Abstract
Background
Ewing sarcoma (EwS) is a rare, aggressive solid tumor of childhood, adolescence and young adulthood associated with pathognomonic EWSR1-ETS fusion oncoproteins altering transcriptional regulation. Genome-wide association studies (GWAS) have identified 6 common germline susceptibility loci but have not investigated low-frequency inherited variants with minor allele frequencies below 5% due to limited genotyped cases of this rare tumor.
Methods
We investigated the contribution of rare and low-frequency variation to EwS susceptibility in the largest EwS genome-wide association study to date (733 EwS cases and 1,346 unaffected controls of European ancestry).
Results
We identified two low-frequency variants, rs112837127 and rs2296730, on chromosome 20 that were associated with EwS risk (OR = 0.186 and 2.038, respectively; P-value < 5×10−8) and located near previously reported common susceptibility loci. After adjusting for the most associated common variant at the locus, only rs112837127 remained a statistically significant independent signal (OR = 0.200, P-value = 5.84×10−8).
Conclusions
These findings suggest rare variation residing on common haplotypes are important contributors to EwS risk.
Impact
Motivate future targeted sequencing studies for a comprehensive evaluation of low-frequency and rare variation around common EwS susceptibility loci.
Background
Ewing sarcoma (EwS) is a rare bone or soft tissue tumor predominantly occurring in the second decade of life [1]. The specific cells of origin leading to EwS tumors are unknown, with current evidence indicating EwS likely arises from mesoderm- or neural crest-derived mesenchymal stem cells [2,3]. The overall age-adjusted incidence of EwS is 0.128 per 100,000 population with individuals of European ancestry at a 9-fold risk relative to African Americans and Asian/Pacific Islanders (0.155 in White, 0.017 in Asians/Pacific islanders, and 0.017 in African Americans) [4]. The reported disparity in EwS incidence by ancestry suggests the importance of germline susceptibility to EwS risk.
A defining feature of EwS tumors is the somatically acquired translocation between EWSR1 (22q12) and a member of the ETS transcription factor family, most commonly FLI1 (11q24) (85% of cases) [5–7]. The resulting fusion oncoprotein produces aberrant and strong transcriptional regulators that bind to GGAA microsatellites and ETS-like motifs, which are thereby converted into potent enhancers, to promote cellular transformation by deregulating key target genes in cell cycle control, migration and apoptosis pathways [7–12]. Aside from recurrent EWSR1-ETS fusions, most EwS tumors display remarkably low somatic mutation rates [1,13–16].
The presence of EwS EWSR1-ETS fusions provides a molecularly distinct phenotype for genomic characterization, despite small case sample sizes. Previous genome-wide association studies (GWAS) have identified 6 common genetic susceptibility loci associated with EwS risk (1p36.22, 6p25.1, 10q21, 15q15, 20p11.22 and 20p11.23) [17]. The number of identified susceptibility loci are notable given small samples, suggesting a homogenous phenotype as defined by the fusion oncoprotein may aid in identifying germline associations. Effect estimates for variants at these loci exhibit elevated odds ratios (OR > 1.7), which is high for cancer GWAS and striking in light of the rarity of EwS in familial cancer predisposition syndromes [18]. Most EwS susceptibility loci reside near GGAA microsatellites and may disrupt local binding of EWSR1-ETS fusion oncoproteins to these microsatellites, suggesting germline-somatic interactions could be important for EwS susceptibility. As a proof-of-concept such germline-somatic interaction has been demonstrated for the chr10 EwS susceptibility gene EGR2 [11].
Despite recent efforts to characterize the genetic architecture of EwS, thus far, no study has investigated the contribution of low-frequency variants (minor allele frequencies (MAF) < 0.05) to EwS risk. The high locus-to-case discovery ratio of previous EwS GWAS and large effect sizes of common EwS susceptibility loci led our group to revisit whether current series of EwS cases would be sufficient to detect associations between rare or low-frequency variants and EwS risk. We systematically scanned across the genome for well-imputed, low-frequency variants associated with EwS susceptibility in the largest collection of genotyped EwS cases to date (733 EwS cases and 1,346 controls) [17].
