Abstract
Background.
Circulating tumor DNA (ctDNA) is a promising technology for treatment selection, prognostication, and surveillance after definitive therapy. Its use in the perioperative setting for patients with metastatic disease has not been well studied. We characterize perioperative plasma ctDNA and its association with progression-free survival (PFS) in patients undergoing surgery for peritoneal metastases.
Patients and Methods.
We recruited 71 patients undergoing surgery for peritoneal metastases and evaluated their plasma with a targeted 73-gene ctDNA next-generation sequencing test before and after surgery. The association between perioperative ctDNA, as well as other patient factors, and PFS was evaluated by Cox regression.
Results.
ctDNA was detectable in 28 patients (39.4%) preoperatively and in 37 patients (52.1%) postoperatively. Patients with high ctDNA [maximum somatic variant allele fraction (MSVAF) ≥ 0.25%] had worse PFS than those with low MSVAF (< 0.25%) in both the pre- and postoperative settings (median 4.8 vs. 19.3 months, p < 0.001, and 9.2 vs.15.0 months, p = 0.049, respectively; log-rank test). On multivariate analysis, high-grade histology [hazard ratio (HR) 3.42, p = 0.001], incomplete resection (HR 2.35, p = 0.010), and high preoperative MSVAF (HR 3.04, p = 0.001) were associated with worse PFS. Patients with new postoperative alterations in the context of preoperative alteration(s) also had a significantly shorter PFS compared with other groups (HR 4.28, p < 0.001).
Conclusions.
High levels of perioperative ctDNA and new postoperative ctDNA alterations in the context of preoperative alterations predict worse outcomes in patients undergoing resection for peritoneal metastases. This may highlight a role for longitudinal ctDNA surveillance in this population.
Keywords: Cancer prognostication, Liquid biopsy, Cancer surveillance
BACKGROUND
Liquid biopsy of circulating tumor DNA (ctDNA) is rapidly emerging as a versatile clinical tool for identification of targetable alterations for precision medicine,1-9 prognostication and stratification,10,11 evaluation of ongoing treatment response or resistance,12,13 and surveillance of cancer recurrence following curative-intent therapy. However, the application of ctDNA to postoperative cancer surveillance in the metastatic cancer setting remains a challenge; For example, while one study of pre- and postoperative ctDNA in patients with colorectal cancer found that, in the localized disease setting, detection of postoperative ctDNA was highly specific and sensitive for recurrence when tracked longitudinally, in the metastatic setting, detection of ctDNA was less informative.11 Additionally, the appropriate timing of liquid biopsies in these settings remains an open question: in a study of gastric cancer patients, patients with metastatic disease experienced a transient increase in ctDNA when measured within the first 3 weeks postoperatively,14 and a study in colorectal cancer found marginal decreases or increases 24 h after incomplete surgical resection and decrease only after complete resection, with an estimated half-life of ctDNA of 114 min.12 It is possible that surgical trauma to microscopic residual tumor tissue and ongoing cancer cell death accounted for these observed counterintuitive changes in postoperative ctDNA or that operative trauma to compartmental barriers such as the blood-peritoneal barrier15 altered ctDNA pharmacokinetics.
By this logic, postoperative ctDNA is potentially relevant in the setting of cytoreductive surgery (CRS) and hyperthermic intraperitoneal chemotherapy (HIPEC). In this procedure, visible peritoneal carcinomatosis is surgically removed and chemotherapy is administered directly into the peritoneum. This approach has demonstrated efficacy in appendiceal cancer with peritoneal metastasis,16 peritoneal mesothelioma,17 and colorectal cancer with peritoneal metastasis.18 In all of these cases, the completeness of resection is a critically important prognostic factor postoperatively.16,17,19 Thus, acute postoperative ctDNA could theoretically validate the completion of CRS and potentially the ongoing surgical/chemotherapeutic cytotoxic effect of CRS and HIPEC.
This study is a prospective trial of pre- and postoperative ctDNA in patients undergoing CRS with or without HIPEC for the treatment of peritoneal carcinomatosis arising from a variety of primary advanced malignancies. We seek to evaluate the prognostic value of postoperative ctDNA and changes in postoperative ctDNA in these patients.
