Abstract
We combine instant structured illumination microscopy (iSIM) with total internal reflection fluorescence microscopy (TIRFM), improving the lateral spatial resolution of TIRFM to 115 +/− 13 nm without compromising speed and enabling imaging frame rates up to 100 Hz over hundreds of time points. We apply instant TIRF-SIM to multiple live samples, achieving rapid, high contrast super-resolution imaging close to the coverslip surface.
Total internal reflection fluorescence microscopy (TIRFM)1 provides unparalleled optical sectioning, exploiting an evanescent field induced at the boundary between high and low refractive index media to selectively excite fluorophores within one wavelength of the coverslip surface. The superb background rejection, low phototoxicity, high speed, and sensitivity of TIRFM has been used to study diverse biological phenomena at the plasma membrane, including endocytosis, exocytosis, and focal adhesion dynamics. TIRFM has also been combined with super-resolution methods, particularly SIM2–5 to enable subdiffractive imaging in living cells3,6. Unfortunately, all previous methods sacrifice temporal resolution to improve spatial resolution, limiting their effectiveness in studying dynamic phenomena.
We and others have developed SIM implementations that improve spatial resolution without compromising speed7–9. These microscopes sharpen the image ‘instantly’ (i.e. during image formation) by optically combining information from excitation- and emission- point-spread functions (PSFs), eliminating the need to acquire and process extra diffraction-limited images that slows classic SIM. Our previous iSIM design7,10 modified a swept field confocal geometry, scanning an array of sharp excitation foci to elicit fluorescence, de-scanning the fluorescence, rejecting out-of-focus fluorescence with a pinhole array, and locally contracting each focus before rescanning to produce a super-resolution image.
TIRFM requires highly inclined illumination impinging upon the boundary between media with different refractive indices. We reasoned that placing an annular mask at a Fourier image plane (optically conjugate to the objective back focal plane) would block all subcritical rays, enabling TIRFM without otherwise perturbing the speed and functionality of our original iSIM. Annular illumination has been used to generate a single spot in diffraction-limited11 and stimulated emission depletion TIRFM12, yet for parallelized iSIM an array of spots is needed.
We created such a pattern by carefully positioning an annulus one focal length away from the foci produced by our excitation microlens array, simultaneously filtering out low angle rays in each excitation focus. The resulting beams were relayed to the sample by iSIM optical components, including a 1.7 numerical aperture (NA) objective lens used for the large range of critical angles, facilitating TIRFM. Emission optics were nearly identical to the original iSIM setup (Methods, Supplementary Fig. 1).
Since annular excitation produces a focused spot with pronounced sidelobes, we were concerned that interference between neighboring foci and transfer of energy from the central intensity maxima to the sidelobes would significantly diminish illumination contrast in the focal plane (Supplementary Note 1). When imaging fluorescent dye in TIRFM, we did observe substantial background fluorescence between excitation foci. However, individual foci were sharply defined and the extraneous background could be readily removed with the pinhole array intrinsic to our setup (Supplementary Fig. 2). We confirmed that TIRF was maintained during the imaging process by measuring the depth of the evanescent field with silica beads (Supplementary Fig. 3), finding this value to be 123 nm +/− 6 nm (95% confidence interval). Imaging fixed microtubule samples also demonstrated the improved sectioning characteristic of TIRFM, as microtubules that were otherwise visible disappeared under TIRFM illumination (Supplementary Fig. 4–7).
We estimated system resolution on 100 nm fluorescent beads (Supplementary Fig. 8). In diffraction-limited TIRFM (removing the pinholes and emission microlenses from our setup) beads were resolved to 249 +/− 11 nm (N = 20 beads, mean +/− standard deviation). Descanning, pinholing, locally contracting, and rescanning reduced the apparent bead diameter to 194 +/− 20 nm, and resolution was further improved after deconvolution (10 iterations, Richardson-Lucy deconvolution) to 115 +/− 13 nm. We observed similar results in iSIM using the same objective lens (Supplementary Table 1), implying that spatial resolution did not degrade with TIRF. Images of fixed cells confirmed this progressive resolution improvement (Fig. 1a–c), as individual microtubules had an apparent width of ~125 nm in instant TIRF-SIM (Fig. 1a, 1d), and we were able to distinguish microtubules spaced 134 nm apart, otherwise unresolved in TIRFM (Fig. 1c, e). We further verified our enhanced spatial resolution by resolving fluorophores spaced 120 nm apart in DNA nanorulers (Fig. 1f) and in live cells observing individual GFP-labeled myosin IIA bipolar filaments13 and void areas within GFP-FCHO2 puncta14 (Supplementary Fig. 9), subdiffractive structural features that have previously been resolved with TIRF-SIM.