Materials and methods
Study populations
The study population for the current association analysis has been described previously [17]. In brief, EwS cases were obtained from five sources: a study published by Postel-Vinay et al. [19], the Institut Curie, the Childhood Cancer Survivor Study (CCSS), the Center for Cancer Research (CCR) at the National Cancer Institute (NCI), and the NCI Bone Disease and Injury Study [20]. Ancestry of these EwS cases was estimated using SNPWEIGHTS based on SNPs found to be suitable for inferring population structure [21]. EwS cases with less than 80% European ancestry were excluded resulting in a combined set of 733 EWS cases. A total of 1,346 principal-component-matched, cancer-free controls were selected from the NCI Prostate Lung Colorectal and Ovarian Cancer Screening trial [22], American Cancer Society Cancer Prevention Study II [23], and the Spanish Bladder Cancer Study [24] for the final analysis and included with controls previously used by Postel-Vinay et al [19]. Each study participant provided informed consent, and approval to conduct this research was granted by the Institution Review Board of Institut Curie, National Cancer Institute, as well as 26 participating institutions for CCSS.
Genotyping and quality control
For the Postel-Vinay study, DNA from tumor tissue, blood, and bone marrow was isolated using proteinase K lysis followed by phenol chloroform extraction. Genomic DNA was genotyped by 610 Quadevl arrays (Illumina). For CCSS samples, blood DNA was isolated using the Gentra PureGene Blood kit (QIAGEN) and saliva DNA was extracted using the Oragene kit (DNA Genotek). Whole genome amplification (WGA) was performed for samples without sufficient DNA. For CCSS samples, genotyping was performed at the NCI Cancer Genomics Research Laboratory (CGR) on the Infinium Human Omni5Exome array (Illumina). The remainder of NCI and Institut Curie samples were genotyped by CGR using the OmniExpress-24 v1.1 array (Illumina).
All genotyping was performed according to standard manufacturer protocols. In brief, WGA was performed on 400 ng DNA, and the amplified DNA was fragmented, precipitated, resuspended, and hybridized to the designated arrays. Single-base extension of probes using captured DNA as template was subsequently carried out with fluorophore-conjugated nucleotides. Arrays were then scanned by iScan (Illumina) and SNPs called by GenomeStudio (Illumina). Our downstream quality control included filtering out samples with abnormal heterozygosity rate, sex discordance, <95% completion rates, and unexpected relatedness (IBD > 10%).
Genotype imputation was performed in three sets: (1) the Postel-Vinay study, (2) the CCSS EwS cases and matched controls, and (3) all remaining NCI and Institut Curie samples. All samples were pre-phased using SHAPEIT [25] and imputed using IMPUTE2 [26]. The 1,000 Genomes Phase 3 was used as the reference [27] resulting in 16,367,531 SNPs. Among these SNPs, 10,216,839 were low-frequency variants with MAF < 0.05.
PCR validation of genotypes
Imputed genotypes for the three EwS-associated low-frequency or rare variants (rs78119607, rs112837127, rs2296730) were validated by allele-specific TaqMan assay (Thermo Fisher Scientific) at CGR following standard manufacturer protocols. The 325 samples used for validation were selected based on imputed genotype, study, and amount of available DNA.
Statistical analysis
For each variant, we report an estimate of the odds ratio (OR), 95% confidence interval (CI), and P-value (pMH) using a Mantel-Haenszel Test where subjects are stratified by study (e.g. CCSS, Postel-Vinay, etc.), and, when stated, the genotype at linked neighboring variant(s). Because we focused on less common variants, we used a dominant model (i.e., genotype defined as presence versus absence of rare variant) and an exact, conditional test (mantelhaen.test(exact = T)) [28,29]. We used pMH < 5 × 10−8 to define initial GWAS significance and pMH < 0.05/1684 = 1.09×10−5 for conditional tests, where 1,684 is the number of SNPs with MAF < 0.05 and R2 > 0.004 with one of 6 previously identified SNPs. Potential interaction between low frequency SNPs and common SNPs were examined by logistic regression models with case-control status as outcome, low frequency and common SNPs as well as an interaction term between them as predictors. All statistical tests were two-sided and performed in R v.3.6.2 [28]. We did not investigate associations with significant variants and clinical data as limited clinical data were available for the participating EwS cases.
