiratory failure requiring mechanical ventilation occurred in a third of the patients in the USA, which was relatively greater than other parts of the world. 30% of the patients required mechanical ventilation (MV) before the requirement of supplemental oxygenation, which indicates that a substantial proportion deteriorates soon after the presentation. The patients needing MV were predominantly male (71% vs. 56%) and obese (43% vs. 32%). They had elevated liver function tests and other inflammatory markers.[83] The other notable complications in those who required ventilatory support include need for vasopressor support (95%), cardiac arrhythmias (19%), bacteremia (12%), and new RRT (13%). Besides respiratory tract, the involvement of other organs such as the kidney, heart, digestive tract, blood, and nervous system also reported as for MERS.[54,55,84]
Kidney specific manifestations
Kidney involvement may be directly due to SARS-CoV-2 mediated injury or, a part of multiorgan dysfunction consequent upon cytokine storm. However, kidney involvement is a strong and independent predictor of mortality with COVID. The incidence of AKI with COVID infection reportedly vary from 3%- 9%.[17,51,82,86]. A larger prospective study reported the overall incidence of 5.1%.[87] Li et al.[79] found that 34% of patients had albuminuria on the first day of admission and that 63% developed proteinuria during the hospital stay. 19% of the people showed an elevated level of plasma creatinine. Each one of those (27/27) who had computerized tomography (CT) scan showed radiographic abnormalities of the kidneys with reduced density suggesting inflammation and edema. The study also emphasized that renal impairment may be an independent factor of mortality.[79] A prospective study showed that on admission, 43.9% of the patients had proteinuria, and 26.7% had haematuria. The prevalence of elevated serum creatinine and estimated glomerular filtration under 60 ml/min/1.73 m2 were 14.4, and 13.1%, respectively. Cox proportional hazard regression confirmed that elevated baseline serum creatinine was an independent predictor of mortality. The hazard ratio also increases with the staging of AKI from 1.9 in stage-1, 3.51 in stage-2, and 4.38 in stage-3 AKI. Patients with kidney disease had a significantly higher risk of in-hospital death.[87]
Diagnosis
Reverse transcriptase polymerase chain reaction (RT-PCR)
The sample collection and storage for the diagnosis in a resource-limited place is also challenging. The WHO recommends collecting specimens from the upper respiratory tract (nasopharyngeal - and/or opharyngeal samples); and lower respiratory tract such as sputum, endotracheal aspirate, or bronchoalveolar lavage (BAL). The collection of BAL samples should only be performed in patients on MV. The samples require storage at four degrees celsius. Lower tract samples may have greater sensitivity than upper respiratory tract specimens.[88]
A study[89] showed that pharyngeal virus shedding was very high during the first week of symptoms with a peak on day 4 and was readily isolated from the throat- and lung-derived samples. However, it was not isolated from stool samples despite high virus RNA concentration. Blood and urine never yielded a virus. Viral replicative RNA intermediates confirmed active replication in throat samples. Independent replication of sequence -distinct virus was also observed in throat and lung samples from the same patient. The shedding of viral RNA from sputum outlasted the end of symptoms. Seroconversion occurred after seven days in 50% of patients and in majority of patients by day 14. However, this event was not universally followed by a rapid decline in viral load.
In the laboratory, a reverse transcriptase polymerase chain reaction (RT-PCR) is used for the amplification of the genetic material extracted from the saliva, mucus, and other samples. It involves the synthesis of a double-stranded DNA molecule from an RNA mold. The search is targeted towards the genetic code of the CoV that is conserved. CDC recommends to assess for the presence of 1 or several nucleic acid targets specific to SARS–CoV-2.[77,90] The probes used are based on the initial gene sequence released by the Shanghai Public Health Clinical Center and School of Public Health, Fudan University, Shanghai, China on Virological.org, and subsequent confirmatory evaluation by additional labs.[77] If the test result is positive, it is recommended that the test is repeated for verification. In patients with confirmed COVID-19 diagnosis, the laboratory evaluation should be repeated to evaluate for viral clearance before being released from observation.