PATIENTS AND METHODS
Patients
This is a single-institution study of pre- and postoperative ctDNA in patients receiving surgical management for malignancies with peritoneal metastases and without extraperitoneal disease. Similar to our previously published study, which prospectively recruited patients for evaluation of preoperative ctDNA and progression, the current study is a single-institution prospective exploratory study of pre- and postoperative ctDNA in patients receiving surgical management for malignancies with peritoneal metastases, without extraperitoneal disease. We estimated a sample size of 70–100 patients would be necessary to identify an adjusted hazard ratio (HR) of 2.1–2.5 in PFS between those with high versus low preoperative or postoperative ctDNA quantities (the adjusted HR in our preoperative study was 2.36), assuming a two-sided alpha of 0.05, 0.80 power, a ratio of 2:1 low:high ctDNA patients, and 2 years of accrual with 1 year minimum follow-up. All patients had potentially resectable disease and were referred to our institution’s peritoneal malignancy program for evaluation for cytoreductive surgery and hyperthermic intraperitoneal chemotherapy. This study received prior approval by our institutional review board, and all patients gave signed consent prior to study enrollment (NCT02478931). Ultimately, in 71 patients, pre- and postoperative ctDNA were assayed, with surgeries taking place between May 2015 and October 2017.
Variable Definitions
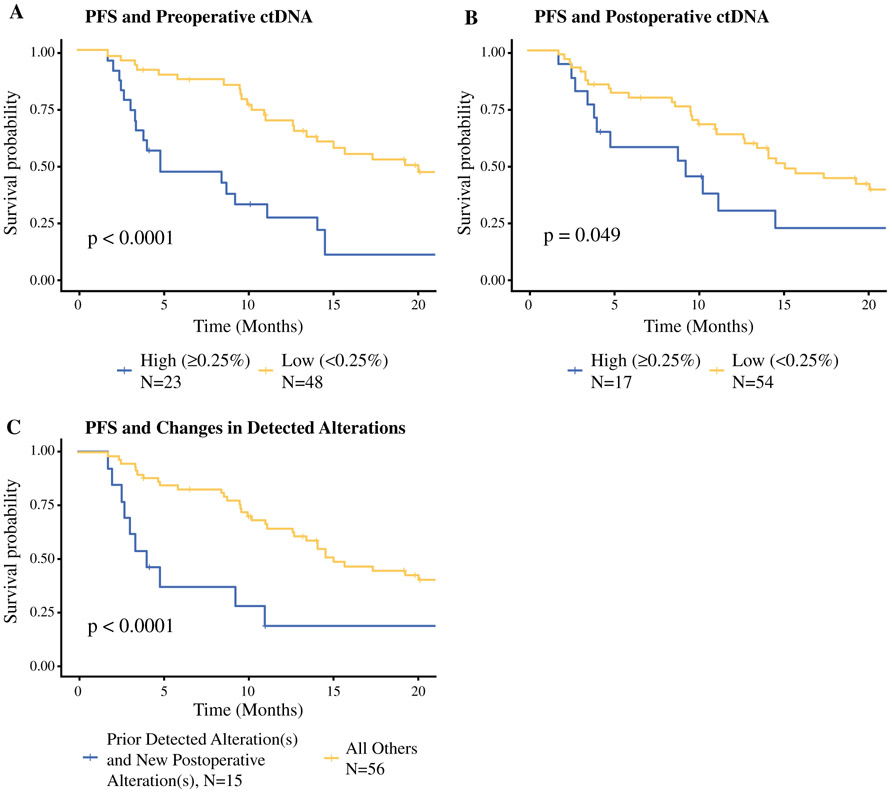
For all analyses in patients with ≥ 1 detected alterations, we defined maximum somatic variant allele fraction (MSVAF) as the highest value (% ctDNA) detected for any allele found in the blood ctDNA [including variants of unknown significance (VUS)]; patients with no detected alterations were assigned a value of 0. We defined high ctDNA as ≥ 0.25% MSVAF in any single detectable alteration in a patient. New detectable alterations in the context of prior detectable alterations were defined as any patient with detectable preoperative alteration(s), and then a new detected postoperatively (i.e., a mutation in a gene not previously found to have a mutation or a new mutation in the same gene) (Fig. 1c).
FIG. 1.
Progression-free survival curves (including VUS). Kaplan–Meier curves for progression-free survival, with p values from log-rank test: a high (≥ 0.25%) MSVAF (blue) versus low (< 0.25%) MSVAF (yellow) preoperatively, b high (≥ 0.25%) MSVAF (blue) versus low (< 0.25%) MSVAF (yellow) postoperatively, and c PFS for patients with newly altered genes detected in ctDNA after surgery in the context of prior detected alterations (blue) versus all other patients (yellow). PFS progression-free survival, ctDNA circulating tumor DNA, VUS variants of unknown significance
VUS are defined as alterations with uncharacterized clinical significance and may be reflective of tumor growth, turnover, size, heterogeneity, vascularization, disease progression, or treatment. Given that the primary aim of this study is to evaluate the use of ctDNA as a postoperative surveillance device rather than a targeted therapeutic aid, including VUS theoretically allowed for increased sensitivity to recurrences.