Results
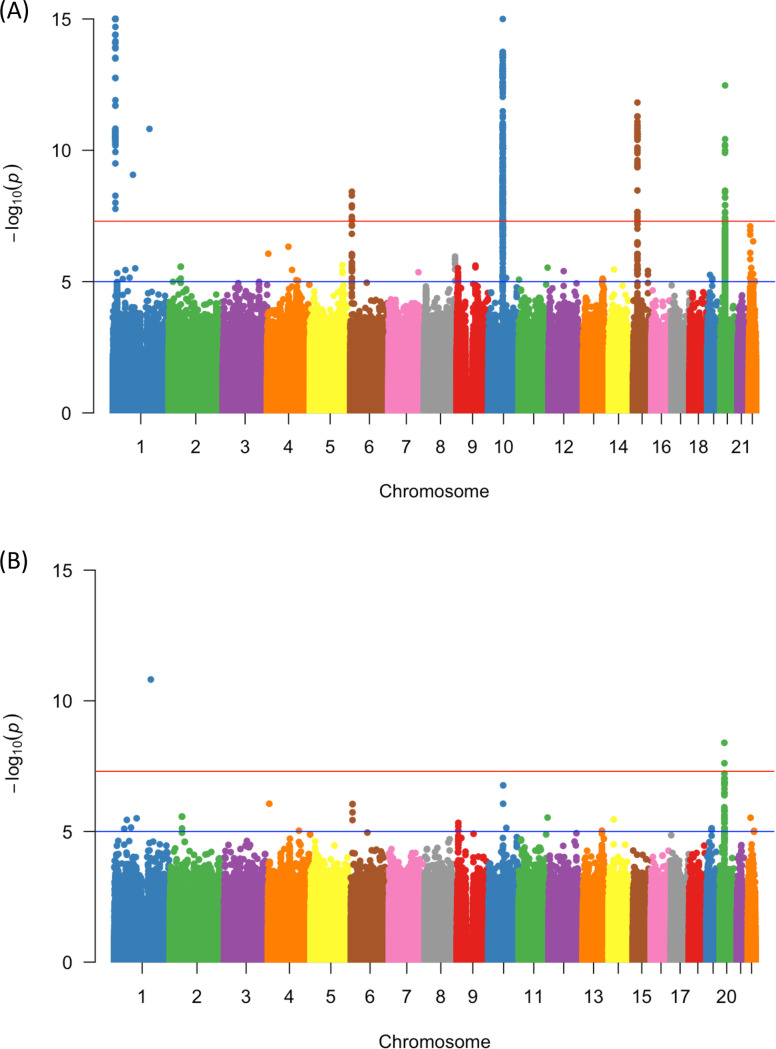
Our analysis identified evidence for associations of three putative low frequency (MAF < 0.05) imputed variants associated with EwS risk, which we advanced to validation studies described below. The variants were located at 1q23.3, 20p11.23, and 20p11.22 (Table 1, Fig 1 and S1 Fig) and tagged by rs78119607, rs112837127, and rs2296730, respectively. The MAF among controls of European ancestry ranged from 0.001 for rs78119607 to 0.046 for rs2296730 with minor allele effect sizes ranging from 0.18 to 16.64 (Table 2). The odds ratio for the minor A allele of rs112837127 suggested a potentially protective effect (OR = 0.18) indicating that in some instances low-frequency variation could reduce susceptibility to EwS.
Table 1. Genome-wide significant associations (P-value < 5×10−8) for identified low-frequency and rare variants with EwS susceptibility using a dominant model stratified by study.
| Region | Coordinate | Variant | Alleles | Minor Allele Counts (Frequency) | MH P-value | ||
|---|---|---|---|---|---|---|---|
| Major | Minor | Controls N = 1,346 | EwS Cases N = 733 | ||||
| 1q23.3 | 163530987 | rs78119607 | G | A | 4 (0.001) | 31 (0.021) | 2.38×10−11 |
| 20p11.23 | 21063508 | rs112837127 | G | A | 87 (0.032) | 9 (0.006) | 6.90×10−9 |
| 20p11.22 | 21367741 | rs2296730 | A | G | 123 (0.046) | 133 (0.091) | 4.92×10−8 |
Fig 1.
Manhattan plots of analyses for all variants (A) and low-frequency and rare variants (MAF < 0.05) (B). Plotted p-values are for allelic tests by chromosome.
Table 2. Estimated odds ratio (OR) for EwS rare variants adjusting for different model covariates.
| Wald method (unadjusted) | Mantel-Haenszel (study) | Mantel-Haenszel (study and variant1) | |||||
|---|---|---|---|---|---|---|---|
| Rare SNP | Common SNP | OR (95% CI) | Fisher’s P-value | OR (95% CI) | P-value | OR (95% CI) | P-value |
| rs112837127 | rs6106336 | 0.19 (0.10 to 0.39) | 1.64×10−8 | 0.18 (0.08 to 0.37) | 6.90×10−9 | 0.20 (0.09 to 0.40) | 5.84×10−8 |
| rs2296730 | rs6106336, rs6047482 | 2.04 (1.58 to 2.69) | 9.78×10−9 | 2.11 (1.60 to 2.77) | 4.92×10−8 | 1.61 (1.16 to 2.24) | 3.50×10−3 |
Models use a dominant allele coding for minor alleles with each individual as the analysis unit.
