Abstract
K. pneumoniae was known as a nosocomial infection that causes human diseases. It is considered as one of the food-borne pathogens as it causes septicemia and diarrhea in humans. This study aims to characterize K. pneumoniae strains isolated from ready to eat processed meat phenotypically and genetically. Three hundred and fifty ready to eat processed meat (Luncheon-meat) samples were collected. Forty-four (12.6%) K. pneumoniae strains were isolated and bio-typed, where the majority were identified to belong to biotype B1. K1 and K2 serotypes were detected and strains were classified as hypermucoviscous K. pneumoniae (HVKP) and classic K. pneumoniae (CKP) (26 and 18 isolates, respectively). The isolates were resistant to several classes of β–lactam antibiotics, ceftazidim and cefotaxime (95.5%), cefoxitin (93.2%), ertapenem (90.9%) and amoxicillin-clavulanic acid (86.4%). They were classified as extended spectrum β–lactamases (ESBLs), AmpC or carbapenemase-producers phenotypically. Eighteen β-lactamase genes were investigated by PCR. The most prominent genes were SHV (63.6%), TEM (52.2%), CTX-M15 (50%), AMPC (47.7%), CIT-M (45.5%) and VIM (43.2%). Co-detection of β–lactam resistance genes revealed 42 gene profiles. Twenty-four isolates had the complete efflux system (AcrAB-ToƖC). Besides, Integrons (I, II, III) were detected in 20 isolates. Molecular typing by ERIC-PCR showed high genetic diversity between isolates as 34 different patterns were identified. Overall, this study confirmed the hazards posed by the presence of multiple resistance genes in the same isolate and this should not be undervalued. Besides, the horizontal transfer of plasmid harboring resistance genes between isolates in food represents potential health risks for consumers in Egypt and so the control and inhibition plans are necessary.
Introduction
Klebsiella pneumoniae is a facultative anaerobic Enterobacteriaceae that presents as normal microbiota of skin, mouth and intestine. However, it is also responsible for pneumonia, urinary, and lower biliary tract diseases [1], intra-abdominal, blood stream infections, meningitis, pyogenic liver abscess and prominent nosocomial infections. Although K. pneumoniae is not recognized as a food born pathogen; it can be isolated from raw vegetables, street foods [1], hospital waste water [2], fish and shrimps [3], farm raised chicken [4], and milk [5].
K. pneumoniae developed different resistance mechanisms to different classes of antimicrobial agents such as efflux pump, changing membrane permeability, production of inactivating enzymes, modification of target site, and acquisition of alternative metabolic pathway that is inhibited by antibiotics [6].
β–lactam is a large class of antibiotics that has different sub-classes including penicillins, cephalosporins, carbapenems, and monobactam. In Gram-negative (G-ve) bacteria, resistance to β–lactams is mediated by different strategies such as production of β–lactamases, efflux pumps, and alteration of penicillin binding proteins. The major mechanism involved in B lactam resistance is the production of β–lactamases [7]. Different β–lactamases have been discovered starting with broad spectrum β–lactamases now designated as extended spectrum β–lactamases (ESBLs), β–lactamases with reduced sensitivity to β–lactamase inhibitors, AmpC and β–lactamases that hydrolyze carbapenems. As β–lactamases are inducible enzymes, the presence of β–lactam antibiotics in food is considered a risk factor that generates the intrinsic resistance of the G-ve bacteria [7]. Consequently, the presence of antimicrobial resistant K. pneumoniae in food is a major health problem.
Bacterial efflux pump is an important system of resistance in G-ve bacteria. Antibiotics are ejected from the cell, thus decreasing antibiotic concentrations inside the cell. Efflux pumps are classified into five major families: the resistance-nodulation-division (RND) family, the major facilitator superfamily (MFS), the ATP (adenosine triphosphate)-binding cassette (ABC) superfamily, the small multidrug resistance (SMR) family, and the multidrug and toxic compound extrusion (MATE) family [8]. ERICs are repetitive intergenic sequences of the Enterobacteriaceae family comprising of 126 base pairs. ERIC-PCR fingerprinting methods are widely used for genetic typing of isolates as they produce fingerprints without the use of endonuclease enzyme activity. They can be used as genetic markers for genetic diversity among bacteria [9].
The present study aims to evaluate the frequency of K. pneumoniae in ready to eat processed meat, resistance to β–lactam antibiotics and to type the isolates serologically, and by molecular method (ERIC-PCR).
Materials and methods
Bacterial isolation and identification
Three hundred and fifty retail ready to eat bovine and/or chicken processed meat (Luncheon) were collected from different supermarkets in Mansoura city, Egypt as illustrated in (S1 Table). Each sample was collected in a sterile plastic bag stored in an icebox and processed within 4–6 hours as previously described [1].
These isolates were identified biochemically as Klebsiella species according to laboratory biochemical standards [10], where 44 K. pneumoniae and 3 K. oxytoca isolates were recovered. K. pneumoniae isolates were bio-typed according to the positive biochemical reactions according to (S2 Table) and used for further studies. The confirmed K. pneumoniae cultures were stored in -80°C for further studies.
The genomic DNA of all K. pneumoniae isolates was extracted as previously described [11]. The confirmation of isolates to be K. pneumoniae was done by PCR as illustrated by Osman et al., [12] using the primer sequence listed in Table 1. K. pneumoniae ATCC 5150 was used as a positive control.
Table 1. The sequence of primers used in the PCR amplification.
| Target gene | Primer sequence 5՝-3՝ | Annealing Temp (ᴼC) | Product size (bp) | Reference |
|---|---|---|---|---|
| K. pneumoniae 16S–23S ITSD | ATTTGAAGAGGTTGCAAACGAT TTCACTCTGAAGTTTTCTTGTGTTC | 57 | 130 | [12] |
| ESBL-genes | ||||
| TEM | GATCTCAACAGCGGTAAG CAGTGAGGCACCTATCTC | 50 | 786 | [11] |
| SHV | ACTATCGCCAGCAGGATC CAGTGAGGCACCTATCTC | 53 | 356 | [11] |
| CTX-M15 | GTGATACCACTTCACCTC AGTAAGTGACCAGAATCAG | 49 | 225 | [11] |
| OXA-1-like | GGCACCAGATTCAACTTTCAAG GACCCCAAGTTTCCTGTAAGTG | 60 | 564 | [13] |
| GES | AGTCGGCTAGACCGGAAAG TTTGTCCGTGCTCAGGAT | 60 | 399 | [13] |
| PER | GCTCCGATAATGAAAGCGT TTCGGCTTGACTCGGCTGA | 60 | 520 | [13] |
| VEB | CATTTCCCGATGCAAAGCGT CGAAGTTTCTTTGGACTCTG | 60 | 648 | [13] |
| AmpC-genes | ||||
| AmpC | ACACGAGTTTGCATCGCCTG CTGAACTTACCGCTAAACAGTGGAAT | 60 | 245 | [14] |
| CIT-M | TGGCCAGAACTGACAGGCAAA TTCTCCTGAACGTGGCTGGC | 60 | 462 | [14] |
| FOX-1 | GCAAACCAGCAATACCATCCA GCTCACCTTGTCATCCAGCTC | 60 | 642 | [14] |
| ACC-1 | AGCTGTTATCCGTGATTACCTGTCT AGCGAACCCACTTCAAATAACG | 60 | 248 | [14] |
| ACT-1 | CATGCTGGATCTGGCAACCT CTTCAGCGTCCAGCATTCCT | 60 | 343 | [14] |
| MOX | GCAACAACGACAATCCATCCT GGGATAGGCGTAACTCTCCCAA | 60 | 895 | [13] |
| Carbapenemase-genes | ||||
| OXA-48 | GCTTGATCGCCCTCGATT GATTTGCTCCGTGGCCGAAA | 57 | 281 | [13] |
| NDM | GGTTTGGCGATCTGGTTTTC CGGAATGGCTCATCACGATC | 52 | 621 | [15] |
| IMP | TTGACACTCCATTTACDG GATYGAGAATTAAGCCACYCT | 55 | 139 | [13] |
| VIM | GATGGTGTTTGGTCGCATA CGAATGCGCAGCACCAG | 55 | 390 | [13] |
| KPC | CATTCAAGGGCTTTCTTGCTGC ACGACGGCATAGTCATTTGC | 55 | 538 | [13] |
| Efflux pump genes | ||||
| AcrA | GTGCCCAACAGTTTCTGATAACG GATGCTCTCAGGCAGCTTAGC | 50 | 150 | [16] |
| AcrB | TGAAAGATGCCATCAGCCGT ATTTTCACGAACGGCGTGGT | 50 | 500 | [16] |
| TOlC | AGAGTTTGATCMTGGCTCAG ACGAGCTGACGACARCCATG | 48 | 700 | [16] |
| Integrons | ||||
| Integron1 | GGTCAAGGATCTGGATTTCG ACATGCGTGTAAATCATCGTC | 60 | 436 | [17] |
| Integron II | CACGGATATGCGACAAAAAGG TGTAGCAAACGAGTGACGAAATG | 60 | 788 | [17] |
| Integron III | AGTGGGTGGCGAATGAGTG TGTTCTTGTATCGGCAGGTG | 60 | 600 | [17] |
Serotyping
The tested isolates were serologically examined for the presence of capsular antigens K1 and K2 by Quelling test [18]. The antigen-antibody reactions were observed microscopically.