Procedure and Follow-Up
Patients underwent resection with the intent of removing all peritoneal metastases similar to prior descriptions from our institution.10,20 In general, patients underwent cytoreductive surgery and intraperitoneal chemotherapy if all or nearly all visible disease could be removed (also known as a complete cytoreduction, defined as removal of all visible or to < 2.5 mm residual peritoneal metastases as documented at the time of surgery). This includes resection of individual or confluent tumor nodules and invaded viscera, followed by hyperthermic peritoneal chemoperfusion with mitomycin C (for colorectal or appendiceal primary tumors) or cisplatin ± doxorubicin (for ovarian cancer and peritoneal mesothelioma) for 90 min. If complete gross resection was not possible, as appropriate, a palliative debulking procedure was performed with the goal of minimizing peritoneal metastasis-related symptoms and future complications.
Histology of the resected specimen(s) was categorized by grade, with low-grade histology defined as acellular mucin and low-grade mucinous carcinoma peritonei and high-grade histology defined as any grade of invasive adenocarcinoma, signet ring cell carcinoma, or high-grade mucinous carcinoma peritonei. We excluded mesothelioma cases in all analyses utilizing grade.
Postoperative treatment and surveillance were at the discretion of the referring oncologist. Due to the nature of referrals at our institution, all surveillance data were not available for every patient. However, patients generally underwent surveillance imaging every 3 to 6 months after surgery. Postoperative systemic treatment was typically reserved for those patients with high-grade malignancies with incomplete resections or without sufficient preoperative systemic treatment.
Sequencing
We used targeted next-generation sequencing (NGS) of ctDNA, using the Guardant Health ctDNA assay (Guardan-t360, www.guardanthealth.com/guardant360/). Guardant Health is a Clinical Laboratory Improvement Amendments (CLIA)-certified, College of American Pathologists (CAP)-accredited, and New York State Department of Health-approved clinical laboratory (Guardant Health, Inc., Redwood City, CA). Their ctDNA assay at the time of sequencing for this study identified specific tumor-related genomic alterations, including point mutations in up to 73 genes, amplifications of 18 genes, 6 gene fusions, and 23 insertion/deletions. This assay has been validated for both analytical and clinical performance; it can detect single molecules of somatic tumor DNA in 10 mL blood samples, has an analytic specificity > 99.9999%, and identifies ≥ 85% of point mutations sequenced from tissue NGS in advanced cancer patients.21 However, among patients with primarily peritoneal disease, even those with advanced cancer may have lower clinical sensitivity related to decreased shedding of tumor DNA into the peripheral circulation.5,10
For our study, we drew preoperative (typically 1–2 weeks before the operation) and postoperative samples (at first follow-up visit, typically 2–5 weeks after the operation). From these blood samples, we had all plasma cell-free DNA sequenced, including both germline cell-free DNA (e.g., from leukocyte lysis) in addition to the ctDNA. Each ctDNA alteration was quantified as a percentage of the mutated DNA fragments at a given nucleotide position divided by the wild-type cell-free DNA at the same nucleotide position. Each sequencing run included a normal control plasma sample from healthy donors (sourced to Guardant Health by AllCells, Inc., Alameda, CA), as previously described.22
Statistics
For descriptive statistics, binary or categorical variables were reported as count and percentage, and continuous variables as mean and standard deviation. The primary endpoint for survival analyses was progression-free survival, which was measured from date of surgery to date of progression. Progression was defined as combination of any of the following: radiographic progression [response evaluation criteria in solid tumors (RECIST) version 1.1],23 progression directly visualized by subsequent surgery or endoscopy, or death. We used Cox proportional hazards models to calculate hazard ratio (HR) and reported these alongside 95% confidence interval (CI), with two-sided p value of ≤ 0.05 considered significant. We visually represented survival outcomes using Kaplan–Meier survival curves. Patients with mesothelioma were eliminated from all survival analyses, as the grading system for these patients is less characterized than the grading of the other cancers in our cohort. We used a univariable screen to select variables for multivariable model selection, followed by backwards selection using a threshold of significance of 0.10. Additionally, we “forced” high/low postoperative ctDNA into all multivariable models regardless of significance, as this was our variable of interest based on our prior study of high/low ctDNA in the preoperative setting.
To study the relationship between post- and preoperative percent ctDNA, we performed a sequential variable analysis. With this approach, we started with the base Cox proportional hazards model for progression-free survival with preoperative percent ctDNA. From this, we sequentially added groups of variables. At each iteration, the hazard ratio and p-value of preoperative percent ctDNA were observed. This approach allows for greater control of the regression process to identify which groups of covariates may be confounding or mediating the relationship between the predictor of interest and outcome, depending on prior knowledge of these variables.
All statistical analyses were performed using SPSS Version 24 (IBM Corp., Armonk, NY) and R Version 3.5.2 (R Foundation for Statistical Computing, Vienna, Austria, http://www.R-project.org/), and figures were designed using the ggplot2 package24 in R.
