Abstract.
Chromobacterium violaceum is an emerging environmental pathogen that causes life-threatening infection in humans and animals. In October 2017, a Bangladeshi farmer was hospitalized with high-grade fever due to an agricultural injury–related wound infection. Bacteriological and 16S rRNA gene investigation detected C. violaceum in the wound discharge. The patient recovered successfully after a combination treatment with meropenem and ciprofloxacin, followed by prolonged medication to avoid recurrence. We strongly propose to incorporate C. violaceum in the differential diagnosis of wound and skin infections occurring in tropical and subtropical regions, especially when the injury was exposed to soil or sluggish water.
INTRODUCTION
Chromobacterium violaceum is a β-proteobacterium that belongs to the Neisseriaceae family. It is a saprophytic bacterium found in tropical and subtropical regions of the world.1 In 1927, Lessler recorded the first human infection case from Malaysia.2 Since then, world over, 154 clinical cases were reported; among them, 23 cases were documented from the Southeast Asian countries, including India, Thailand, Sri Lanka, and Nepal.3 This bacterium rarely causes human infection; if infected, it causes severe systemic disease with a high fatality rate (∼60%).4 Chromobacterium violaceum infection outcome may range from cellulitis and skin abscesses to septic shock and systemic abscesses including vital organs, such as lungs, liver, and spleen.5,6 Herein, we describe a case of agricultural injury–related wound infection caused by C. violaceum. To our knowledge, this is the first successfully treated C. violaceum infection case from Bangladesh.
CASE PRESENTATION
A 40-year-old male farmer from Gazipur district of Dhaka Division in Bangladesh was admitted to the local hospital on October 3, 2017. He was suffering from high fever, chills, pain in the lower abdomen, insufficient urination, and excessive sweating. He had a laceration (4 cm in length, 1 cm diameter, and 1.5 cm deep) on the ankle of his left leg. The patient recollected the precise details of an incident that happened 15 days back where he met with a tractor accident during work in a paddy field, and the wound immediately was exposed to stagnant muddy water. He took immediate treatment from the village physician, medicated with oral cefixime (400 mg) twice a day along with paracetamol (500 mg) for 1 week. However, the infection did not respond to the treatment and persisted with continuous pus discharges. On hospital admission, the patient’s general condition was found to be satisfactory. However, he was febrile with 102°F body temperature, and his pulse, respiratory rate, and blood pressure were detected to be 115 pulses/minute, 21 breaths/minute, and 145/95 mmHg, respectively. The patient’s leukocyte count was 15,000 cells/mm3, with 84% neutrophils, 25% lymphocytes, 2% eosinophils, and 2% monocytes. Blood hemoglobin was 9.6 g/dL, the erythrocyte sedimentation rate was 49 mm/hour, aspartate aminotransferase was 45 U/L, and alanine aminotransferase was 49 U/L. Moreover, absolute platelet counts, random blood glucose, blood urea, electrolytes, and other blood cell parameters remained normal. Abdominal ultrasonography inspection revealed a regular abdomen. After physical examination, surgical debridement of the wound was performed, and intravenous meropenem (500 mg/8 hours) was started immediately.