String test
For the detection of mucoviscosity of the isolates, string test was performed as described previously [19]. The isolates were classified as hypermucoviscous K. pneumoniae (HVKP) isolates give positive results or classic K. pneumoniae (CKP) isolates give negative results.
Determination of antibiogram
Antibiotic sensitivity testing against β-lactams antibiotics [Aztreonam (30), Ceftazidime (30), Cefotaxime (30), Cefoxitin (30), Ceftriaxone (30), Cefepime (30), Ertapenem (10), Imipenem(10), Meropenem (10) and amoxicillin-clavulanic acid (30)] was performed via the disc diffusion method as outlined by CLSI [20]. The MAR index (Multiple Antibiotic Resistance index) was calculated by dividing the number of antibiotics to which the specific isolate is drug-resistant by the total number of multiple antibiotics to which the specific isolate has been exposed [21].
Phenotypic detection of β–lactamases enzymes
Phenotypic detection of ESBLs in K. pneumoniae isolates was performed by a double-disc synergy test (DDST) according to CLSI guidelines [20]. For AmpC β–lactamases activity, the test was conducted as reported by Barwa et al. [14]. The isolates showing distortion of the cefoxitin inhibition zone were considered as AmpC producers, while isolates with no distortion of the inhibition zone were identified as AmpC non-producers. Modified Hodges test (MHT) was used to test for carbapenemases enzymes according to CLSI [20].
Molecular characterization of encoding genes
Eighteen uniplex PCR amplifications were done to detect the presence of different β–lactamases genes (TEM, SHV, CTX-M15, OXA-1-like, OXA-48, GES, PER, VEB, AmpC, CIT-M, FOX-1, ACC-1, ACT-1, MOX and NDM) [11, 13–15] beside efflux pump genes (AcrA, AcrB and TOlC) [16].
In addition, two multiplex PCR tests were performed. The first one was used to detect metallo-β–lactamases genes (IMP, VIM, and KPC) [13]. The second one detected the integron genes I, II, and III [17]. Table 1 showed the primer sequence, annealing temperature, and the product size of the used primers. The amplified PCR products were analyzed on 1.2% agarose gel stained with ethidium bromide, compared with 100 bp plus DNA ladder (Thermofisher Scientific) and scanned in gel documentation and analysis system (Model Gel Doc 1.4, 1189; AccuLab, USA).
ERIC-PCR
Genomic DNA of each isolate was subjected to ERIC-PCR using the primers ERIC1 (5’-ATGTAAGCTCCTGGGGATTCAC-3’) and ERIC2 (5’-AAGTAAGTGACTGGGGTGAGCG-3’) and the PCR condition was followed as described elsewhere [1]. PCR products were analyzed by 2% agarose gel electrophoresis system. A similarity matrix was measured using Dice’s coefficient and the resultant dendrogram was made via the unweighted-pair group method with arithmetic averages (UPGMA).
Statistical analysis
Graph-pad instate was used to analyze the results statistically. Fisher’s exact and Chi-square tests were used and P-value ≤ 0.05 was considered statistically significant.
Results
Biochemical and molecular identification of K. pneumoniae
In this study, 350 ready to eat processed meat specimens (luncheon meat) were collected, forty-four K. pneumoniae isolates were isolated, purified, and biochemically identified in addition to identification using PCR by amplification of K. pneumoniae 16S–23S ITSD with an incidence of 12.6%. Three different biotypes profiles were found among isolates using biochemical activities (S2 Table). The profile B1 was the predominant one, as it was found in 32 isolates (72.7%) with a P-value < 0.0001. The other profiles B3 and B4 were found in seven (15.9%) and five (11.3%) isolates, respectively. No isolates were classified as biotype B2 or B5.
Phenotypic detection of capsular serotypes and hypermucoviscosity
K. pneumoniae serotypes was investigated using Quelling test. Two capsular types (K1 and K2) were identified among the tested isolates. Both serotypes account for 95.5% of isolates. 37 isolates (84%) were K1 with P-value < 0.0001 and only 5 isolates (11.4%) were K2. Besides that, two isolates (4.6%) were untypable (non K1/K2 serotype).
Hypermucoviscosity of isolates was measured using string test. It was found that 26 of the isolates were HVKP (P = 0.0109) while 18 isolates were CKP. The association of capsular serotypes and the hypermucoviscosity of isolates was investigated. It was found that K1 serotype was significantly associated with HVKP (P = 0.0364).
Determination of antibiogram
The antimicrobial sensitivity test of K. pneumoniae isolates was measured using the disc diffusion method. The results showed that 42/44 (95.5%) isolates were resistant to cefoxitin and ceftazidim. In addition, 41(93.2%), 38(86.4%), and 40(90.9%) isolates were resistant to cefotaxime, amoxicillin-clavulanic acid, and ertapenem respectively. Moderate resistance level to aztreonam (63.6%), cefepime (43.2%), imipenem and meropenem (36.4%), and ceftriaxone (27.3%) was recorded. All isolates which showed resistance to imipenem and meropenem; were also resistant to ertapenem. Table 2 showed the antibiotic resistance pattern of the tested isolates. A5, A11, and A12 were the major antibiotypes being represented by 8, 6, and 7 isolates, respectively. Most of the isolates (93.2%) give MAR index of ≥ 0.5.
Table 2. Antibiotic resistance patterns of the tested K. pneumoniae isolates.
| Antibiotype | No of antibiotics | MAR index | No. of isolates/pattern | Resistance pattern |
|---|---|---|---|---|
| A1 | 10 | 1 | 2 | FOX, CAZ, CTX, CRO, FEP, AMC, ATM, ERT, IPM, MEM. |
| A2 | 9 | 0.9 | 1 | FOX, CAZ, CTX, CRO, FEP, ATM, ERT, IPM, MEM. |
| A3 | 3 | FOX, CAZ, CTX, FEP, AMC, ATM, ERT, IPM, MEM. | ||
| A4 | 8 | 0.8 | 3 | FOX, CAZ, CTX, AMC, ATM, ERT, IPM, MEM. |
| A5 | 8 | FOX, CAZ, CTX, CRO, FEP, AMC, ATM, ERT. | ||
| A6 | 7 | 0.7 | 1 | FOX, CAZ, CTX, CRO, FEP, ATM, ERT. |
| A7 | 2 | FOX, CAZ, CTX, FEP, AMC, ATM, ERT. | ||
| A8 | 4 | FOX, CAZ, CTX, AMC, ERT, IPM, MEM. | ||
| A9 | 6 | 0.6 | 1 | FOX, CAZ, CTX, FEP, ATM, MEM. |
| A10 | 3 | FOX, CAZ, CTX, AMC, IPM, MEM. | ||
| A11 | 6 | FOX, CAZ, CTX, AMC, ATM, ERT. | ||
| A12 | 5 | 0.5 | 7 | FOX, CAZ, CTX, CRO, AMC, ERT. |
| A13 | 2 | 0.2 | 1 | FOX, ERT. |
| A14 | 1 | 0.1 | 1 | CAZ. |
| A15 | 1 | ERT. |
FOX: Cefoxitin; CAZ: ceftazidim; CTX: cefotaxime; CRO: ceftriaxone; FEP: cefepime; AMC: amoxicillin-clavulanic acid; ATM: aztreonam; ERT: ertapenem; IPM: imipenem; MEM: meropenem, MAR: multiple antibiotic resistance.