In the United States, the CDC has developed the most widely used SARS–CoV-2 assay. The kit contains PCR primer-probe sets for 2 regions of the viral nucleocapsid gene (N1 and N2) and for the human RNase P gene to ensure the RNA extraction was successful. This assay differs from the WHO primer-probe sets, which target the SARS–CoV-2 RNA-dependent RNA polymerase (RdRP) and envelope (E) genes.[89] Both assays have high analytic sensitivity and specificity for SARS–CoV-2, with minimal cross-reactivity with other circulating strains of coronaviruses, and both use a cycle threshold (CT) of less than 40 as the criterion for positivity.
The lack of an established reference standard, use of differing sample collection and preparation methods, and an incomplete understanding of viral dynamics across the time course of infection, hamper the rigorous assessment of the diagnostic accuracy of the many newly introduced SARS–CoV-2 assays.[91] Conversely, after a patient has had a positive test result, several authorities have recommended obtaining at least 2 negative upper respiratory tract samples, collected at intervals of 24 hours or longer, to document SARS–CoV-2 clearance.[92]
Viral culture
Although viral culture is an important method to evaluate viral infectivity and activity, it is not commonly used in clinical practice because of its low sensitivity and long turn-around time for virus detection.[93] virus isolation in a culture in the laboratory with a facility for viral culture using Vero-CCL-81 cells is possible. However, this facilities are limited within the country.
Rapid antigen detection tests
Rapid antigen detection test (RDT) detects the presence of viral antigens expressed by the COVID-19 virus in a sample from the respiratory tract. It detects the target antigen as it binds to specific antibodies fixed to a paper strip enclosed in a plastic casing. It generates a visually detectable signal, typically within 30 minutes. However, this test has a limitation in the form of expression occurring only when the virus is actively replicating. It can be used to identify acute or early infection. The test result varies with time from onset of illness, the concentration and the quality of the specimen and the precise formulation of the reagents in the test kits.[94,95]
From previous experience of the use of these types of kits in Influenza, the sensitivity of these tests is expected to vary from 34-80% for COVID-19 as well.[96] False-positive results may also occur with the antibodies on the test strip also recognize antigens of viruses other than COVID-19, such as human CoV causing common cold. Prototypes of such tests for other novel coronaviruses have not received regulatory approval[97,98] but are under development.[99] Monoclonal antibodies against the nucleocapsid protein of SARS–CoV-2 have been developed which might form the basis of a future rapid antigen detection test.[100]
Rapid antibody diagnostic test
The principle of the test is based on the detection of the IgM and IgG antibodies, in the blood of patients with COVID-19.[101,102,103,104] The test can be performed with enzyme-linked immunosorbent assays (ELISA). The test is relatively less complex than other molecular tests and primarily used for the epidemiological purpose in limited situations.[105] The IgG antibodies appear late in the second week after onset of symptoms, while the majority of them show futures of recovery.[106,107]
Thus the use of this test in making clinical intervention and in the prevention of transmission of the disease remains limited. The negative result also does not exclude recent SARS–CoV-2 exposure and infection.[105] Several factors, like age, malnutrition, the severity of the disease, and immunosuppressed state because of medications or HIV like disease, affects the formation of the antibodies.[106,107] However, the test may be reported in combination with RT-PCR report with the presence or absence of antibody response.[106,107] The possibility of cross-reactivity and false positivity of COVID specific antibody with other human CoV can not be excluded.[88,107,108]
The information about the protective nature of the antibody is still emerging; however, antibodies against S-protein may be protective, and plasma from recovered patients show neutralizing activity.[38] The test may help in analyzing antibody responses to COVID-19, a critical response in the development of vaccines. The test can be used for detecting the epidemiological extent of infection missed during active surveillance efforts, analyzing the attack rate, and infection fatality rate.