1Adjustment for contributing study and nearby common SNP(s).
To validate the imputed genotypes of the three associated low-frequency and rare variants, we first examined the imputation quality score (S1 Table) and distribution of alleles (S2 Table) across three studies populations, and we did not observe significant heterogeneity among the study populations. To further confirm the findings, an allele-specific TaqMan assay was designed for the three variants and carried out in a subset of 325 samples from the EwS GWAS with available remaining DNA. As shown in S2 Fig, we were able to replicate the imputed genotypes for rs112837127 and rs2296730 with 98.46% and 100% concordance rate. The imputed genotype for rs78119607 did not replicate as no minor alleles were called by the TaqMan assay, suggesting poor imputation of this variant using the 1000 Genomes Project reference set despite imputation scores of over 0.43 (S1 Table).
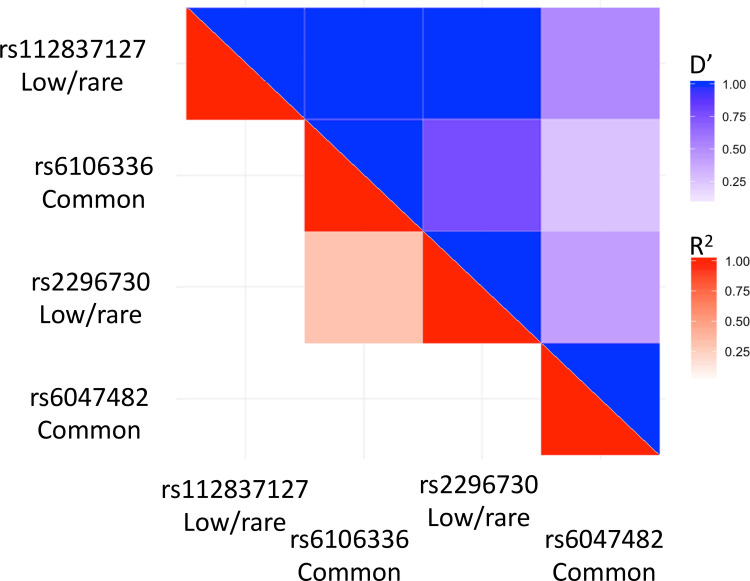
The two validated low frequency variants, rs112837127 and rs2296730, associated with EwS on chromosome 20 are in proximity to two previously identified EwS common susceptibility variants, rs6106336 and rs6047482. The identified low-frequency variants were tested for linkage disequilibrium (LD) with the common variants in 1000 Genomes Project European populations using the LDmatrix tool in LDlink (Fig 2) [30,31]. rs112837127 did not display evidence for LD with either the nearby common variant (R2EUR rs6106336 = 0.005, R2EUR rs6047482 = 0.023) or the other low-frequency variant (R2EUR rs2296730 = 0.003). However, rs2296730 displayed evidence for moderate levels of LD with the common rs6106336 variant (R2EUR = 0.311), but not the common rs6047482 variant (R2EUR = 0.006). Estimates of D′, a measure of allelic transmission, suggest the two associated low-frequency variants (rs112837127 and rs2296730) are transmitted on haplotypes of the common rs6106336 variant (S3 Fig), with the minor A allele of rs112837127 being transmitted with the major T allele of rs6106336 (D′EUR = 1.0) and the minor G allele of rs2296730 being transmitted with the minor G allele of the rs6106336 (D′EUR = 0.772).
Fig 2. Patterns of Linkage Disequilibrium (LD) for rare, low-frequency and common variants associated with EwS at the chromosome 20p11.22–23 susceptibility locus.
R2 values are in shades of red while D’ values are in shades of blue, with darker values indicating higher degree of LD. All LD measures were estimated in LDlink using 1,000 Genomes Project European populations as the reference panel.