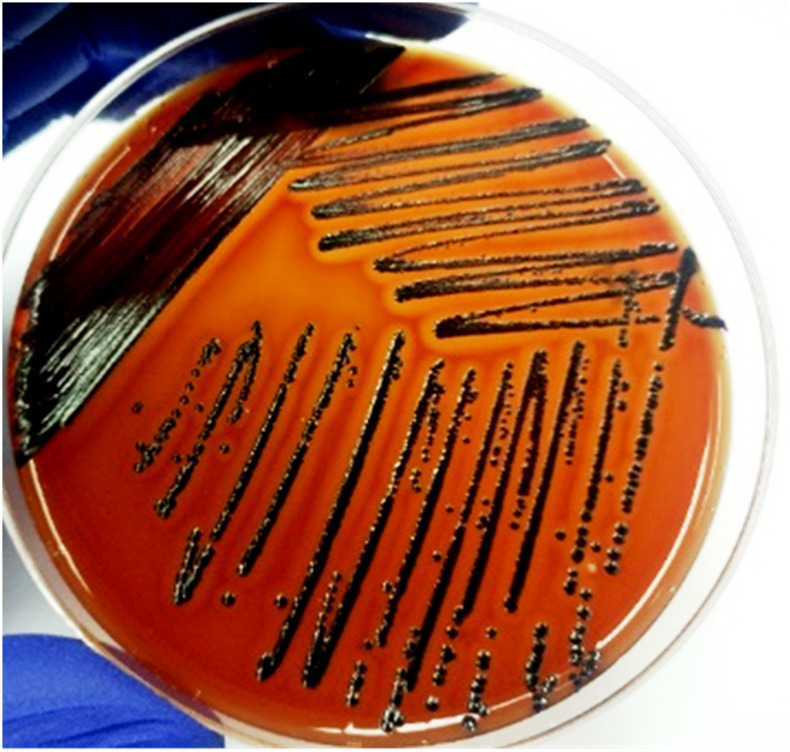
The aseptically collected pus specimen was subjected to bacteriological investigation. We cultured the specimen on MacConkey agar and 5% sheep blood agar plates as described previously.5 After overnight incubation, significant growth of distinguishable violet-pigmented colonies were detected which produced β-hemolysis reaction on a blood agar plate (Figure 1). Gram staining of the culture revealed Gram-negative coccobacilli. The pathogen was identified as C. violaceum through traditional biochemical analysis and reconfirmed with both the API 20 NE test and VITEK 2 compact system using GN card (BioMérieux, Marcy-l’Etoile, France); both these identification systems were able to detect C. violaceum with 99% probability (Table 1). We further subjected the bacterium for molecular identification and extracted the genomic DNA from a pure culture using a QIAamp DNA Mini Kit (Qiagen, Dusseldorf, Germany), as per the manufacturer’s instructions. We then performed amplification and sequencing of the partial 16S rRNA gene as described previously,7 and the sequence was submitted to GenBank (accession number MH061318). Furthermore, we constructed the phylogenetic tree to show genetic relationships between C. violaceum-RDN09 isolate and other Chromobacterium species using sequences from the GenBank database. The neighbor-joining method and the pair-wise deletion of the gap were used to construct the phylogenetic tree; the evolutionary distance was calculated by the p-distance implementation in MEGA 6.8 The phylogeny analysis indicated high bootstrap score between the studied strain and the sequences of C. violaceum ATCC 12472, whereas other Chromobacterium species demonstrated low bootstrap scores (Figure 2). Antibiotic susceptibility was tested by the disk diffusion method using 21 different antibiotics. Results were interpreted referring Clinical & Laboratory Standards Institute guideline for non-lactose fermenting Gram-negative bacteria.9 The isolate C. violaceum RDN09 was found to be resistant to ampicillin, Augmentin, cefixime, colistin, cefotaxime, polymyxin B, ceftriaxone, and ceftazidime. However, the isolate was susceptible to the following 13 antimicrobial agents: amikacin, azithromycin, ciprofloxacin, cotrimoxazole, ertapenem, gentamicin, imipenem, levofloxacin, meropenem, nitrofurantoin, netilmicin, piperacillin plus tobramycin, and tobramycin. After the culture identification and antimicrobial susceptibility results, the clinician changed the course of therapy to a combination of meropenem (500 mg/8 hours) and ciprofloxacin (500 mg/12 hours) for 14 days. During hospital discharge, the patient was afebrile and was advised to continue oral ciprofloxacin (500 mg/12 hours) for 2 months to avoid the recurrence of infection. After 2 months, the wound healed completely and demonstrated a negative blood culture.
Figure 1.
Chromobacterium violaceum RDN09 produces violet-pigmented colonies with β-hemolysis reaction on a blood agar plate. This figure appears in color at www.ajtmh.org.
Table 1.