Ancillary diagnostic test
Radiographic imaging: The plain chest radiography is still the early and easy ancillary supportive test in COVID-19 management. The bilateral pneumonia is the most common finding ranging from 11.8-100%. The bilateral findings are more common than a unilateral focus.[82,109]
Computed tomography of thorax appears more sensitive than plain radiography. A large study have shown that typical imaging features, include ground-glass opacities (86.1%) or mixed ground-glass opacities and consolidation (64.4%), vascular involvement in the lesion (71.3%), and traction bronchiectasis (52.5%). The lesion on CT images had more peripheral distribution (87.1%), bilateral involvement (82.2%), lower lung predominant, and multifocal each in 54.5%.[110] Studies reported that CT chest might be more sensitive than serial nasopharyngeal sampling and RT-PCR test at a single-point diagnosis of COVID-19.[111,112] Although artificial intelligence may help in distinguishing COVID-19 from other etiologic agents of community-acquired pneumonia,[113] but the CT findings, do not exclude a co-infection or an alternative diagnosis.[114]
Other Biomarkers Associated with COVID-19
Decreased albumin, elevated C-reactive protein, and elevated lactate dehydrogenase levels, and lymphopenia were other laboratory parameters associated with COVID-19.[115] Increased erythrocyte sedimentation rates, elevated aspartate aminotransferase, alanine aminotransferase, and creatinine kinase levels, leukopenia, leukocytosis increased bilirubin and creatinine levels were associated with severe cases and multi-organ involvements.[50,51,116] These biomarkers are an indication of the inflammatory host response to SARS–CoV-2, as observed in any patients with sepsis.[117] It is difficult to predict clinical outcomes with any identified single or combination of biomarkers currently exists.
In summary, COVID-19 is a novel Beta CoV infection which has genomic homology with Bat CoV and transmitted to human beings through intermediate host. Person to person transmission in the human being is a major mode of transmission, which lead to pandemic from a small cluster outbreak from Wuhan city of China. The lung is primarily involved. However, kidney involvement is frequent and is an independent risk factor for mortality with this novel CoV infection. With the increasing stage and severity of AKI, the hazard ratio of death of patients with COVID also increases. The overall case-fatality rate is reportedly 2.3% in confirmed cases, about 15% in elderly patients, in particular those aged ≥80 years, and 8% in people who are 70-79 years of age.[118] The mainstay of therapy of COVID-19 is supportive and preventive requiring quarantine and waiting self recoverty.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Coleman CM, Frieman MB. Coronaviruses: Important emerging human pathogens. J Virol. 2014;88:5209–12. doi: 10.1128/JVI.03488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin Y, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23:130–7. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–3. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–9. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 5.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet. 2020;395:565–74. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO. Coronavirus disease (COVID-2019) situation reports. 2020. [Last accessed on 2020 Mar 05]. Available from: https://wwwwhoint/emergencies/diseases/novel-coronavirus-2019/situationreports .
- 8.World Health Organization. Director-General's remarks at the media briefing on 2019-nCoV on 11 February 2020. [Last accessed on 2020 Feb 12]. Available from: https://wwwwhoint/dg/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-o n-2019-ncov-on-11-february-2020 .
- 9.Centers for Disease Control and Prevention. First travel-related case of 2019 novel coronavirus detected in United States, January 21, 2020. [Last accessed on 2020 Jan 21]. Available from: https://wwwcdcgov/media/releases /2020/p0121-novel-coronavirus-travel-casehtml .
- 10.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–33. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO. Coronavirus disease (COVID-2019) situation reports. 2020. [Last accessed on 2020 Mar 21]. Available from: https://wwwwhoint/emergencies/diseases/no vel-coronavirus-2019/situationreports .
- 12.European Centre for Disease Prevention and Control. Novel coronavirus in China. [Last accessed on Jan 23]. Available from: https://wwwecdceuropaeu/en/no vel-coronavirus-china .
- 13. [Last accessed on 2020 Apr 26]. Available from: https://www.who.int/docs/default-source/coronavi ruse/situation-reports/20200422-sitrep-93-cov id-19.pdf?sfvrsn=35cf80d7_4 .
- 14.CDC. [Last accessed on 2020 Mar 23]. Available from: https://wwwcdcgov/coronavirus/2019-ncov/cases-updates/world-maphtml .