To further test if the two low-frequency variants tagged independent EwS association signals, odds ratios and P-values for the association with EwS were calculated with and without conditioning on the neighboring common variants. Conditional analyses indicated that rs112837127 was statistically associated with EwS (OR = 0.20, 95%CI = 0.09–0.40, P-value = 5.84×10−8; Table 2) independent from neighboring common variants. As in the R2 analyses, the low-frequency rs22966730 variant demonstrated evidence for a correlation with the common rs6106336 variant as observed in the attenuated odds ratio estimate and increase in p-value in the conditional analysis (OR = 1.61, 95%CI = 1.16–2.24, P-value = 3.50×10−3; Table 2). Finally, we examined potential interaction between rs2296730 and rs6106336 (p = 0.568), rs2296730 and rs6047482 (p = 0.319), as well as rs6106336 and rs112837127 (p = 0.538) and found no significant evidence for SNP-SNP interactions.
Discussion
We report an analysis of well-imputed low-frequency variants based on common genotyped variants in a large EwS case series to investigate the contribution of low-frequency variants to the underlying genetic architecture of EwS susceptibility. We found evidence for associations of two low-frequency variants (rs112837127 and rs22966730) with EwS risk, and one of the variants, rs112837127, demonstrated an association independent of a nearby common germline susceptibility variant. Our findings suggest that in addition to common germline susceptibility variants, low-frequency variants are important for genetic susceptibility to EwS. Germline variants associated with lower cancer risk are less commonly reported, but not unheard of. Previously, three SNPs located near base excision repair genes were found to be negatively associated with Wilms tumor risk [32]. SNPs in the vitamin D receptor gene have also been linked to decreased risk in prostate cancer in African American men [33] and rs1866074 near the thymine DNA glycosylase gene were reported to be correlated with lower colorectal cancer risk [34]. The minor allele of rs112837127 is most prevalent in British and Finnish populations where the allele frequency could be > 5% while no African or east Asian population carries this allele [35]. This SNP is located in a long terminal repeat region 2.7 Kb upstream of a non-coding RNA, LINC00237, which has been found to drive self-renewal of tumor initiating cells by binding and promoting stability of β-catenin [36]. Interestingly, the activation of Wnt/β-catenin pathway has been shown to antagonize transcription activities of EWS/ETS fusion gene in Ewing sarcoma cells [37]. Whether the minor allele of rs112837127 tags a haplotype with modified LINC00237 expression remains to be investigated.
As EwS is a rare sarcoma of young people, it is not unexpected that low-frequency variation contributes to EwS susceptibility. Although EwS may be an exceptional case of a rare, well-defined malignancy with high associated odds ratios, our study suggests that efforts to examine low-frequency and rare germline associations in existing samples of rare cancer sets could be fruitful, even despite limited sample sizes. Additionally, our study provides an example in which common germline susceptibility loci discovered by GWAS may harbor synthetic associations with rare and low-frequency variants [28]. These synthetic associations may be of particular importance for EwS susceptibility as it is plausible common, low-frequency and rare variation at GGAA microsatellites may interact to impact binding of EWSR1-FLI1 fusion oncoproteins and alter regulation of downstream genes in core EwS regulatory pathways. In the case of EwS, common variant associations may highlight important EwS germline susceptibility regions where low-frequency and rare variation have important roles altering EwS risk. A limitation of our study is the lack of validation in an independent cohort as well as a lack of regional EwS sequencing of the relevant region to identify potential causal variants which can be functionally examined through in vitro experiments. Another limitation is the absence of clinical and demographic data which limited our ability to describe possible associations with the variants identified. As EwS is a rare tumor, few large case series exist for genomic investigation. Larger study populations will be essential for further confirmation of this new association. As future germline association studies investigate the genetic architecture of EwS, improved efforts to systematically interrogate low-frequency variant associations through a variety of sequencing and statistical methods are essential for accelerating understanding of the underlying genetic architecture of EwS susceptibility.
Supporting information
Plots are for rs78119607 (A), rs112837127 (B), and rs2296730 (C).
(DOCX)
(DOCX)
Linkage disequilibrium between the common variant rs6106336 and the two identified low frequency variants (A) rs112837127 and (B) rs2296730 using LDpair and all European 1,000 Genomes populations as a reference.