Phenotypic detection of β-lactamases enzymes
In this study, the isolates were investigated for the production of ESBL, AmpC and carbapenemase enzymes phenotypically. The results revealed that 22, 21, and 16 isolates were ESBL-producers, AmpC-producers, and carbapenemase-producers, respectively. One isolate (4.8%) was classified as an ESBL and AmpC co-producer. Nine isolates were AmpC- and carbapenemase co-producers. Seven isolates were ESBL and carbapenemase co-producers. Two isolates were not classified as any of the aforementioned classes.
The correlation between the different β-lactamse classes and biotyping was studied. Biotype B1 was significantly present in ESBL-producers (P = 0.0455) and carbapenemase-producers (P<0.0001) while B2 was detected significantly in AmpC-producers (P = 0.0075). The serotype K1 was present in all classes, significantly (P<0.0001).
Molecular characterization of resistance encoding genes and integrons
Examination of resistance encoding genes of the 44 isolates using PCR revealed that ESBLs genes, SHV, TEM, CTX-M15, GES, VEB, and OXA-1-like were harbored by 28, 23, 22, 17, eight, and three isolates, respectively. PER gene wasn’t harbored by any of the tested isolates. For AmpC genes, AmpC, CIT-M, FOX-1, and ACT-1 were harbored by 21, 20, 11, and five isolates, respectively. ACC-1 and MOX genes were not be detected in the tested isolates. Carbapenemases genes VIM, NDM, OXA-48, IMP, and KPC were detected in 18, 12, 10, 9, and five isolates, respectively. Therefore, 41, 35, and 31 isolates could be classified as ESBL, AmpC, and carbapenemase producers, respectively, as they harbored the genes specific for each class. For ESBL genes, the most predominant were SHV, TEM, and CTX-M15 as they were harbored by 68.3%, 56.1%, and 53.7%, of isolates, respectively. For AmpC genes, AmpC, CIT-M were the most abundant genes being carried by 60% and 57.1%, respectively. Regarding Carbapenemases genes, VIM and KPC were the highest (58.1%) and the lowest (16.1%) genes detected, respectively. Among phenotypically classified ESBL-producers, detection of ESBL genes revealed that the most prominent genes were SHV (72.7%), TEM (59.1%), and CTX-M15 (54.5%). In addition, they carried AmpC and carbapenemases genes (AmpC, CIT-M, FOX-1, IMP, VIM, KPC, NDM, and OXA-48 in 8, 11, four, five, 11, four, five, and five isolates, respectively). Both VEB and KPC were significantly associated with ESBL-producers (P<0.0001 and P = 0.0143, respectively). Besides, it was found that TEM was co-detected significantly with SHV (P = 0.002).
Regarding phenotypically identified AmpC producers, AmpC genes (AmpC, CIT-M, FOX-1, and ACT-1) were detected in 61.9%, 47.6%, 38.1%, and 23.8%, respectively. The present study found that ACT-1 and FOX-1 genes were significantly presented in AmpC-producers (P<0.0016 and P = 0.0143, respectively). The other ESBL genes (TEM, SHV, CTX-M15, OXA-1-Like, and GES) were present in 11, 13, 10, one and 8 isolates, respectively, while carbapenemase genes (IMP, VIM, KPC, NDM, and OXA-48) were harbored by four, 8, one, 7, and five isolates, respectively. Moreover, AmpC gene was significantly associated with CIT-M (P = 0.039). Concerning the presence of carbapenemase genes among phenotypic carbapenemase-producers, VIM (62.5%), NDM (56.3%), and IMP (37.5%) were the most prevalent genes. ESBL genes (TEM, SHV, CTX-M15 GES, and VEB) were detected in 9, 10, 7, 6, and two of the tested isolates, respectively. On the other hand, AmpC genes (AmpC, CIT-M, Fox-1, and ACT-1) were found in 12, 8, three, and two isolates respectively. IMP gene was associated with VIM gene (P = 0.0324) and KPC was associated with VIM and NDM (P = 0.0455). In addition, VIM gene was correlated with HVKP (P = 0.0231) while OXA-1-Like and ACT-1 genes were correlated with CKP isolates significantly.
The co-detection of β-lactam resistance genes in the tested isolates was shown in Table 3. For example, TEM was significantly associated with GES, AmpC, FOX-1, ACT-1, and IMP. CIT-M was co-detected significantly with FOX-1 and VIM. Besides, IMP was significantly associated with VIM and CTX-M15.
Table 3. Co-detection rates of β-lactam resistance encoding genes amongst 44 K. pneumoniae isolates.
| Observed frequency (%) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Resistance encoding genes | ||||||||||||||
| TEM | SHV | CTX-M15 | OXA-1-like | GES | VEB | AmpC | CIT-M | FOX-1 | ACT-1 | OXA-48 | NDM | IMP | VIM | |
|
TEM |
||||||||||||||
| SHV | 61 | |||||||||||||
| CTX-M15 | 43 | 46 | ||||||||||||
| OXA-1-like | 9 | 7 | 5 | |||||||||||
| GES | 39*** | 39 | 50** | 33 | ||||||||||
| VEB | 17 | 18 | 22 | 33* | 12 | |||||||||
| AmpC | 39** | 32*** | 55 | 67* | 47 | 50*** | ||||||||
| CIT-M | 52 | 50 | 55*** | 67** | 53 | 50 | 52 | |||||||
| FOX-1 | 35* | 27 | 32 | 33 | 29 | 38 | 29 | 40*** | ||||||
| ACT-1 | 17* | 17* | 9 | 33*** | 6 | 0*** | 14 | 10 | 0*** | |||||
| OXA-48 | 90 | 18* | 22 | 33 | 30 | 20 | 50 | 60* | 40* | 10 | ||||
| NDM | 22 | 29 | 23 | 0*** | 41** | 0*** | 33 | 25 | 25 | 20 | 30 | |||
| IMP | 35*** | 27 | 23 | 33 | 12** | 13* | 29 | 30 | 25 | 0*** | 40*** | 25 | ||
| VIM | 43 | 46 | 22*** | 33 | 47 | 38 | 48 | 60*** | 67*** | 20*** | 50 | 50 | 70*** | |
| KPC | 9 | 7 | 14* | 0** | 24*** | 13 | 14* | 10 | 8 | 0** | 10 | 17* | 10 | 75*** |
***P, 0.0001
**P, 0.001
*P, 0.05, Numbers in Bold are statistically significant associations.
Investigation of the gene profile of the tested isolates revealed that there were 42 different gene profiles among the 44 isolates as illustrated in Table 4. There was only one profile (Pr1) carried by two isolates while each of the other profiles was represented by one isolate. Two isolates harbored nine genes, 10 isolates carried four genes and five and six genes were carried by eight and six isolates, respectively. Resistance genes profiles were unique as each profile was represented by only one isolate.