- 15. [Last accessed on 2020 Mar 23];Indian Council of Medical Research Update: COVID.19 (23/03/2020 10:00 AM) [Google Scholar]
- 16.Ramanan Laxminarayan, Director of the Center for Disease Dynamics, Economics and Policy (CDDEP) a public health research group based in Washington D.C. and New Delhi. Available from: https://www.indiatoday.in/india/story/coronavirus-cases-india-covid-19-ramanan-laxminarayan-interview-1658087-2020-03-21 .
- 17.Cheng Y, Luo R, Wang K, Zhang M, Wang Z, Dong L, et al. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020;97:829–38. doi: 10.1016/j.kint.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.2020 coronavirus lockdown in India. [Last accessed on 2020 Apr 28]. Available from: https://enwikipediaorg/wiki/2020_coronavirus_lockdown_in_India .
- 19.COVID-19 and India. [Last accessed on 2020 Apr 27]. Available from: https://wwwmohfwgovin/
- 20.India under COVID-19 lockdown. The Lancet. [Last accessed on2020 Apr 27]. Editorial. 25th April 2020. doi: 10.1016/S0140-6736 (20) 30938-7. Available from: https://www.thelancet.com/journals/lan cet/article/PIIS0140-6736(20)30938-7/fu lltext .
- 21.The Center For Disease Dynamics, Economics and Policy. COVID-19 in India: Potential Impact of the Lockdown and Other Longer-Term Policies. [Last accessed on 2020 Apr 28]. Available from: https://cddep.org/publications/covid-19-india-pot ential-impact-of-the-lockdown-and-other-longer-term-policies/
- 22.Gursel M, Gursel I. Is global BCG vaccination coverage relevant to the progression of SARS-CoV-2 pandemic [published online ahead of print, 2020 Apr 6] Med Hypotheses. 2020:109707. doi: 101016/jmehy 2020109707. [Google Scholar]
- 23.Ma Y, Zhao Y, Liu J, He X, Wang B, Fu S, et al. Effects of temperature variation and humidity on the death of COVID-19 in Wuhan, China. Sci Total Environ. 2020;724:138226. doi: 10.1016/j.scitotenv.2020.138226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gasmi A, Noor S, Tippairote T, Dadar M, Menzel A, Bjørklund G. Individual risk management strategy and potential therapeutic options for the COVID-19 pandemic [published online ahead of print, 2020 Apr 7] Clin Immunol. 2020:108409. doi: 10.1016/j.clim.2020.108409. doi: 101016/jclim 2020108409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.South Asia. [Last accessed on 2020 Apr 29]. Available from: https://enwikipediaorg/wiki/South_Asia .
- 26.Lipsitch M, Swerdlow DL, Finelli L. Defining the epidemiology of Covid-19-studies needed. N Engl J Med. 2020;382:1194–6. doi: 10.1056/NEJMp2002125. [DOI] [PubMed] [Google Scholar]
- 27.World Health Organization. Novel coronavirus situation report -2 January 22, 2020. [Last accessed on 2020 Jan 23]. Available from: https://wwwwhoint/docs/default-source/corona viruse/situation-reports/20200122-sitr ep-2-2019-ncovpdf .
- 28.Van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, et al. Aerosol and surface stability of SARS-CoV-2 as Compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–7. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–9. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu P, Zhu J, Zhang Z, Han Y, Huang L. A familial cluster of infection associated with the 2019 novel coronavirus indicating potential person-to-person transmission during the incubation period. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa077. doi: 101093/infdis/jiaa077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Report of the WHO-China Joint Mission on Coronavirus DIsease 2019 (COVID-2019). February 16-24, 2020. [Last accessed on 2020 Mar 04]. Available from: http://www.who.int/docs/default-source/corona viruse/who-china-joint-mission-on-covid-19-final-report.pdf .