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank the following clinicians for providing samples used in this study: C. Alenda, F. Almazán, D. Ansoborlo, L. Aymerich, L. Benboukbher, C. Beléndez, C. Berger, C. Bergeron, P. Biron, J. Y. Blay, E. Bompas, H. Bonnefoi, P. Boutard, B. Bui-Nguyen, D. Chauveaux, C. Calvo, A. Carboné, C. Clement, T. Contra, N. Corradini, A. S. Defachelles, V. Gandemer-Delignieres, A. Deville, A. Echevarria, J. Fayette, M. Fraga, D. Frappaz, J. L. Fuster, P. García-Miguel, J. C. Gentet, P. Kerbrat, V. Laithier, V. Laurence, P. Leblond, O. Lejars, R. López-Almaraz, B. López-Ibor, P. Lutz, J. F. Mallet, L. Mansuy, P. Marec Bérard, G. Margueritte, A. Marie Cardine, C. Melero, L. Mignot, F. Millot, O. Minckes, G. Margueritte, C. Mata, M. E. Mateos, M. Melo, C. Moscardó, M. Munzer, B. Narciso, A. Navajas, D. Orbach, C. Oudot, H. Pacquement, C. Paillard, Y. Perel, T. Philip, C. Piguet, M. I. Pintor, D. Plantaz, E. Plouvier, S. Ramirez-Del-Villar, I. Ray-Coquard, Y. Reguerre, M. Rios, P. Rohrlich, H. Rubie, A. Sastre, G. Schleiermacher, C. Schmitt, P. Schneider, L. Sierrasesumaga, C. Soler, N. Sirvent, S. Taque, E. Thebaud, A. Thyss, R. Tichit, J. J. Uriz, J. P. Vannier, F. Watelle-Pichon.
Data Availability
EWS GWAS is available on dbGaP under accession number phs001549.v1.p1 Data from CCSS is available on dbGaP under accession number phs001327.v1.p1.
Funding Statement
This work was supported by the National Cancer Institute (CA55727, G.T. Armstrong, Principal Investigator), with additional funding for genotyping from the Intramural Research Program of the National Institutes of Health, National Cancer Institute and the Intramural Research Program of the American Cancer Society. This work was supported by grants from the Institut Curie, the Inserm, the Ligue Nationale Contre le Cancer (Equipe labellisée, Carte d’Identité des Tumeurs program and Recherche Epidémiologique 2009 program), the ANR-10-EQPX-03 from the Agence Nationale de la Recherche, the European PROVABES (ERA-649 NET TRANSCAN JTC-2011), and ASSET (FP7-HEALTH-2010-259348) projects. This research was supported by FP7 grant “EURO EWING Consortium” No. 602856 and the following associations: Courir pour Mathieu, Dans les pas du Géant, Les Bagouzamanon, Enfants et Santé, M la vie avec Lisa, Lulu et les petites bouilles de lune, les Amis de Claire, l’Etoile de Martin and the Société Française de lutte contre les Cancers et les leucémies de l’Enfant et de l’adolescent. The laboratory of T. G. P. Grünewald is supported by grants from the ‘Verein zur Förderung von Wissenschaft und Forschung an der Medizinischen Fakultät der LMU München (WiFoMed)’, by LMU Munich’s Institutional Strategy LMU excellent within the framework of the German Excellence Initiative, the ‘Mehr LEBEN für krebskranke Kinder—Bettina-Bräu-Stiftung’, the Wilhelm Sander-Foundation (2016.167.1), the Barbara and Hubertus Trettner foundation, the Gert and Susanna Mayer foundation, the Matthias-Lackas foundation, the Friedrich-Baur foundation, the Dr. Leopold and Carmen Ellinger foundation, the Dr. Rolf M. Schwiete foundation, the Deutsche Forschungsgemeinschaft (DFG 391665916), the Barbara and Wilfried Mohr foundation, the SMARCB1 e.V. assoication, and by the German Cancer Aid (DKH-70112257). D. Surdez is supported by SiRIC (Grant « INCa-DGOS-4654). The Metzler lab received grants from the European Commission Seventh Framework Program FP7-HEALTH “Euro Ewing Consortium EEC”, project number EU-FP7 602856, the “Schornsteinfeger helfen krebskranken Kindern” Foundation and the Trettner Foundation. The group of U. Dirksen is supported by the German Cancer Aid grant 108128, the Barbara and Hubertus Trettner foundation, the Gert and Susanna Mayer foundation; ERA-Net-TRANSCAN consortium ´PROVABES´ (01KT1310), and Euro Ewing Consortium EEC, project number EU-FP7 602856, both funded under the European Commission Seventh Framework Program FP7-HEALTH (http://cordis.europa.eu/); This work was supported by the Instituto de Salud Carlos III (PI16CIII/00026) and the Asociación Pablo Ugarte, Fundación Sonrisa de Alex, ASION-La Hucha de Tomás, Sociedad Española de Hematología y Oncología Pediátricas. Support