Table 4. Relationship between ERIC typing and β-lactam resistance encoding genes profiles.
| ERIC pattern | No. of genes | β-lactam resistance encoding genes profile | β-lactam resistance encoding genes | No. of total isolates | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TEM | SHV | CTX-M15 | OXA-1-like | GES | VEB | AmpC | CIT-M | FOX-1 | ACT-1 | OXA-48 | NDM | IMP | VIM | KPC | ||||
| 1 | 1 | Pr1 | - | - | - | - | - | - | + | - | - | - | - | - | - | - | - | 2 |
| 2 | ||||||||||||||||||
| 6 | 2 | Pr2 | - | + | - | - | - | - | - | - | + | - | - | - | - | - | - | 1 |
| 13 | Pr3 | - | - | + | - | - | - | + | - | - | - | - | - | - | - | - | 1 | |
| 5 | 3 | Pr4 | - | + | + | - | - | - | - | - | - | + | - | - | - | - | - | 1 |
| 14 | Pr5 | - | - | - | - | - | + | + | - | - | - | - | - | - | + | - | 1 | |
| 28 | Pr6 | + | - | - | - | - | + | - | - | - | - | - | - | - | + | - | 1 | |
| 24 | Pr7 | - | + | - | - | - | - | + | - | - | - | - | + | - | - | - | 1 | |
| 34 | Pr8 | + | - | - | - | - | - | - | - | - | - | + | + | - | - | - | 1 | |
| 1 | 4 | Pr9 | + | - | - | - | + | - | - | - | + | - | - | - | - | + | - | 1 |
| 7 | Pr10 | - | + | + | - | + | - | + | - | - | - | - | - | - | - | - | 1 | |
| 9 | Pr11 | + | - | + | - | - | - | + | - | - | - | - | - | + | - | - | 1 | |
| 11 | Pr12 | - | + | + | - | + | - | + | - | - | - | - | - | - | - | 1 | ||
| 16 | Pr13 | - | + | + | - | + | - | - | - | - | - | - | - | - | + | - | 1 | |
| 18 | Pr14 | + | + | + | - | - | - | - | + | - | - | - | - | - | - | - | 1 | |
| 19 | Pr15 | - | + | + | - | - | + | - | + | - | - | - | - | - | - | - | 1 | |
| 27 | Pr16 | + | + | - | - | - | + | - | + | - | - | - | - | - | - | - | 1 | |
| 33 | Pr17 | + | + | + | - | - | - | - | - | - | + | - | - | - | - | - | 1 | |
| untypable | Pr18 | + | - | + | - | - | - | - | - | + | - | + | - | - | - | - | 1 | |
| 3 | 5 | Pr19 | + | + | + | - | - | - | - | + | - | - | - | - | + | - | - | 1 |
| 13 | Pr20 | - | - | + | - | + | - | + | + | - | - | - | + | - | - | - | 1 | |
| 14 | Pr21 | + | + | - | - | + | + | - | - | - | - | + | - | - | - | - | 1 | |
| 15 | Pr22 | + | + | - | + | - | - | - | - | - | - | - | - | + | + | - | 1 | |
| 21 | Pr23 | + | + | - | - | - | - | + | - | - | + | - | + | - | - | - | 1 | |
| 26 | Pr24 | + | + | + | - | + | - | - | - | - | - | - | + | - | - | - | 1 | |
| 29 | Pr25 | - | + | - | - | + | - | - | - | - | - | - | + | - | + | + | 1 | |
| 31 | Pr26 | - | + | - | - | - | - | + | - | - | - | - | + | + | + | - | 1 | |
| 1 | 6 | Pr27 | + | + | - | - | + | - | - | + | + | - | - | - | - | + | - | 1 |
| 4 | Pr28 | + | + | - | - | - | - | - | + | - | - | + | - | + | + | - | 1 | |
| 8 | Pr29 | - | + | + | - | + | - | + | + | - | - | - | - | - | + | - | 1 | |
| 13 | Pr30 | + | + | - | - | + | - | - | - | - | - | + | + | + | - | - | 1 | |
| 25 | Pr31 | + | + | - | - | - | - | + | + | - | + | - | - | - | + | - | 1 | |
| 22 | Pr32 | + | + | - | - | - | - | - | + | + | - | - | - | + | + | - | 1 | |
| 1 | 7 | Pr33 | - | + | + | + | - | + | + | + | + | - | - | - | - | - | - | 1 |
| 2 | Pr34 | + | - | + | - | + | + | + | - | + | - | - | - | - | - | + | 1 | |
| 17 | Pr35 | - | + | + | - | + | - | - | + | + | - | - | + | - | + | - | 1 | |
| 30 | Pr36 | + | - | - | + | + | - | + | + | - | + | + | - | - | - | - | 1 | |
| 32 | Pr37 | - | + | - | - | - | - | + | + | + | - | + | + | - | + | - | 1 | |
| 10 | 8 | Pr38 | + | - | + | - | - | - | + | + | + | - | + | - | + | + | - | 1 |
| 12 | Pr39 | + | - | + | - | + | - | + | + | + | - | - | + | - | + | - | 1 | |
| 20 | Pr40 | - | - | + | - | + | - | + | + | - | - | - | + | + | + | + | 1 | |
| 3 | 9 |
Pr41 | + | + | + | - | + | - | + | + | - | - | + | - | - | + | + | 1 |
| 23 | Pr42 | + | - | + | - | - | + | + | + | + | - | + | - | + | + | - | 1 | |
Pr: profile.
For efflux pump genes, AcrA, AcrB and TOlC were amplified in 42 (95.5%), 28(63.6%) and 37(84.1%) isolates, respectively. Twenty-four isolates harbored the three genes. Six of them were ESBL-producers, four isolates were ESBL and carbapenemase-coproducers, five were AmpC-producers and eight were AmpC and carbapenemase-coproducers. Isolate No. 39 (ESBL and AmpC-coproducer) harbored the three efflux genes.
Investigation of the distribution of integrons between isolates revealed that integrons (I, II, III) were detected in 20/44 isolates. Eleven, two, and four isolates were found to carry Int I, Int II, and Int III, respectively. Two isolates harbored both Int I and Int III and one isolate carried Int II and Int III. ESBL-producers were shown to be significantly associated with Int I (P = 0.033). In AmpC-producers, only 9/21 of isolates harbored Int I (three isolates), Int III (three isolates), each of Int II, Int I+Int III, and Int II+Int III was carried by one isolate. Six isolates in carbapenemase-producers harbored Int I (two isolates), Int III (three isolates), and Int II+ Int III (one isolate). Int I and Int III were present significantly in K1 serotyped isolates (P = 0.02 and 0.07, respectively). It was found that isolates carrying integrons were resistant to 5 or more antimicrobials (P< 0.0001), harbored 5 or more of resistance genes (P = 0.0256), and carried the complete efflux system (P = 0.0038).
ERIC
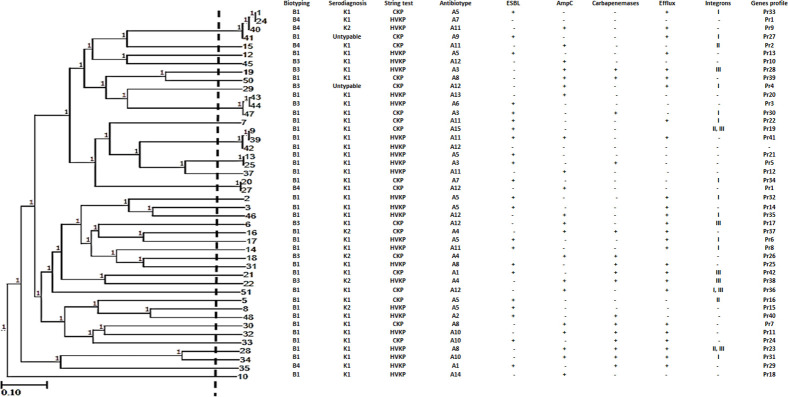
ERIC molecular typing gave high diversity among the tested isolates. As seen in Fig 1, ERIC-PCR classified the isolates into 34 different patterns. Pattern 1 included four isolates, patterns 3 and 13 each enclosed three isolates, and patterns 2 and 14 each contained two isolates while the other patterns, each one represented by one isolate. The number of bands varied between one and 12 bands, which ranged between 100 and >1500 bp. Distribution of ERIC patterns among isolates harboring β-lactam resistance encoding genes was illustrated in Table 3 where 42 different resistance gene profiles were distributed among 34 ERIC patterns that represent all tested K. pneumoniae isolates, where ERIC pattern 1 and 2 have the same gene profile (Pr1).