Phenotypic characteristics of Chromobacterium violaceum RDN09, isolated from infected wound
| Bacteriological tests performed | Test results |
|---|---|
| Gram stain | Gram-negative bacilli |
| Biochemical reaction | |
| Catalase test | Positive |
| Oxidase test | Positive |
| Kligler iron agar | |
| a. Acid production in slant | Negative |
| b. Acid production in butt | Positive |
| c. Hydrogen sulfide production (H2S) | Negative |
| d. Gas production | Negative |
| e. Alkali production in slant | Positive |
| Motility indole urea test | |
| a. Motility | Positive |
| b. Indole production | Negative |
| c. Urea hydrolysis | Negative |
| Simmons citrate reaction test | Positive |
| Nitrate reduction | Positive |
| Gelatin liquefaction | Positive |
| Esculin | Negative |
| Lysine decarboxylase | Negative |
| Ornithine decarboxylase | Negative |
| Arginine dihydrolase | Positive |
| Sugar fermentation | |
| a. Glucose | Positive |
| b. Lactose | Negative |
| c. Sucrose | Negative |
| d. Maltose | Negative |
| e. Mannose | Negative |
| f. Arabinose | Negative |
| g. Salicin | Negative |
| h. Mannitol | Negative |
| i. Inositol | Negative |
| Culture media type | |
| a. Chocolate agar | Dark black color colony |
| b. Blood agar | Dark violet colony with β hemolysis |
| c. MacConkey | Violet-pigmented colony |
| d. Mueller–Hinton | Violet-pigmented colony |
| e. Gelatin agar plate | Violet-pigmented colony |
| Salt (NaCl) tolerance (%) | |
| 0 | Positive |
| 1 | Positive |
| 2 | Positive |
| 3 | Negative |
| 4 | Negative |
| 5 | Negative |
| 6 | Negative |
| 8 | Negative |
| Growth temperature | 26–42°C (aerobic) 35°C in 5% CO2 |
| VITEK 2 (bio-number) | 0067000140541210 |
| API 20 NE (number) | 5150555 |
Figure 2.
Phylogenetic tree based on 16S rRNA partial sequences, illustrating the homology between the 16S rRNA gene sequence of Chromobacterium violaceum RDN09 (black box) and other reported 16S rRNA gene sequences of C. violaceum retrieved from the National Center for Biotechnology Information gene bank (black diamond). Mega 6 software package was used to construct the phylogenetic tree following the neighbor-joining method with 1,000 bootstraps. Percentage bootstrap support is indicated by values at each node (values < 50 are omitted). As an out-group, we used Staphylococcus aureus strain Jaa1.
DISCUSSION
Chromobacterium violaceum is the only species of its genus that is associated with human infections presenting high morbidity and mortality rates. It escapes the human defense mechanism because of the elevated levels of superoxide dismutase and catalase activity.10 In the recent decade, the worldwide incidence of C. violaceum infections has increased dramatically, particularly, in Southeast Asia because of the recent global climate dynamics.11 The subject of this case was infected when the average temperature in Dhaka was around 31°C12; such a weather condition supports C. violaceum growth in the environment.
As the patient reported a recent incident of an agricultural injury, after which he developed the infection. Perhaps, the farmland stagnant water could be the source of C. violaceum, and the injury acted as a portal of entry for infection.13 Previous case studies suggest several predisposing factors for C. violaceum infection such as neutrophil dysfunction, immunocompromised state, chronic granulomatous disease, and glucose-6-phosphate dehydrogenase deficiency.11 However, the subject in our case was unaffected by such factors. He was healthy with no history of immunocompromised illnesses, and this could also be a reason for his complete recovery without any complication. Diagnosis of C. violaceum infection is completely dependent on the bacteriological culture followed by biochemical characterization.3 We found that the conventional biochemical identification process is time-consuming and requires expertise in result interpretation, whereas the API 20 NE and VITEK 2 systems have reduced turnaround time compared with the traditional biochemical methods.
We also evaluated the utility of partial 16S rRNA gene sequencing for precise molecular detection of a clinical pathogen that cannot be easily identified by phenotypic methods.14 Analysis of partial 16S rRNA gene sequence of the strain showed 99% homology with the C. violaceum ATCC 12472 type strain. All C. violaceum strains analyzed shared the same clade in the phylogenetic tree (Figure 2). We suggest that the 16S rRNA gene sequencing technique described in this study can be used alternatively to phenotypic methods for detecting C. violaceum, in case advanced bacteriological identification systems such as API 20NE and VITEK 2 are unavailable. Antibiotic susceptibility trends on C. violaceum remain sparse because of fewer reports from clinical specimens.15 However, as per the available reports, the bacterium is susceptible to aminoglycosides, chloramphenicol, fluoroquinolone, and carbapenems and extremely resistant to penicillin and broad-spectrum cephalosporins.16 In this case, the combination of meropenem and ciprofloxacin regimen showed the best treatment outcome when used together with oral ciprofloxacin as prolonged maintenance therapy.15,17 Chromobacterium violaceum may become a significant human pathogen because of the ongoing global climate dynamics. This work is expected to sensitize clinicians from tropical and subtropical areas on rapid diagnosis and appropriate antimicrobial treatment for the remission of this life-threatening infection. Therefore, we suggest incorporating C. violaceum in the differential diagnosis of wound and skin infections, especially when the injury comes in contact with soil and/or sluggish water.