to St. Jude Children’s Research Hospital also provided by the Cancer Center Support (CORE) grant (CA21765, C. Roberts, Principal Investigator) and the American Lebanese-Syrian Associated Charities (ALSAC). The KORA study was initiated and financed by the Helmholtz Zentrum München—German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. Furthermore, KORA research was supported within the Munich Center of Health Sciences (MC-Health), Ludwig-Maximilians-Universität, as part of LMUinnovativ. The laboratory of A.P. Garcia is supported by Gobierno de Navarra, Proyectos de Biomedicina 2018. Ref. 54/2018 and Fundación Caja Navarra/La Caixa to Niños Contra el Cáncer. Leidos Biomedical Research Inc. and Information Management Services, Inc. provided support in the form of salaries for authors J.M., E.K., C.L.D., L.B., K.J., M.M., K.W., W.Z., and M.Y., but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Grünewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Álava E, Kovar H, et al. Ewing sarcoma. Nat Rev Dis Primer. 2018;4: 5 10.1038/s41572-018-0003-x [DOI] [PubMed] [Google Scholar]
- 2.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11: 421–429. 10.1016/j.ccr.2007.02.027 [DOI] [PubMed] [Google Scholar]
- 3.von Levetzow C, Jiang X, Gwye Y, von Levetzow G, Hung L, Cooper A, et al. Modeling initiation of Ewing sarcoma in human neural crest cells. PloS One. 2011;6: e19305 10.1371/journal.pone.0019305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jawad MU, Cheung MC, Min ES, Schneiderbauer MM, Koniaris LG, Scully SP. Ewing sarcoma demonstrates racial disparities in incidence-related and sex-related differences in outcome: an analysis of 1631 cases from the SEER database, 1973–2005. Cancer. 2009;115: 3526–3536. 10.1002/cncr.24388 [DOI] [PubMed] [Google Scholar]
- 5.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359: 162–165. 10.1038/359162a0 [DOI] [PubMed] [Google Scholar]
- 6.Aurias A. Chromosomal translocations in Ewing’s sarcoma. N Engl J Med. 1983;309: 496–498. [PubMed] [Google Scholar]
- 7.Toomey EC, Schiffman JD, Lessnick SL. Recent advances in the molecular pathogenesis of Ewing’s sarcoma. Oncogene. 2010;29: 4504–4516. 10.1038/onc.2010.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gangwal K, Sankar S, Hollenhorst PC, Kinsey M, Haroldsen SC, Shah AA, et al. Microsatellites as EWS/FLI response elements in Ewing’s sarcoma. Proc Natl Acad Sci U S A. 2008;105: 10149–10154. 10.1073/pnas.0801073105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guillon N, Tirode F, Boeva V, Zynovyev A, Barillot E, Delattre O. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PloS One. 2009;4: e4932 10.1371/journal.pone.0004932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell. 2017;171: 163–178.e19. 10.1016/j.cell.2017.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grünewald TGP, Bernard V, Gilardi-Hebenstreit P, Raynal V, Surdez D, Aynaud M-M, et al. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat Genet. 2015;47: 1073–1078. 10.1038/ng.3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musa J, Cidre-Aranaz F, Aynaud M-M, Orth MF, Knott MML, Mirabeau O, et al. Cooperation of cancer drivers with regulatory germline variants shapes clinical outcomes. Nat Commun. 2019;10: 4128 10.1038/s41467-019-12071-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 2014;10: e1004475 10.1371/journal.pgen.1004475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014;4: 1342–1353. 10.1158/2159-8290.CD-14-0622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014;4: 1326–1341. 10.1158/2159-8290.CD-13-1037 [DOI] [PubMed] [Google Scholar]
- 16.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499: 214–218. 10.1038/nature12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machiela MJ, Grünewald TGP, Surdez D, Reynaud S, Mirabeau O, Karlins E, et al. Genome-wide association study identifies multiple new loci associated with Ewing sarcoma susceptibility. Nat Commun. 2018;9: 3184 10.1038/s41467-018-05537-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart BW, Wild C, International Agency for Research on Cancer, World Health Organization, editors. World cancer report 2014 Lyon, France: International Agency for Research on Cancer; 2014. [Google Scholar]