Fig 1. Genotyping of K. pneumoniae isolates using ERIC-PCR method.
The dendrogram was constructed using ERIC-PCR patterns of K. pneumoniae isolates. Similarity clustering analysis was performed using the UPGMA and Dice coefficient. The dashed line is a hypothetical line showing 85% similarity. Biotyping, serodiagnosis, hypermucoviscosity, ESBL-producers, AmpC-producers, Carbapenemase-producers, efflux system, integrons, and genes profile among K. pneumoniae isolates were reported. B: biotype, CKP: classical K. pneumoniae, HVKP: hypermucoviscous K. pneumoniae, U: untypable, Pr: profile, ERIC-PCR, enterobacterial repetitive intergenic consensus-PCR. UPGMA, unweighted-pair group method with arithmetic averages.
Discussion
K. pneumoniae is an opportunistic pathogen that is found not only in clinical specimens but also in food. Foodborne pathogens are widely distributed but research about K. pneumoniae as a food pathogen is infrequent. Recently, K. pneumoniae is the main cause of foodborne outbreaks in different countries [1]. In this study, K. pneumoniae isolates were isolated from ready to eat processed meat (Luncheon) with an incidence of 12.6% which indicates that contaminated food with K. pneumoniae is common in Egypt. Similar results were reported in china where the incidence of K. pneumoniae in 998 food samples was 9.9% [4]. Another study conducted in Spain isolated only 9 strains (5.6%) of K. pneumoniae from 160 vegetables [22].
One of the important virulence factors of K. pneumoniae is the production of a capsule that gives it a mucoid appearance on an agar plate beside that it protects it against phagocytosis and serum bactericidal effect [1]. In addition, classifying K. pneumoniae isolates as virulent ones are associated with serotypes K1 and K2 [23, 24]. Two capsular types (K1 and K2) were identified by Quelling test. Two isolates (4.6%) were untypable (non K1/K2 serotype). Yu et al. had described that among 50 K. pneumoniae isolates, 26 (52%) were K1, 14 (28%) were K2 and 10 (20%) isolates were non K1/K2 serotype [20]. Other studies reported different prevalence rates for K1 and K2, where four capsular types (K1, K2, K20, and K54) were identified in 8 isolates only and the rest of the isolates were non K1/K2 serotypes [1]. Besides that, another study illustrated that K1 and K2 serotypes were found in 28.5% and 7.14% of the tested K. pneumoniae isolates, respectively [25].
Another virulence factor of K. pneumoniae, hypermucoviscosity, was measured using the string test. The results showed that 59% of isolates were HVKP and 41% of isolates were CKP. In contrast, Gharrah et al. [11] found that 33% of isolates were HVKP. Diverse capsular ingredients and an increased amount of capsular material have been described in hypervirulent K. pneumoniae isolates [26]. Similar to other studies that reported that HVKP isolates are associated with K1 or K2 serotype [23], the current results illustrated that K1 and K2 serotypes were found as both HVKP (59.1%; 23 and 3 isolates respectively) and CKP (36.4%; 14 and 2 isolates respectively) but significantly associated with HVKP (P = 0.0018). However, little work elucidating the role of the hypermucoviscous (HMV) phenotype in the pathogenicity of K. pneumoniae exists [11].
The extensive use of antimicrobials in the ecosystems resulted in the emergence of antimicrobial-resistant K. pneumoniae. In the current study, most of the isolates (93.2%) were resistant to cefotaxime. These isolates usually produce ESBL and show resistance to other β-lactam antibiotics [27, 28]. In addition, the studied isolates show high resistance to ceftazidim, cefoxitin amoxicillin-clavulanic acid, and ertapenem, and moderate resistance level to aztreonam, cefepime, imipenem, meropenem, and ceftriaxone. There was a wide assortment of resistance patterns among isolates, where 15 antibiotypes were found. The predominant antibiotype was A5 which includes isolates resistant to 8 antimicrobials. 93.2% of isolates were resistant to five or more antimicrobials with MAR index ≥0.5. The presence of such resistant isolates in food represents a risk factor to the food consumers as they are considered as high-risk sources of antimicrobial contamination, also, they cause infections that can’t be treated by these agents and result in an increase in the morbidity and mortality rates. The isolates that show resistance to ertapenem, meropenem, and imipenem (36.4%) are more likely to produce carbapenemase enzymes. These findings are contrary to what was reported by another study which found that 3.2% of isolates were resistant to amoxicillin-clavulanic acid, cefotaxime, and cefoxitin. Besides that, resistance to ceftriaxone was only 1.6% [1]. The high rates of antimicrobial resistance detected in this study can be attributed to the lack of strict policies that govern the use of antibiotics in Egypt.
In this study 22 isolates were ESBL-producers, 21 isolates were AmpC-producers and 16 isolates were carbapenemase-producers. Two isolates were not categorized as any of the previous classes. The percentage of K. pneumoniae isolates that produce ESBLs differs between countries. Their percent in Arabian countries is high (62.5% and 50% of isolates) as reported by Aljanaby and Alhasani [29] and Gharrah et al. [11], respectively. This is similar to the present results as 50% of isolates were categorized as ESBL-producers. This may have a significant effect on the treatment strategies using β-lactam antibiotics which increase the morbidity and mortality among food consumers. AmpC enzymes are β-lactamases that hydrolyze penicillins, cephalosporins, and cephamycins with low hydrolysis rates for cefepime, cefpirome, and carbapenems [30]. They aren’t inactivated by available β-lactamases inhibitors in contrast to ESBLs [31]. The present results showed that 47.7% of isolates were AmpC-producers with 4.8% classified as ESBL and AmpC co-producer. Barwa et al. showed that 31.6% of isolates were AmpC-producers [14], moreover, 65 K. pneumoniae isolates were ESBL-producers and 7.7% of them were AmpC-producers as reported by Zorgani et al. [32].
Among used antibiotics, carbapenem is considered the choice for the treatment of critical infections due to multidrug-resistant K. pneumoniae. However, there is an increasing incidence of carbapenem resistance through K. pneumoniae isolates in many countries due to the extensive use of the carbapenems [33]. In the present work, 38.6% of isolates were found to be carbapenemase-producers, 56.3% of them were AmpC-coproducer while the rest were ESBL-producers too. Japoni-Nejad et al. showed that 12 isolates were carbapenemases-producers and 8 of them were AmpC-producers [34]. Besides, Yazgan et al. showed that 51% of collected K. pneumoniae were ESBL-producers, 49% of them were carbapenemase-producers [8]. These results may be attributed to the excessive use of these antibiotics. There is a significant increase in carbapenem resistance K. pneumoniae isolates in Egypt as it increased from 13.9% to 44.4% [25, 35]. Several mechanisms are responsible for this type of resistance including AmpC or ESBL production with porin loss, carbapenemase production, or metallo-β-lactamase production [36].
All phenotypic methods used in this study detects isolates as positive ESBL, AmpC, or carbapenemase producer but they were unable to differentiate the different types or families of each class. Therefore, it is necessary to use other techniques such as PCR for confirmation of phenotypic results and discrimination of different genes. Examination using PCR classified 41, 35, and 31 isolates as ESBL, AmpC, and carbapenemase producers, respectively. Sutandhio et al. showed that 12 isolates harbored carbapenemase genes but only 6 isolates of them were Modified Hodges test positive [37]. Another study showed that 37.5% of K. pneumoniae isolates were ESBL or AmpC-producers phenotypically while 43% of them were ESBL only, 11% were AmpC and 36% were ESBL and AmpC genotypically [38]. Similar results were observed in other studies [14, 39]. This variation between phenotypic and PCR results may be because these genes are present but are not usually expressed [14]. Therefore, not all isolates detected as ESBL, AmpC, and carbapenemases producers by PCR can be detected by phenotypic methods.