Acknowledgments:
This research study was funded by core donors who provide unrestricted support to icddr, b for its operations and research. Current donors providing unrestricted support include the governments of Bangladesh, Canada, Sweden, and the United Kingdom. We gratefully thank our core donors for their support and commitment to icddr, b’s research efforts. We would like to thank the study participant for sharing the case details. We are thankful to Niyaz Ahmed for his critical review of the manuscript. The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
REFERENCES
- 1.de Vasconcelos ATR, De Almeida DF, Hungria M, Guimaraes CT, Antônio RV, Almeida FC, De Almeida LG, De Almeida R, Alves-Gomes JA, Andrade EM, 2003. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc Natl Acad Sci U S A 100: 11660–11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sneath P, Singh RB, Whelan J, Edwards D, 1953. Fatal infection by Chromobacterium violaceum. Lancet 262: 276–277. [DOI] [PubMed] [Google Scholar]
- 3.dan Lin Y, Majumdar SS, Hennessy J, Baird RW, 2016. The spectrum of Chromobacterium violaceum infections from a single geographic location. Am J Tro Med Hyg 94: 710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Siqueira IC, Dias J, Ruf H, Ramos EAG, Maciel EAP, Rolim A, Jabur L, Vasconcelos L, Silvany C, 2005. Chromobacterium violaceum in siblings, Brazil. Emerg Infect Dis 11: 1443–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker S, Campbell JI, Stabler R, Nguyen HV, To DS, Nguyen DV, Farrar J, 2008. Fatal wound infection caused by Chromobacterium violaceum in Ho Chi Minh city, Vietnam. J Clin Microbiol 46: 3853–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaufman SC, Ceraso D, Schugurensky A, 1986. First case report from Argentina of fatal septicemia caused by Chromobacterium violaceum. J Clin Microbiol 23: 956–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu Y, Lee C, Kim J, Hwang S, 2005. Group‐specific primer and probe sets to detect methanogenic communities using quantitative real‐time polymerase chain reaction. Biotechnol Bioeng 89: 670–679. [DOI] [PubMed] [Google Scholar]
- 8.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S, 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wayne P, 2011. CLSI Document M100-S21 Performance Standards for Antimicrobial Susceptibility Testing Twenty-First Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- 10.Miller DP, Blevins W, Steele D, Stowers M, 1988. A comparative study of virulent and avirulent strains of Chromobacterium violaceum. Can J Microbiol 34: 249–255. [DOI] [PubMed] [Google Scholar]
- 11.Yang CH, Li YH, 2011. Chromobacterium violaceum infection: a clinical review of an important but neglected infection. J Chin Med Assoc 74: 435–441. [DOI] [PubMed] [Google Scholar]
- 12.Istiaque A, Khan SI, 2018. Impact of ambient temperature on electricity demand of Dhaka city of Bangladesh. Energy Power Eng 10: 319–331. [Google Scholar]
- 13.Ponte R, Jenkins SG, 1992. Fatal Chromobacterium violaceum infections associated with exposure to stagnant waters. Pediatr Infect Dis J 11: 583–586. [DOI] [PubMed] [Google Scholar]
- 14.Mignard S, Flandrois JP, 2006. 16S rRNA sequencing in routine bacterial identification: a 30-month experiment. J Microbiol Methods 67: 574–581. [DOI] [PubMed] [Google Scholar]
- 15.Campbell JI, Lan NPH, Qui PT, Farrar JJ, Baker S, 2013. A successful antimicrobial regime for Chromobacterium violaceum induced bacteremia. BMC Infect Dis 13: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore CC, Lane JE, Stephens JL, 2001. Successful treatment of an infant with Chromobacterium violaceum sepsis. Clin Infect Dis 32: e107–e110. [DOI] [PubMed] [Google Scholar]
- 17.Madi DR, Vidyalakshmi K, Ramapuram J, Shetty AK, 2015. Successful treatment of Chromobacterium violaceum sepsis in a south Indian adult. Am J Trop Med Hyg 93: 1066–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]