RESULTS
Patient Demographics
Our cohort included 71 patients followed for a median of 11 months (interquartile range 4.8–20.8 months) postoperatively. The mean age at diagnosis was 55 years, and the cohort included 46 (64.8%) patients with primary appendiceal cancer, 16 (22.5%) with colorectal cancer, 3 (4.2%) with peritoneal mesothelioma, 3 (4.2%) with small bowel cancer, and 3 (4.2%) with other malignancies. Forty-three (60.6%) had a high-grade histology (excluding mesothelioma). Ultimately, 54 patients (76.1%) underwent CRS/HIPEC and 17 (23.9%) underwent palliative debulking. Fifty-four (76.1%) had a complete cytoreduction. Overall, 28 patients (39.4%) had detectable alterations preoperatively, of whom 23 had a high ctDNA quantity (defined as any alteration ≥ 0.25% of total ctDNA) (Table 1).
TABLE 1.
Patient characteristics
| Characteristic | Value | N (%) or mean (SD) |
|---|---|---|
| Patient count | 71 | |
| Sex | Men | 25 (35.2%) |
| Women | 46 (64.8%) | |
| Age at diagnosis (years) | 54.97 (11.37) | |
| Primary cancer | Appendix | 46 (64.8%) |
| Colon | 16 (22.5%) | |
| Mesothelioma | 3 (4.2%) | |
| Ovary | 1 (1.4%) | |
| Small bowel | 3 (4.2%) | |
| Testes | 1 (1.4%) | |
| Unknown | 1 (1.4%) | |
| Histology category | Low grade | 25 (35.2%) |
| High grade | 43 (60.6%) | |
| MPM | 3 (4.2%) | |
| Surgical procedure | CRS/HIPEC | 54 (76.1%) |
| Debulking | 17 (23.9%) | |
| Complete resection | 54 (76.1%) | |
| Detectable preoperative ctDNA | 28 (39.4%) | |
| Preoperative MSVAF | 0.81 (2.19) | |
| Preoperative MSVAF | Low (< 0.25%) | 48 (67.6%) |
| High (≥ 0.25%) | 23 (32.4%) | |
| Number of preoperative alterations | 1.11 (2.62) | |
| Detectable postoperative ctDNA | 37 (52.1%) | |
| Postoperative MSVAF | 0.46 (1.13) | |
| Postoperative MSVAF | Low (< 0.25%) | 54 (76.1) |
| High (≥ 0.25%) | 17 (23.9%) | |
| Number of preoperative alterations | 1.31 (3.93) | |
| ctDNA slope* | −0.08 (0.44) | |
| Preoperative systemic therapy | Chemotherapy | 18 (25.4%) |
| Chemotherapy and targeted therapy | 18 (25.4%) | |
| None | 35 (49.3%) |
Difference in MSVAF pre- and postoperatively, divided by days between ctDNA biopsy collection
ctDNA circulating tumor DNA, CRS cytoreductive surgery, HIPEC hyperthermic intraperitoneal chemotherapy, SD standard deviation, MSVAF maximum somatic variant allele fraction, MPM malignant pleural mesothelioma, N number for count variables
Postoperatively, 37 patients had detectable ctDNA, of whom 17 had high ctDNA. Overall, patients with detectable preoperative alterations were more likely to have deleterious mutation(s) detected than VUS (N = 20 versus 15, respectively), but in the postoperative setting, deleterious mutation(s) were found in patients at the same rate as VUS (N = 22 for both) (Supplementary Fig. 1). Of patients with disease progression, 66.6% (N = 32) progressed within the peritoneum, 18.8% (N = 9) progressed outside of the peritoneum, and 14.6% (N = 7) had progressive disease both within and outside of the peritoneum. Among patients with progressive disease, the site of recurrence did not correlate with detected pre- or postoperative ctDNA (p = 0.243 and 0.726, respectively, by Kruskal–Wallis test).
Patients with Colon Cancer were More Likely than Those with Appendiceal Cancer to Show Detectable Preoperative ctDNA
Previous work had demonstrated that advanced cancers have different propensities towards detectable ctDNA, with appendiceal cancer specifically having a low yield.5 This was consistent with our preoperative findings, where colon cancer patients were about twice as likely as appendiceal cancer patients to demonstrate detectable alterations (62.5% vs. 32.6%, respectively, p = 0.07). Interestingly, in the postoperative setting, this gap significantly narrowed (62.5% vs. 50%, p = 0.57).