- 19.Postel-Vinay S, Véron AS, Tirode F, Pierron G, Reynaud S, Kovar H, et al. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet. 2012;44: 323–327. 10.1038/ng.1085 [DOI] [PubMed] [Google Scholar]
- 20.Troisi R, Masters MN, Joshipura K, Douglass C, Cole BF, Hoover RN. Perinatal factors, growth and development, and osteosarcoma risk. Br J Cancer. 2006;95: 1603–1607. 10.1038/sj.bjc.6603474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu K, Wang Z, Li Q, Wacholder S, Hunter DJ, Hoover RN, et al. Population substructure and control selection in genome-wide association studies. PloS One. 2008;3: e2551 10.1371/journal.pone.0002551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prorok PC, Andriole GL, Bresalier RS, Buys SS, Chia D, Crawford ED, et al. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin Trials. 2000;21: 273S–309S. 10.1016/s0197-2456(00)00098-2 [DOI] [PubMed] [Google Scholar]
- 23.Calle EE, Rodriguez C, Jacobs EJ, Almon ML, Chao A, McCullough ML, et al. The American Cancer Society Cancer Prevention Study II Nutrition Cohort: rationale, study design, and baseline characteristics. Cancer. 2002;94: 2490–2501. 10.1002/cncr.101970 [DOI] [PubMed] [Google Scholar]
- 24.Castaño-Vinyals G, Cantor KP, Malats N, Tardon A, Garcia-Closas R, Serra C, et al. Air pollution and risk of urinary bladder cancer in a case-control study in Spain. Occup Environ Med. 2008;65: 56–60. 10.1136/oem.2007.034348 [DOI] [PubMed] [Google Scholar]
- 25.O’Connell J, Gurdasani D, Delaneau O, Pirastu N, Ulivi S, Cocca M, et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 2014;10: e1004234 10.1371/journal.pgen.1004234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5: e1000529 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Core Team. R: A language and environment for statistical computing. Vienna, Austria; 2014. Available: http://www.R-project.org/
- 29.Mehta CR, Patel NR, Gray R. Computing an Exact Confidence Interval for the Common Odds Ratio in Several 2 × 2 Contingency Tables. J Am Stat Assoc. 1985;80: 969–973. 10.2307/2288562 [DOI] [Google Scholar]
- 30.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinforma Oxf Engl. 2015;31: 3555–3557. 10.1093/bioinformatics/btv402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machiela MJ, Chanock SJ. LDassoc: an online tool for interactively exploring genome-wide association study results and prioritizing variants for functional investigation. Bioinforma Oxf Engl. 2018;34: 887–889. 10.1093/bioinformatics/btx561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu J, Jia W, Wu C, Fu W, Xia H, Liu G, et al. Base Excision Repair Gene Polymorphisms and Wilms Tumor Susceptibility. EBioMedicine. 2018;33: 88–93. 10.1016/j.ebiom.2018.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daremipouran MR, Beyene D, Apprey V, Naab TJ, Kassim OO, Copeland RL, et al. The Association of a Novel Identified VDR SNP With Prostate Cancer in African American Men. Cancer Genomics—Proteomics. 2019;16: 245–255. 10.21873/cgp.20129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reddy Parine N, Alanazi IO, Shaik JP, Aldhaian S, Aljebreen AM, Alharbi O, et al. TDG Gene Polymorphisms and Their Possible Association with Colorectal Cancer: A Case Control Study. In: Journal of Oncology [Internet]. 2019 [cited 19 Feb 2020]. 10.1155/2019/7091815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexander TA, Machiela MJ. LDpop: an interactive online tool to calculate and visualize geographic LD patterns. BMC Bioinformatics. 2020;21: 14 10.1186/s12859-020-3340-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Z, Yao L, Liu Y, Zhu P. LncTIC1 interacts with β-catenin to drive liver TIC self-renewal and liver tumorigenesis. Cancer Lett. 2018;430: 88–96. 10.1016/j.canlet.2018.05.023 [DOI] [PubMed] [Google Scholar]
- 37.Pedersen EA, Menon R, Bailey KM, Thomas DG, Van Noord RA, Tran J, et al. Activation of Wnt/β-Catenin in Ewing Sarcoma Cells Antagonizes EWS/ETS Function and Promotes Phenotypic Transition to More Metastatic Cell States. Cancer Res. 2016;76: 5040–5053. 10.1158/0008-5472.CAN-15-3422 [DOI] [PMC free article] [PubMed] [Google Scholar]