- 32.Burke RM, Midgley CM, Dratch A, Fenstersheib M, Haupt T, Holshue M, et al. Active monitoring of persons exposed to patients with confirmed COVID-19-United States, January-February 2020. MMWR Morb Mortal Wkly Rep. 2020;69:245–6. doi: 10.15585/mmwr.mm6909e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu J, Gu J, Li K, Xu C, Su W, Lai Z, et al. COVID-19 outbreak associated with air conditioning in restaurant, Guangzhou, China, 2020. Emerg Infect Dis. 2020:26. doi: 10.3201/eid2607.200764. doi: 103201/eid2607200764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng K, Poon BH, Kiat Puar TH, Shan Quah JL, Loh WJ, Wong YJ, et al. COVID-19 and the Risk to Health Care Workers: A case report. Ann Intern Med. 2020 doi: 10.7326/L20-0175. doi: 107326/L20-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong SC, Kwong RT, Wu TC, Chan JWM, Chu MY, Lee SY, et al. Risk of nosocomial transmission of coronavirus disease 2019: An experience in a general ward setting in Hong Kong. J Hosp Infect. 2020 doi: 10.1016/j.jhin.2020.03.036. doi: 101016/jjhin 202003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen W, Lan Y, Yuan X, Deng X, Li Y, Cai X, et al. Detec[table 2019]-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg Microbes Infect. 2020;9:469–73. doi: 10.1080/22221751.2020.1732837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. doi: 101001/jama 20203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu F, Yan L, Wang N, Yang S, Wang L, Tang Y, et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa345. doi: 101093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.AABB. AABB's Coronavirus Resources. [Last accessed on 2020 Apr 21]. Available from: http://wwwaabborg/advocacy/regulatorygovernment/Pages/AABB-Coronavirus-Resou rcesaspx .
- 40.Hu Z, Song C, Xu C, Jin G, Chen Y, Xu X, et al. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci. 2020;63:706–11. doi: 10.1007/s11427-020-1661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian G, Yang N, Ma AHY, Wang L, Li G, Chen X, et al. A COVID-19 Transmission within a family cluster by presymptomatic infectors in China [published online ahead of print, 2020 Mar 23] Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa316. ciaa316 doi: 101093/cid/ciaa316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR, et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility [published online ahead of print, 2020 Apr 24] N Engl J Med. 2020 doi: 10.1056/NEJMoa2008457. 101056/NEJMoa2008457 doi: 101056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park SY, Kim YM, Yi S, Lee S, Na BJ, Kim CB, et al. Coronavirus disease outbreak in call center, South Korea [published online ahead of print, 2020 Apr 23] Emerg Infect Dis. 2020:26. doi: 10.3201/eid2608.201274. doi: 103201/eid2608201274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghinai I, Woods S, Ritger KA, McPherson TD, Black SR, Sparrow L, et al. Community transmission of SARS-CoV-2 at two family gatherings-Chicago, Illinois, February-March 2020. MMWR Morb Mortal Wkly Rep. 2020;69:446–50. doi: 10.15585/mmwr.mm6915e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kampf G, Todt D, Pfaender S, Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hosp Infect. 2020;104:246–51. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2 [published online ahead of print, 2020 Apr 8] Science. 2020 doi: 10.1126/science.abb7015. eabb7015 doi: 101126/scienceabb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coronavirus Disease- update 2019. [Last accessed on 2020 Apr 28]. Available from: https://wwwcdcgov/coronavirus/2019-ncov/daily-life-coping/animalshtml .