KRAS Status and Treatment Factors Do Not Account for Differences in Detection of Postoperative ctDNA
KRAS (as measured from tissue pathology) status is not associated with increased likelihood of detecting postoperative ctDNA (OR 0.44, 95% CI 0.10–1.85, p = 0.22 by Fischer’s exact test), nor does it have any effect on PFS (HR 1.51 95% CI 0.74–3.1, p = 0.259). Of the patients who received postoperative systemic therapy (i.e., chemotherapy, N =8; biological/targeted therapy, N = 4; immunotherapy, N = 3, or combination chemotherapy–targeted therapy, N = 16), only one patient started systemic therapy prior to collection of blood for postoperative ctDNA evaluation.
High MSVAF and New Alterations Postoperatively are Associated with Shorter PFS
High (≥ 0.25) MSVAF in both the pre- and postoperative setting were associated with a decreased PFS (Fig. 1a, b) on Kaplan–Meier analysis. Additionally, the patients who had detectable alterations prior to surgery and new detectable alterations postoperatively did worse (HR 4.28, 95% CI 2.19–8.37, p < 0.0001) than all others (Fig. 1c). When comparing these patients who had detectable alterations prior to surgery and new detectable alterations specifically with the 15 patients with prior detectable alterations but no new detectable alterations postoperatively, we found a similar, albeit statistically insignificant (p = 0.18), trend.
Preoperative High MSVAF, High Tumor Grade, and Incomplete Resection Status are Associated Independently with Shorter PFS
In an analysis of PFS, a univariate screen identified high tumor grade, incomplete resection, presence of preoperative ctDNA, the number of pre- and postoperative detected alterations, and high pre- and postoperative ctDNA to be associated with progression (Table 2). Multivariable analysis with backwards selection identified tumor grade (HR 3.43, 95% CI 1.71–6.90, p = 0.001), incomplete resection (HR 2.35, 95% CI 1.23–4.49, p = 0.010), and high preoperative ctDNA (HR 3.04, 95% CI 1.55–5.98, p = 0.001) as significant predictors of progression (Table 2). High postoperative ctDNA was nonsignificant when adjusted for high preoperative ctDNA, but was significant (HR 2.15, 95% CI 1.10–4.21, p = 0.025) when in a multivariable model with grade and complete resection. A sequential variable analysis found that adding postoperative ctDNA quantity to a model already containing high/low preoperative ctDNA did little to change the PFS HR associated with the preoperative value (HR 4.54 to 3.80, 16.3% reduction), suggesting that high preoperative ctDNA is a strong prognosticator, independently of its effect on postoperative ctDNA (Supplementary Table 1).
TABLE 2.
Progression-free survival after surgery
| Variable (n) | Median PFS | Univariate |
Multivariated |
||
|---|---|---|---|---|---|
| Months (95% CI) | HR (95% CI) | p value | HR (95% CI) | p value | |
| Age (71)a | 1.00 (0.98–1.03) | 0.970 | – | ||
| Sex (71) | – | ||||
| Women (46) | 14.6 (9.6–NR) | Reference | |||
| Men (25) | 12.7 (10.2–20.0) | 1.41 (0.78–2.57) | 0.258 | ||
| Gradeb | |||||
| Low (25) | 24.9 (NR) | Reference | Reference | ||
| High (43) | 9.6 (7.8–11.4) | 3.47 (1.74–6.92) | < 0.001 | 3.42 (1.70–6.91) | 0.001 |
| Complete resectionc | |||||
| Yes (54) | 15.0 (11.0–19.1) | Reference | Reference | ||
| No (17) | 8.8 (1.3–16.2) | 2.06 (1.09–3.88) | 0.025 | 2.35 (1.23–4.49) | 0.010 |
| Preoperative ctDNA alterations | – | ||||
| No (43) | 20.0 (13.3–26.8) | Reference | |||
| Yes (28) | 8.4 (3.3–13.6) | 3.25 (1.80–5.87) | < 0.001 | ||
| Preoperative MSVAFd | |||||
| < 0.25% (48) | 19.3 (10.9–27. 6) | Reference | Reference | ||
| ≥ 0.25% (23) | 4.8 (0.0–11.3) | 3.73 (2.06–6.77) | < 0.001 | 3.04 (1.55–5.98) | 0.001 |
| Number of preoperative ctDNA alt. (71)a | 1.16 (1.07–1.25) | < 0.001 | – | ||
| Postoperative ctDNA alteration | – | ||||
| No (34) | 14.6 (11.7–17.4) | Reference | |||
| Yes (37) | 11.1 (5.7–15.6) | 1.26 (0.71–2.26) | 0.431 | ||
| Postoperative MSVAF (71)a | 1.25 (1.04–1.50) | 0.019 | – | ||
| Postoperative MSVAF | |||||
| < 0.25% (54) | 15.0 (10.9–19.2) | Reference | Reference | ||
| ≥ 0.25% (17) | 9.2 (2.8–15.6) | 1.90 (0.99–3.62) | 0.053 | 1.25 (0.60–2.64) | 0.551 |
| No. postoperative alterations (71)a | 1.07 (1.01–1.13) | 0.016 | – | ||
Cox proportional hazards regression for progression-free survival showing median survival time, univariable hazard ratios, and final multivariable hazard ratios resulting from backwards stepwise selection including variables with p ≤ 0.10, and with postoperative high/low MSVAF “forced” into final model as variable of interest. All alteration counts include variants of unknown significance in addition to characterized alterations