PCR is considered the gold standard method for the detection of different classes of β-lactamase enzymes [40]. The PCR amplification of β-lactam resistance genes was performed on all isolates. For ESBL genes, the most predominant were SHV, TEM, and CTX-M15 as harbored by 63.6%, 52.3%, and 50%, of isolates, respectively. Similarly, Hasani et al. illustrated that SHV gene was the predominant gene (80.9%) in the isolates followed by CTX-M and TEM (73% and 58%, respectively) [41]. For AmpC genes, AmpC, CIT-M were the most abundant genes as were carried by 47.7% and 45.5%, respectively. In contrast, Ghonaim and Moaety showed that CIT and MOX genes were present in 18.9% and 6.1%, respectively, and ACC, FOX, and ACT genes were not detected [42]. In Carbapenemases genes, VIM and KPC were the highest (43.2%) and the lowest (9.1%) genes detected respectively. In contrary to other studies that reported that OXA-48, NDM, and VIM were the predominant genes in this order [26]. ESBL and AmpC-coproducer (isolate No. 39) harbors 9 genes (Pr41). For efflux pump genes, AcrA, AcrB and TOlC were amplified in 95.5%, 63.6% and 84.1% of isolates, respectively. The coexistence of several genes of ESBL, AmpC, and carbapenemases in the same isolate revealed serious epidemiological, clinical and public health threats.
The association between β-lactam resistance encoding genes was investigated amongst the 44 K. pneumoniae isolates as illustrated in Table 3 to determine if there was a non-random association between genes that may be due to genes co-location. Some combinations of β-lactam resistance encoding genes were detected significantly. In contrast to our results, Lalzampuia et al., [43] found that for eight isolates, 7 harbored CTX-M-1 gene and/or TEM gene. The CTX-M and TEM were the most common gene combinations (33.33%) in E. coli isolates from broiler farms in the Philippines [44]. Benmahmod et al. reported that one isolate had both IMP and NDM, and one isolate co-carried IMP and VIM [45].
The investigation of the β-lactam resistance genes profile revealed that 42 different profiles were distributed among 44 K. pneumoniae isolates. The gene profile of isolates showed that there is a great genetic diversity between isolates and that the plasmids encoding for β-lactamases can be easily spread by horizontal transfer among Enterobacteriaceae including K. pneumoniae [33].
Efflux pump systems in K. pneumoniae include AcrAB and mdtK systems, that belong to (RND) and (MATE) family efflux pumps, respectively. The AcrAB-TolC pump is composed of an outer-membrane channel (TolC), a secondary transporter located in the inner membrane (AcrB), and a periplasmic component (AcrA) [25, 46]. In this study, the Efflux pump system consisting of AcrAB-ToƖC was determined by PCR. Twenty-four isolates (54.5%) harbored the three genes. On the other hand, 20 isolates were missing either the AcrA or AcrB efflux pump or the ToƖC outer membrane protein. Other studies showed that 82.14% of isolates harbored AcrAB while only 5 isolates showed an incomplete AcrAB efflux system [25]. In addition, another study showed that 100% of isolates carried AcrAB while 96% carried ToƖC genes [23]. Multidrug efflux system (AcrAB- ToƖC) is responsible for the resistance of K. pneumoniae to β-lactam, tetracyclines, and quinolones [23].
Investigation of integrons distribution among isolates revealed that Integrons (I, II, III) were detected in 45.5% of isolates, of which 55%, 10%, and 20% of isolates carried Int I, Int II, and Int III, respectively. Both Int I and Int III were co-founded in 20% of isolates and 10% of isolate co-carried Int II and Int III. A study conducted by Sedighi et al. showed that Int I was present in 8% of isolates while Int II and Int III were absent from all isolates [47]. Rezai et al. reported that Int I was detected in 79.3% while Int II was harbored by 10.3% of isolates[48]. ESBL-producers were significantly associated with Int I (P = 0.033), similar to what reported by Elsherif et al., as Int 1 was associated with CTX-M gene (P = 0.039) [49]. In contrary to what reported about the association of Int III with GES gene [49], current results showed that they coexist only in one isolate out of seven Int III positive isolates. Additionally, IMP gene coexisted with Int III in 42.9% of isolates which was unlike the study conducted by Elsherif et al. who reported that Int III is of low occurrence with IMP gene [49].
In this study, the characterization of NDM-encoding isolates was examined where NDM can break down all the β-lactam antibiotics except aztreonam. This gene is located on a transmissible plasmid and is associated with other resistant genes. This may result in a broad drug resistance, which makes the treatment options limited [33]. Herein, these isolates were significantly classified as K1 (P = 0.0011) and B1 (P<0.0001). Half of them were classified as HVKP. Only four isolates harbored integrons, three of them carry Int I and one isolate carry IntII+IntIII. Nine out of the 12 isolates were confirmed as carbapenemase-producers by phenotypic and PCR methods, while three isolates were identified only by PCR. This may be attributed to one of the Modified Hodges test drawbacks in that it gives false-negative results for NDM producers [50]. NDM gene was present as sole carbapenemase gene in four (33%) isolates, significantly associated with VIM gene in six isolates (P<0.0001), with IMP gene in three isolates (25%), with OXA-48 in four isolates (33%) and with KPC in two isolates (16.7%)(P = 0.0249), in addition, it was significantly associated with ESBL genes (GES and OXA-1-like) with P = 0.0011 and <0.0001, respectively. Khalil et al. also reported the association between NDM and other carbapenemase genes as KPC, IMP, VIM, and OXA-48 where they were detected in NDM-1 producing K. pneumoniae with the frequency of 80%, 30%, 50%, and 30% respectively [33]. Eight isolates harbored the three efflux genes (66.7%) with P = 0.0214. Although NDM is a broad spectrum carbapenemase that can inactivate β-lactam except for aztreonam [25], four isolates in this study showed resistance to aztreonam and harbored NDM indicating that a new antimicrobial resistance pattern in Egyptian isolates may exist.
Recently, ERIC-PCR is one of the most potent tools used for the analysis of genetic relatedness in K. pneumoniae [33]. ERIC molecular typing results gave high diversity among the tested isolates as it classified the isolates into 34 different patterns. The association between phenotypic and molecular typing gave no clear relation between isolates except in the case of isolates No. 47 and 41among ESBL-producers and isolates No. 22 and 19 among carbapenemase-producers. The relation between ERIC patterns and β-lactam resistance encoding genes profiles was studied where 42 different resistance gene profiles were distributed among 34 ERIC patterns that represent all tested K. pneumoniae isolates. Similar results were reported by Kazemian et al. [51] Our results indicated the significance and convenience of ERIC-PCR and β-lactam resistance encoding genes profiles in differentiating isolates based on genetic relatedness. The diversity in our isolates represented a problem in the treatment of K. pneumoniae infections. Similar genetic diversity represented by ERIC was described in previous studies [9, 33].
In conclusion, the present study indicated that food (ready to eat processed meat; Luncheon) is a good reservoir of resistant K. pneumoniae isolates. This represents a public health problem and good control of the emergence and the transmission of these isolates is needed. This can be done by developing more prevention strategies on making and selling this food in supermarkets.