- 48.Rabenau HF, Cinatl J, Morgenstern B, Bauer G, Preiser W, Doerr HW. Stability and inactivation of SARS coronavirus. Med Microbiol Immunol. 2005;194:1–6. doi: 10.1007/s00430-004-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimball A, Hatfield KM, Arons M, James A, Taylor J, Spicer K, et al. Asymptomatic and presymptomatic SARS-CoV-2 infections in residents of a long-term care skilled nursing facility-King County, Washington, March 2020. MMWR Morb Mortal Wkly Rep. 2020;69:377–81. doi: 10.15585/mmwr.mm6913e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395:507–13. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Onder G, Rezza G, Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy [published online ahead of print, 2020 Mar 23] JAMA. 2020 doi: 10.1001/jama.2020.4683. doi: 101001/jama 20204683. [DOI] [PubMed] [Google Scholar]
- 53.Liang W, Guan W, Chen R, Wang W, Li J, Xu K, et al. Cancer patients in SARS-CoV-2 infection: A nationwide analysis in China. Lancet Oncol. 2020;21:335–7. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: Recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–34. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y, et al. From SARS to MERS, thrusting coronaviruses into the spotlight? Viruses. 2019;11:E59. doi: 10.3390/v11010059. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak-An update on the status. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–92. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–9. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–36. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–8. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Angeletti S, Benvenuto D, Bianchi M, Giovanetti M, Pascarella S, Ciccozzi M. COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis. J Med Virol. 2020 doi: 10.1002/jmv.25719. doi: 101002/jmv 25719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang L, Shen FM, Chen F, Lin Z. Origin and evolution of the 2019 novelcoronavirus. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa112. doi: 101093/cid/ciaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang X, Wu C, Li X, Song Y, Yao X, Wu X, et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020 doi: 10.1093/nsr/nwaa036. doi: 101093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tortorici MA, Veesler D. Structural insights into coronavirus entry. Adv Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jia HP, Look DC, Shi L, Hickey M, Pewe L, Netland J, et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol. 2005;79:14614–21. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS? J Virol. 2020;94:e00127–20. doi: 10.1128/JVI.00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang N, Jiang S, Du L. Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev Vaccines. 2014;13:761–74. doi: 10.1586/14760584.2014.912134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xia S, Zhu Y, Liu M, Lan Q, Xu W, Wu Y, et al. Fusion mechanism of 2019- nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020 doi: 10.1038/s41423-020-0374-2. doi: 101038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu F, Du L, Ojcius DM, Pan C, Jiang S. Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect. 2020 doi: 10.1016/j.micinf.2020.01.003. doi: 101016/j micinf 202001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Song W, Gui M, Wang X, Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Millet JK, Whittaker GR. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–34. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA. 2005;102:11876–81. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Wilde AH, Snijder EJ, Kikkert M, van Hemert MJ. Host factors in coronavirus replication. Curr Top Microbiol Immunol. 2018;419:1–42. doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sawicki SG, Sawicki DL. Coronavirus transcription: A perspective. Curr Top Microbiol Immunol. 2005;287:31–55. doi: 10.1007/3-540-26765-4_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hussain S, Pan J, Chen Y, Yang Y, Xu J, Peng Y, et al. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J Virol. 2005;79:5288–95. doi: 10.1128/JVI.79.9.5288-5295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Perrier A, Bonnin A, Desmarets L, Danneels A, Goffard A, Rouille Y, et al. The C-terminal domain of the MERS coronavirus M protein contains a trans-Golgi network localization signal. J Biol Chem. 2019;294:14406–21. doi: 10.1074/jbc.RA119.008964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. StatPearls [Internet] Treasure Island (FL): StatPearls Publishing; 2020. Jan, Features, Evaluation and Treatment Coronavirus (COVID-19) [Updated 2020 Mar 8] Available from: https://wwwncbinlmnihgov/books/NBK554776/ [PubMed] [Google Scholar]
- 78.Peiris JSM, Chu CM, Cheng VCC, Chan KS, Hung IF, Poon LL, et al. HKU/UCH SARS Study Group. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: A prospective study. Lancet. 2003;361:1767–72. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Z, Wu M, Guo J, Guo J, Liao X, Song S, et al. Caution on kidney dysfunctions of 2019-nCoV patients. MedRxiv Preprint. 2020 doi: 101101/2020020820021212. [Google Scholar]
- 80.Chu KH, Tsang WK, Tang CS, Lam MF, Lai FM, To KF, et al. Acute renal impairment in coronavirus-associated severe acute respiratory syndrome. Kidney Int. 2005;67:698–705. doi: 10.1111/j.1523-1755.2005.67130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3) JAMA. 2016;315:801–10. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–9. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goyal P, Choi JJ, Pinheiro LC, Schenck EJ, Chen R, Jabri A, et al. Clinical characteristics of Covid-19 in New York City. N Engl J Med. 2020 doi: 10.1056/NEJMc2010419. doi: 101056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.CDC-USA. [Last accessed on 2020 Apr 28]. Last accessed on 2020 Apr 28. Available from: https://wwwcdcgov/coronavirus/2019-ncov/sy mptoms-testing/sympto mshtml .