ctDNA circulating tumor DNA, MSVAF maximum somatic variant allele fraction, NR not reached, NS not significant by backwards selection, PFS progression-free survival
Continuous variable
Excluding patients with mesothelioma
Resection of all or nearly all (< 2.5 mm) gross disease
ctDNA as the highest maximum somatic variant allele fraction (MSVAF)
Pre- and Postoperative MSVAF are Correlated
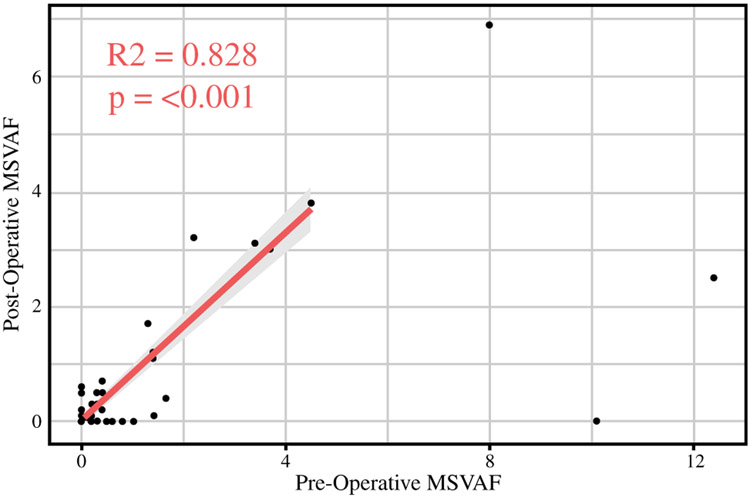
To further study the relationship between pre- and postoperative ctDNA, we performed a study of correlation between pre- and postoperative ctDNA and found significant correlation between these two values. Specifically, after removing three patients with exceptionally elevated preoperative ctDNA, the Pearson R2 was 0.828 (p < 0.001) (Fig. 2).
FIG. 2.
Correlation between pre- and postoperative circulating tumor DNA. Correlation plot of pre- and postoperative MSVAF in all patients. Eliminating three high-leverage points results in a high degree of correlation between these values (R2 = 0.828; p < 0.001). ctDNA circulating tumor DNA
No Correlation Between Disease Burden and MSVAF in Patients Undergoing Cytoreductive Surgery
We found no correlation between pre-/postoperative MSVAF and peritoneal cancer index (a measure of disease burden) (Spearman’s correlation coefficient −0.14 and 0.03, respectively) in the 53 patients who had this value reported intraoperatively.
DISCUSSION
Prediction of cancer recurrence and progression after CRS/HIPEC in the setting of peritoneal metastases is an area of active research, given the morbidity associated with these procedures and the limitations of imaging in this setting, as well as the paucity of specific biomarkers. Circulating tumor DNA may serve as a universal biomarker specific to individual patients’ tumors that can be followed over time. The present study seeks to evaluate the use of ctDNA as a possible predictor of treatment response and recurrence in peritoneal disease. Our findings demonstrate that, on multivariate analysis, high MSVAF preoperatively (≥ 0.25%)10 as well as high-grade tumor and incomplete resection are significantly and independently associated with shorter time to recurrence. Unlike in the setting of localized disease,11 we found that postoperative ctDNA quantity was not an independent predictor for progression. These findings are consistent with other literature findings on postoperative ctDNA in the metastatic setting, suggesting that there is a postoperative window in which ongoing cell necrosis in microscopic residual disease may indeed be a sign of treatment response, which may confound the implications of high MSVAF in the postoperative setting.14
While preoperative detection of ctDNA was closely correlated with tissue of origin, with colon cancer having higher levels than appendiceal cancer, this was not the case for the postoperative ctDNA, where 50% or more patients with either disease had detectable ctDNA. A potential explanation is that surgical intervention results in substantial tumor cell lysis and shedding of DNA, which may be detected in postoperative ctDNA assessments.12 Additionally, the relatively low yield of ctDNA alterations in malignancies of the peritoneum5,10 suggests that the same peritoneal-plasma barrier15 which limits systemic chemotherapy delivery to peritoneal disease and malignant ascites (necessitating intraabdominal chemotherapy such as HIPEC) may limit ctDNA release into systemic circulation, and that CRS may be disrupting this barrier. Of note, in the setting of gastroesophageal adenocarcinoma, disease limited to the peritoneum (despite being clinically more aggressive) has been reported to have a much lower mutation detection rate by and MSVAF ctDNA than disease involving other sites.25 Regardless of the mechanism, the relatively low detection rate of mutations by ctDNA, in particular with mucinous appendiceal tumors in the preoperative setting, may limit its clinical utility. Similarly, the kinetics of ctDNA remain an area of active investigation, and it is possible that interstitial ctDNA in the peritoneum would demonstrate an increased half-life as compared with tumor shedding directly into venous circulation or in other body compartments.26 Separately, the finding that new alterations detected postoperatively in the context of prior detected alterations associates with a poor prognosis (Fig. 1c), suggests that, in these patients, a new aggressive cancer clone may be evolving or that surgical handling of tissues has permitted previously undetectable clones to now be detected. Interestingly, our ctDNA detection rate in patients with CRC was significantly lower than what has been published in series of patients with primary CRC,27 suggesting that peritoneal carcinomatosis may present a unique scenario for ctDNA analysis, regardless of primary histology.