Supporting information
(DOCX)
(DOCX)
Acknowledgments
Author is grateful to Dr. Youssif M. Ali (School of Biological Science, University of Cambridge, United Kingdom) who kindly copyedited the manuscript for language usage, spelling, and grammar. This study was performed at the Microbiology and Immunology Department, Faculty of Pharmacy, Mansoura University, Mansoura, Egypt.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Zhang S, Yang G, Ye Q, Wu Q, Zhang J, Huang Y. Phenotypic and Genotypic Characterization of Klebsiella pneumoniae Isolated From Retail Foods in China. Front Microbiol. 2018;9:289 Epub 2018/03/17. 10.3389/fmicb.2018.00289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakkas H, Bozidis P, Ilia A, Mpekoulis G, Papadopoulou C. Antimicrobial Resistance in Bacterial Pathogens and Detection of Carbapenemases in Klebsiella pneumoniae Isolates from Hospital Wastewater. Antibiotics (Basel). 2019;8(3). Epub 2019/06/30. 10.3390/antibiotics8030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almeida MVA, Cangussu IM, Carvalho ALS, Brito ILP, Costa RA. Drug resistance, AmpC-beta-lactamase and extended-spectrum beta-lactamase-producing Enterobacteriaceae isolated from fish and shrimp. Rev Inst Med Trop Sao Paulo. 2017;59:e70 Epub 2017/11/09. 10.1590/S1678-9946201759070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo Y, Zhou H, Qin L, Pang Z, Qin T, Ren H, et al. Frequency, Antimicrobial Resistance and Genetic Diversity of Klebsiella pneumoniae in Food Samples. PLoS One. 2016;11(4):e0153561 Epub 2016/04/15. 10.1371/journal.pone.0153561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munoz MA, Welcome FL, Schukken YH, Zadoks RN. Molecular epidemiology of two Klebsiella pneumoniae mastitis outbreaks on a dairy farm in New York State. J Clin Microbiol. 2007;45(12):3964–71. Epub 2007/10/12. 10.1128/JCM.00795-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato S, Aoyama T, Uejima Y, Furuichi M, Suganuma E, Takano T, et al. Pyogenic liver abscess due to hypervirulent Klebsiella pneumoniae in a 14-year-old boy. J Infect Chemother. 2019;25(2):137–40. Epub 2018/08/06. 10.1016/j.jiac.2018.07.006 . [DOI] [PubMed] [Google Scholar]
- 7.Chavez MV, Caicedo LD, Castillo JE. Occurrence of beta-Lactamase-Producing Gram-Negative Bacterial Isolates in Water Sources in Cali City, Colombia. Int J Microbiol. 2019;2019:1375060 Epub 2019/10/05. 10.1155/2019/1375060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yazgan B, Turkel I, Guckan R, Kilinc K, Yildirim T. Comparison of biofilm formation and efflux pumps in ESBL and carbapenemase producing Klebsiella pneumoniae. J Infect Dev Ctries. 2018;12(3):156–63. Epub 2018/03/31. 10.3855/jidc.9677 . [DOI] [PubMed] [Google Scholar]
- 9.Mehr VP, Shokoohizadeh Leili, Mirzaee M, Savari M. Molecular Typing of Klebsiella pneumoniae Isolates by Enterobacterial Repetitive Intergenic Consensus (ERIC)–PCR. Infect EpidemiolMicrobiol. 2017;3(4):112–6. 10.18869/modares.iem.3.4.112 [DOI] [Google Scholar]
- 10.Krieg N, Holt J. Bergey's Manual of systemic bacteriology Baltimore, M.D.21202, USA: William and Wilkins; 1984. [Google Scholar]
- 11.Gharrah MM, El-Mahdy AM, Barwa aRF. Association between Virulence Factors and Extended Spectrum Beta-Lactamase Producing Klebsiella pneumoniae Compared to Nonproducing Isolates. Interdisciplinary Perspectives on Infectious Diseases. 2017;2017 10.1155/2017/7279830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osman KM, Hassan HM, Orabi A, Abdelhafez AS. Phenotypic, antimicrobial susceptibility profile and virulence factors of Klebsiella pneumoniae isolated from buffalo and cow mastitic milk. Pathog Glob Health. 2014;108(4):191–9. Epub 2014/06/11. 10.1179/2047773214Y.0000000141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–5. Epub 2010/01/15. 10.1093/jac/dkp498 . [DOI] [PubMed] [Google Scholar]
- 14.Barwa R, Abdelmegeed E, Galil aKAE. Occurrence and detection of AmpC β-lactamases among some clinical isolates of Enterobacteriaceae obtained from Mansoura University Hospitals, Egypt. African Journal of Microbiology Research. 2012;6(41):6924–30. [Google Scholar]
- 15.Nordmann P, Poirel L, Carrer A, Toleman MA, Walsh TR. How to detect NDM-1 producers. J Clin Microbiol. 2011;49(2):718–21. Epub 2010/12/03. 10.1128/JCM.01773-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdel-Rhman S, Naggar WE, El-Sokkary MA, Barwa aR. Phenotypic and genotypic characteristics in relation to some efflux pump systems in Escherichia coli and Klebsiella pneumonia clinical isolates. Egyptian Journal of Medical Microbiology. 2011;20(3):1–14. [Google Scholar]
- 17.Rizk DE, El-Mahdy AM. Emergence of class 1 to 3 integrons among members of Enterobacteriaceae in Egypt. Microb Pathog. 2017;112:50–6. Epub 2017/09/25. 10.1016/j.micpath.2017.09.023 . [DOI] [PubMed] [Google Scholar]
- 18.Edmondson AS, Cooke EM. The production of antisera to the Klebsiella capsular antigens. J Appl Bacteriol. 1979;46(3):579–84. Epub 1979/06/01. 10.1111/j.1365-2672.1979.tb00858.x . [DOI] [PubMed] [Google Scholar]
- 19.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–18. Epub 2013/01/11. 10.4161/viru.22718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu WL, Ko WC, Cheng KC, Lee CC, Lai CC, Chuang YC. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis. 2008;62(1):1–6. Epub 2008/05/20. 10.1016/j.diagmicrobio.2008.04.007 . [DOI] [PubMed] [Google Scholar]
- 21.Hu Y, Li F, Zheng Y, Jiao X, Guo L. Isolation, Molecular Characterization and Antibiotic Susceptibility Pattern of Vibrio parahaemolyticus from Aquatic Products in the Southern Fujian Coast, China. J Microbiol Biotechnol. 2020;30(6):856–67. Epub 2020/03/12. 10.4014/jmb.2001.01005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falomir MP, Rico H, Gozalbo D. Enterobacter and Klebsiella species isolated from fresh vegetables marketed in Valencia (Spain) and their clinically relevant resistances to chemotherapeutic agents. Foodborne Pathog Dis. 2013;10(12):1002–7. Epub 2013/08/29. 10.1089/fpd.2013.1552 . [DOI] [PubMed] [Google Scholar]
- 23.Ferreira RL, da Silva BCM, Rezende GS, Nakamura-Silva R, Pitondo-Silva A, Campanini EB, et al. High Prevalence of Multidrug-Resistant Klebsiella pneumoniae Harboring Several Virulence and beta-Lactamase Encoding Genes in a Brazilian Intensive Care Unit. Front Microbiol. 2018;9:3198 Epub 2019/02/07. 10.3389/fmicb.2018.03198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang B, Pan F, Wang C, Zhao W, Sun Y, Zhang T, et al. Molecular epidemiology of Carbapenem-resistant Klebsiella pneumoniae in a paediatric hospital in China. Int J Infect Dis. 2020;93:311–9. Epub 2020/02/19. 10.1016/j.ijid.2020.02.009 . [DOI] [PubMed] [Google Scholar]
- 25.Wasfi R, Elkhatib WF, Ashour HM. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci Rep. 2016;6:38929 Epub 2016/12/23. 10.1038/srep38929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamel NA, El-Tayeb WN, El-Ansary MR, Mansour MT, Aboshanab KM. Phenotypic screening and molecular characterization of carbapenemase-producing Gram-negative bacilli recovered from febrile neutropenic pediatric cancer patients in Egypt. PLoS One. 2018;13(8):e0202119 Epub 2018/08/30. 10.1371/journal.pone.0202119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rezai MS, Salehifar E, Rafiei A, Langaee T, Rafati M, Shafahi K, et al. Characterization of Multidrug Resistant Extended-Spectrum Beta-Lactamase-Producing Escherichia coli among Uropathogens of Pediatrics in North of Iran. Biomed Res Int. 2015;2015:309478 Epub 2015/06/13. 10.1155/2015/309478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vinueza-Burgos C, Ortega-Paredes D, Narvaez C, De Zutter L, Zurita J. Characterization of cefotaxime resistant Escherichia coli isolated from broiler farms in Ecuador. PLoS One. 2019;14(4):e0207567 Epub 2019/04/06. 10.1371/journal.pone.0207567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aljanaby AAJ, Alhasani A.H.A. Virulence factors and antibiotic susceptibility patterns of multidrug resistance Klebsiella pneumoniae isolated from different clinical infections. African Journal of Microbiology Research. 2016;10(22):829–43. [Google Scholar]