- 85.WHO- Interim Guidance. Clinical management of severe acute respiratory infection (SARI) when COVID-19 disease is suspected Interim guidance 13 March 2020 [Google Scholar]
- 86.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical characteristics of 2019 novel coronavirus infection in China [e-pub ahead of print] N Engl J Med. [Last accessed on 2020 Mar 02]. Available from: https://doiorg/101056/NE JMoa2002032 .
- 87.Cheng Y, Luo R, Wang K, Zhang M, Wang Z, Dong L, et al. Kidney impairment is associated with in-hospital death of COVID-19 patients [e-pub ahead of print] medRxiv. [Last accessed on 2020 Mar 02]. 2020021820023242. Available from: https://doiorg/101101/2020021820023242 .
- 88.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019 [published online ahead of print, 2020 Apr 1] Nature. 2020 doi: 10.1038/s41586-020-2196-x. doi: 101038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 89.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. doi: 102807/1560-7917ES20202532000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. [Last accessed on 2020 Apr 28]. Available from: https://wwwfdagov/media/134922/download .
- 91.Liu Y, Yan LM, Wan L, Xiang TX, Le A, Liu JM, et al. Viral dynamics in mild and severe cases of COVID-19 [Letter] Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30232-2. doi: 101016/S1473-3099 (20) 30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Centers for Disease Control and Prevention. Discontinuation of Transmission-Based Precautions and Disposition of Patients with COVID-19 in Healthcare Settings (Interim Guidance) [Last accessed on 2020 Apr 04]. Available from: wwwcdcgov/coronavirus/2019-ncov/hcp/disposition-hospitalized-patientshtml .
- 93.Charlton CL, Babady E, Ginocchio CC, Hatchette TF, Jerris RC, Li Y, et al. Practical guidance for clinical microbiology laboratories: Viruses causing acute respiratory tract infections. Clin Microbiol Rev. 2019;32:e00042–118. doi: 10.1128/CMR.00042-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Advice on the use of point-of-care immunodiagnostic tests for COVID-19. [Last accessed 2020 Apr 8]. Available from: https://wwwwhoint/news-room/commentaries/detail/advice-on-the-use-of-point-of-care- immunodiagnostic-tests-for-covid-19 .
- 95.Cheng MP, Papenburg J, Desjardins M, Kanjilal S, Quach C, Libman M, et al. Diagnostic testing for severe acute respiratory syndrome-related coronavirus-2: A narrative review. Ann Intern Med. 2020 doi: 10.7326/M20-1301. doi: 107326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bruning AHL, Leeflang MMG, Vos JMBW, Spijker R, de Jong MD, Wolthers KC, et al. Rapid tests for influenza, respiratory syncytial virus, and other respiratory viruses: A systematic review and meta-analysis. Clin Infect Dis. 2017;65:1026–32. doi: 10.1093/cid/cix461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lau SK, Woo PC, Wong BH, Tsoi HW, Woo GK, Poon RW, et al. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in SARS patients by enzyme-linked immunosorbent assay. J Clin Microbiol. 2004;42:2884–9. doi: 10.1128/JCM.42.7.2884-2889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen Y, Chan KH, Kang Y, Chen H, Luk HK, Poon RW, et al. A sensitive and specific antigen detection assay for Middle East respiratory syndrome coronavirus. Emerg Microbes Infect. 2015;4:e26. doi: 10.1038/emi.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.FIND. SARS-CoV-2 Diagnostic Pipeline. [Last accessed on 2020 Mar 23]. Available from: wwwfinddxorg/covid-19/pipeline .