A major limitation of the present study is the restricted number of postoperative time points, which failed to characterize longer-term ctDNA dynamics in these postoperative patients. Future studies should evaluate the prognostic ability of postoperative ctDNA in the metastatic setting using a broader variety of time points to better characterize the use of this technology. Additionally, it is possible that a larger cohort would be better powered to find prognostic value in the acute postoperative ctDNA level, though the effect size we observed was somewhat marginal. Finally, the size of the cohort also limited our ability to perform subset analyses based on histology; future studies should evaluate the primary tumor-specific effect of postoperative ctDNA on PFS and cancer-specific survival. Without such confirmation with a larger (and possibly more homogeneous) cohort across several time-points, our findings should be considered to be hypothesis generating.
Overall, ctDNA is a promising biomarker for patients undergoing cancer surgery. Given the ease to access peripheral blood and its potential as a prognostic and predictive marker, further studies are needed to define its role in the context of surgical management of various malignancies. Because it is a tissue-agnostic marker, its potential utility is broad. At this time, in the setting of peritoneal surface metastases, it appears that preoperative ctDNA may hold considerable prognostic significance, while postoperative ctDNA prognostic ability is less clear. It is certainly clear that additional studies are warranted to define a role for ctDNA in surgical decision-making for patients with peritoneal metastatic disease.
Supplementary Material
Acknowledgments
FINANCIAL SUPPORT Funded in part by the National Cancer Institute (grant P30 CA023100) and the Joan and Irwin Jacobs Fund philanthropic fund. Study also funded in part by the Guardant Health (Guardant Health, Inc., Redwood City, CA). The project described was partially supported by the National Institutes of Health (grant TL1TR001443 of CTSA funding beginning August 13, 2015 and beyond). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
DISCLOSURES Razelle Kurzrock discloses stock and other equity interests (IDbyDNA, CureMatch, Inc., and Soluventis); consulting or advisory role (Gaido, LOXO, X-Biotech, Actuate Therapeutics, Roche, NeoMed, Soluventis, and Pfizer); speaker’s fee (Roche); research Fundingf [Incyte, Genentech, Merck Serono, Pfizer, Sequenom, Foundation Medicine, Guardant Health, Grifols, Konica Minolta, DeBiopharm, Boerhringer Ingelheim, and OmniSeq (all institutional)]; board member (CureMatch, Inc). Paul Riviere discloses consulting fees from Peptide Logic, LLC. Richard Lanman is an employee of Guardant Health, Inc.
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material The online version of this article (https://doi.org/10.1245/s10434-020-08331-x) contains supplementary material, which is available to authorized users.