- 30.Coudron PE, Moland ES, Thomson KS. Occurrence and detection of AmpC beta-lactamases among Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a veterans medical center. J Clin Microbiol. 2000;38(5):1791–6. Epub 2000/05/02. 10.1128/JCM.38.5.1791-1796.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manchanda V, Singh NP. Occurrence and detection of AmpC beta-lactamases among Gram-negative clinical isolates using a modified three-dimensional test at Guru Tegh Bahadur Hospital, Delhi, India. J Antimicrob Chemother. 2003;51(2):415–8. Epub 2003/02/04. 10.1093/jac/dkg098 . [DOI] [PubMed] [Google Scholar]
- 32.Zorgani A, Daw H, Sufya N, Bashein A, Elahmer O, Chouchani C. Co-Occurrence of Plasmid-Mediated AmpC beta-Lactamase Activity Among Klebsiella pneumoniae and Escherichia Coli. Open Microbiol J. 2017;11:195–202. Epub 2017/11/21. 10.2174/1874285801711010195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khalil MAF, Elgaml A, El-Mowafy M. Emergence of Multidrug-Resistant New Delhi Metallo-beta-Lactamase-1-Producing Klebsiella pneumoniae in Egypt. Microb Drug Resist. 2017;23(4):480–7. Epub 2016/08/31. 10.1089/mdr.2016.0003 . [DOI] [PubMed] [Google Scholar]
- 34.Japoni-Nejad A, Ghaznavi-Rad E, van Belkum A. Characterization of Plasmid-Mediated AmpC and Carbapenemases among Iranain Nosocomial Isolates of Klebsiella pneumoniae Using Phenotyping and Genotyping Methods. Osong Public Health Res Perspect. 2014;5(6):333–8. Epub 2015/01/07. 10.1016/j.phrp.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kotb S, Lyman M, Ismail G, Abd El Fattah M, Girgis SA, Etman A, et al. Epidemiology of Carbapenem-resistant Enterobacteriaceae in Egyptian intensive care units using National Healthcare-associated Infections Surveillance Data, 2011–2017. Antimicrob Resist Infect Control. 2020;9:2 Epub 2020/01/09. 10.1186/s13756-019-0639-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Messaoudi A, Mansour W, Jaidane N, Chaouch C, Boujaafar N, Bouallegue O. Epidemiology of resistance and phenotypic characterization of carbapenem resistance mechanisms in Klebsiella pneumoniae isolates at Sahloul University Hospital-Sousse, Tunisia. Afr Health Sci. 2019;19(2):2008–20. Epub 2019/10/28. 10.4314/ahs.v19i2.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutandhio S, Budiono, Hardiono, Kuntaman, Wasito EB, Lusida aMI. Comparation of phenotypic and genotypic profile of carbapenemase producing Escherichia coli. Folia Medica Indonesiana. 2018;54(1):10–5. [Google Scholar]
- 38.Chirindze LM, Zimba TF, Sekyere JO, Govinden U, Chenia HY, Sundsfjord A, et al. Faecal colonization of E. coli and Klebsiella spp. producing extended-spectrum beta-lactamases and plasmid-mediated AmpC in Mozambican university students. BMC Infect Dis. 2018;18(1):244 Epub 2018/05/31. 10.1186/s12879-018-3154-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.AlTamimi M, AlSalamah A, AlKhulaifi M, AlAjlan H. Comparison of phenotypic and PCR methods for detection of carbapenemases production by Enterobacteriaceae. Saudi J Biol Sci. 2017;24(1):155–61. Epub 2017/01/06. 10.1016/j.sjbs.2016.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abouelfetouh A, Torky AS, Aboulmagd E. Phenotypic and genotypic characterization of carbapenem-resistant Acinetobacter baumannii isolates from Egypt. Antimicrob Resist Infect Control. 2019;8:185 Epub 2019/12/14. 10.1186/s13756-019-0611-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hasani A, Purmohammad A, Rezaee MA, Hasani A, Masoud a. Integron-Mediated Multidrug and Quinolone Resistance in Extended-Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae. Archives of Pediatric Infectious Diseases. 2016;5(2). 10.5812/pedinfect.36616 [DOI] [Google Scholar]
- 42.Ghonaim RA, Moaety aHA. Comparison between Multiplex PCR and Phenotypic Detection Methods for Identifying AmpC B-lactamases Among Clinical Isolates of Enterobacteriaceae in Zagazig University Hospitals, Egypt. Clinical Microbiology. 2018;7(3). 10.4172/2327-5073.1000313 [DOI] [Google Scholar]
- 43.Lalzampuia H, Dutta TK, Warjri I, Chandra R. PCR-Based Detection of Extended-Spectrum beta-Lactamases (bla CTX-M-1 and bla TEM) in Escherichia coli, Salmonella spp. and Klebsiella pneumoniae Isolated from Pigs in North Eastern India (Mizoram). Indian J Microbiol. 2013;53(3):291–6. Epub 2014/01/16. 10.1007/s12088-013-0378-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gundran RS, Cardenio PA, Villanueva MA, Sison FB, Benigno CC, Kreausukon K, et al. Prevalence and distribution of blaCTX-M, blaSHV, blaTEM genes in extended- spectrum beta- lactamase- producing E. coli isolates from broiler farms in the Philippines. BMC Vet Res. 2019;15(1):227 Epub 2019/07/07. 10.1186/s12917-019-1975-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benmahmod AB, Said HS, Ibrahim RH. Prevalence and Mechanisms of Carbapenem Resistance Among Acinetobacter baumannii Clinical Isolates in Egypt. Microb Drug Resist. 2019;25(4):480–8. Epub 2018/11/06. 10.1089/mdr.2018.0141 . [DOI] [PubMed] [Google Scholar]
- 46.Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, et al. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509(7501):512–5. Epub 2014/04/22. 10.1038/nature13205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sedighi M, Halajzadeh M, Ramazanzadeh R, Amirmozafari N, Heidary M, Pirouzi S. Molecular detection of beta-lactamase and integron genes in clinical strains of Klebsiella pneumoniae by multiplex polymerase chain reaction. Rev Soc Bras Med Trop. 2017;50(3):321–8. Epub 2017/07/13. 10.1590/0037-8682-0001-2017 . [DOI] [PubMed] [Google Scholar]
- 48.Sadegh RM, Alireza Rafiei, Fatemeh Ahangarkani, Masoumeh Bagheri-Nesami, Attieh Nikkhah, Khaironesa Shafahi, et al. Emergence of Extensively Drug Resistant Acinetobacter baumannii-Encoding Integrons and Extended-Spectrum Beta-Lactamase Genes Isolated from Ventilator-Associated Pneumonia Patients. Jundishapur J Microbiol 2017;10(7):e14377. [Google Scholar]
- 49.Elsherif RH, Ismail DK, El-Kholy YS, Gohar NM, Elnagdy SM, Elkraly OA. Integron-mediated multidrug resistance in extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolated from fecal specimens in Egypt. J Egypt Public Health Assoc. 2016;91(2):73–9. Epub 2016/07/28. 10.1097/01.EPX.0000483165.56114.d8 . [DOI] [PubMed] [Google Scholar]
- 50.Zhou M, Wang D, Kudinha T, Yang Q, Yu S, Xu YC. Comparative Evaluation of Four Phenotypic Methods for Detection of Class A and B Carbapenemase-Producing Enterobacteriaceae in China. J Clin Microbiol. 2018;56(8). Epub 2018/05/18. 10.1128/JCM.00395-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kazemian H, Heidari H, Ghanavati R, Ghafourian S, Yazdani F, Sadeghifard N, et al. Phenotypic and Genotypic Characterization of ESBL-, AmpC-, and Carbapenemase-Producing Klebsiella pneumoniae and Escherichia coli Isolates. Med Princ Pract. 2019;28(6):547–51. Epub 2019/04/18. 10.1159/000500311 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.