- 100.Sheridan C. Fast, portable tests come online to curb coronavirus pandemic. Nat Biotechnol. 2020 doi: 10.1038/d41587-020-00010-2. doi: 101038/d41587-020-00010-2. [DOI] [PubMed] [Google Scholar]
- 101.Liu Y, Liu Y, Diao B, Ren F, Wang Y, Ding J, et al. Diagnostic indexes of a rapid IgG/IgM combined antibody test for SARS-CoV-2 medxriv [Internet] 2020. [Last accessed 2020 Mar 26]. Available from: https://doiorg/101101/2020032620044883 .
- 102.Zhang P, Gao Q, Wang T, Ke Y, Mo F, Jia R, et al. Evaluation of recombinant nucleocapsid and spice protein serological diagnosis of novel coronavirus disease 2019 (COVID-19) medxriv [Internet] 2020. Available from: https://wwwmedrxivorg/content/101101/2020031720036954v1 .
- 103.Pan Y, Li X, Yang G, Fan J, Tang Y, Zhao J, et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients medxriv [Internet] 2020. Available from: https://doiorg/101101/2020031320035428 . [DOI] [PMC free article] [PubMed]
- 104.Li Z, Yi Y, Luo X, Xion N, Liu Y, Li S, et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020. Available from: https://onlinelibrarywileycom/doi/a bs/101002/jmv25727 . [DOI] [PMC free article] [PubMed]
- 105.Guo L, Ren L, Yang S, Xiao M, Chang, Yang F, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa310. doi: 101093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019 medxriv [Internet] 2020. Available from: https://wwwmedrxivorg/conte nt/101101/2020030220030189v1fullpdf . [DOI] [PMC free article] [PubMed]
- 107.Okba NMA, Muller MA, Li W, Wang C, Geurtsvan Kessel CH, Corman VM, et al. SARS-COV-2 specific antibody responses in COVID-19 patients medxriv [Internet] 2020. Available from: https://wwwmedrxivorg/cont ent/101101/2020031820038059v1 .
- 108.Lin D, Liu L, Zhang M, Hu Y, Yang Q, Guo J, et al. Evaluation of serological tests in the diagnosis of 2019 novel coronavirus (SARS-CoV-2) infections during the COVID-19 outbreak medxriv [Internet] 2020. Available from: https://doiorg/101101/2020032720045153 . [DOI] [PMC free article] [PubMed]
- 109.Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X, et al. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295:202–7. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhao W, Zhong Z, Xie X, Yu Q, Liu J. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: A multicenter study. AJR Am J Roentgenol. 2020;214:1072–7. doi: 10.2214/AJR.20.22976. [DOI] [PubMed] [Google Scholar]
- 111.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases? Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. doi: 10.1148/radiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, et al. Sensitivity of chest CT for COVID-19: Comparison to RT-PCR? Radiology. 2020:200432. doi: 10.1148/radiol.2020200432. doi: 10.1148/radiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT? Radiology. 2020:200905. doi: 10.1148/radiol.2020200905. doi: 10.1148/radiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu X, Cai Y, Huang X, Yu X, Zhao L, Wang F, et al. Co-infection with SARS-CoV-2 and influenza A virus in patient with pneumonia, China. Emerg Infect Dis. 2020:26. doi: 10.3201/eid2606.200299. doi: 103201/eid2606200299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rodriguez-Morales AJ, Cardona-Ospina JA, Gutiérrez-Ocampo E, Villamizar-Peña R, Holguin-Rivera Y, Escalera-Antezana JP, et al. Latin American Network of Coronavirus Disease 2019-COVID-19 Research (LANCOVID-19). Clinical, laboratory and imaging features of COVID-19: A systematic review and meta-analysis? Travel Med Infect Dis. 2020:101623. doi: 10.1016/j.tmaid.2020.101623. doi: 10.1016/j.tmaid. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen L, Liu HG, Liu W, Liu J, Liu K, Shang J, et al. [Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia] Zhonghua Jie He He Hu Xi Za Zhi. 2020;43:203–8. doi: 10.3760/cma.j.issn.1001-0939.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 117.Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa248. doi: 101093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. doi: 101001/jama 20202648. [DOI] [PubMed] [Google Scholar]