REFERENCES
- 1.Goodall J, Mateo J, Yuan W, et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov. 2017;7(9):1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikeda S, Schwaederle M, Mohindra M, Fontes Jardim DL, Kurzrock R. MET alterations detected in blood-derived circulating tumor DNA correlate with bone metastases and poor prognosis. J Hematol Oncol. 2018;11(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kato S, Okamura R, Baumgartner JM, et al. Analysis of circulating tumor DNA and clinical correlates in patients with esophageal, gastroesophageal junction, and gastric adenocarcinoma. Clin Cancer Res. 2018;24(24):6248–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato S, Okamura R, Mareboina M, et al. Revisiting epidermal growth factor feceptor (EGFR) amplification as a target for anti-EGFR therapy: analysis of cell-free circulating tumor DNA in patients with advanced malignancies. JCO Precis Oncol. 2019. 10.1200/PO.18.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato S, Schwaederle MC, Fanta PT, et al. Genomic assessment of blood-derived circulating tumor DNA in patients with colorectal cancers: correlation with tissue sequencing, therapeutic response, and survival. JCO Precis Oncol. 2019. 10.1200/PO.18.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riviere P, Fanta PT, Ikeda S, Baumgartner J, Heestand GM, Kurzrock R. The mutational landscape of gastrointestinal malignancies as reflected by circulating tumor DNA. Mol Cancer Ther. 2018;17(1):297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shatsky R, Parker BA, Bui NQ, et al. Next-Generation sequencing of tissue and circulating tumor DNA: the UC San Diego Moores Center for personalized cancer therapy experience with breast malignancies. Mol Cancer Ther. 2019;18(5):1001–1011. [DOI] [PubMed] [Google Scholar]
- 8.Tjensvoll K, Lapin M, Buhl T, et al. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. Mol Oncol. 2016;10(4):635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mardinian K, Okamura R, Kato S, Kurzrock R. Temporal and spatial effects and survival outcomes associated with concordance between tissue and blood KRAS alterations in the pancancer setting. Int J Cancer. 2020;146(2):566–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumgartner JM, Raymond VM, Lanman RB, et al. Preoperative circulating tumor DNA in patients with peritoneal carcinomatosis is an independent predictor of progression-free survival. Ann Surg Oncol. 2018;25(8):2400–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scholer LV, Reinert T, Orntoft MW, et al. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin Cancer Res. 2017;23(18):5437–5445. [DOI] [PubMed] [Google Scholar]
- 12.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21(7):827. [DOI] [PubMed] [Google Scholar]
- 14.Pu WY, Zhang R, Xiao L, et al. Prediction of cancer progression in a group of 73 gastric cancer patients by circulating cell-free DNA. BMC Cancer. 2016;16(1):943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugarbaker PH, Stuart OA, Vidal-Jove J, Pessagno AM, DeBruijn EA. Pharmacokinetics of the peritoneal-plasma barrier after systemic mitomycin C administration. Cancer Treat Res. 1996;82:41–52. [DOI] [PubMed] [Google Scholar]
- 16.Chua TC, Moran BJ, Sugarbaker PH, et al. Early- and long-term outcome data of patients with pseudomyxoma peritonei from appendiceal origin treated by a strategy of cytoreductive surgery and hyperthermic intraperitoneal chemotherapy. J Clin Oncol. 2012;30(20):2449–2456. [DOI] [PubMed] [Google Scholar]
- 17.Yan TD, Deraco M, Baratti D, et al. Cytoreductive surgery and hyperthermic intraperitoneal chemotherapy for malignant peritoneal mesothelioma: multi-institutional experience. J Clin Oncol. 2009;27(36):6237–6242. [DOI] [PubMed] [Google Scholar]
- 18.Franko J, Ibrahim Z, Gusani NJ, Holtzman MP, Bartlett DL, Zeh HJ 3rd. Cytoreductive surgery and hyperthermic intraperitoneal chemoperfusion versus systemic chemotherapy alone for colorectal peritoneal carcinomatosis. Cancer. 2010;116(16):3756–3762. [DOI] [PubMed] [Google Scholar]
- 19.Verwaal VJ, Bruin S, Boot H, van Slooten G, van Tinteren H. 8-year follow-up of randomized trial: cytoreduction and hyperthermic intraperitoneal chemotherapy versus systemic chemotherapy in patients with peritoneal carcinomatosis of colorectal cancer. Ann Surg Oncol. 2008;15(9):2426–2432. [DOI] [PubMed] [Google Scholar]
- 20.Baumgartner JM, Tobin L, Heavey SF, Kelly KJ, Roeland EJ, Lowy AM. Predictors of progression in high-grade appendiceal or colorectal peritoneal carcinomatosis after cytoreductive surgery and hyperthermic intraperitoneal chemotherapy. Ann Surg Oncol. 2015;22(5):1716–1721. [DOI] [PubMed] [Google Scholar]
- 21.Lanman RB, Mortimer SA, Zill OA, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One. 2015;10(10):e0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Odegaard JI, Vincent JJ, Mortimer S, et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res. 2018;24(15):3539–3549. [DOI] [PubMed] [Google Scholar]
- 23.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. [DOI] [PubMed] [Google Scholar]
- 24.Wickham H Easily Install and Load the “Tidyverse.” tidyverse;2017. [Google Scholar]
- 25.Maron SB, Chase LM, Lomnicki S, et al. Circulating tumor DNA sequencing analysis of gastroesophageal adenocarcinoma. Clin Cancer Res. 2019;25(23):7098–7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khier S, Lohan L. Kinetics of circulating cell-free DNA for biomedical applications: critical appraisal of the literature. Future Sci OA. 2018;4(4):FSO295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang YC, Wang D, Jin L, et al. Circulating tumor DNA detectable in early- and late-stage colorectal cancer patients. Biosci Rep. 2018. 10.1042/BSR20180322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.