Figure 1, Resolution enhancement via instant TIRF-SIM.
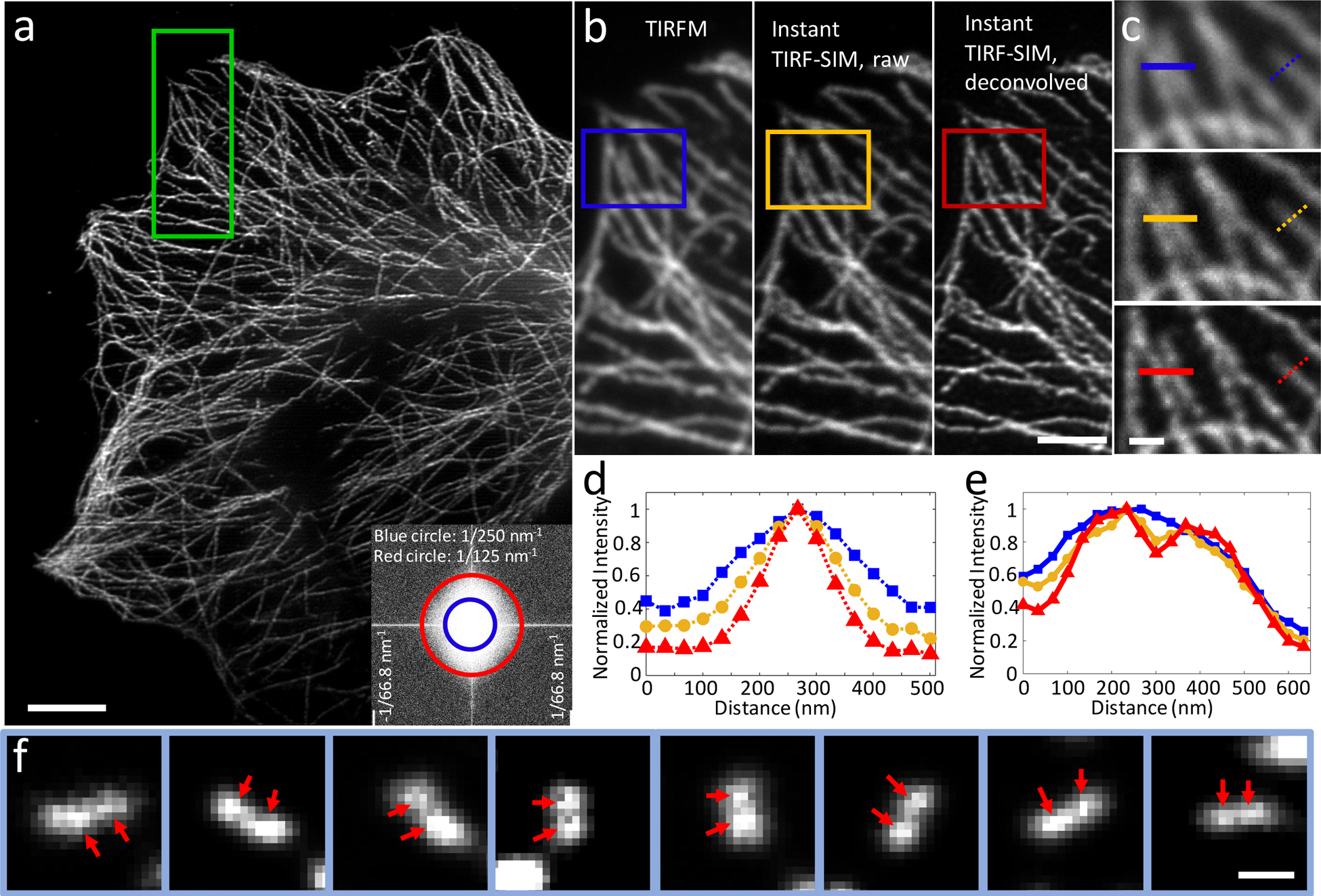
a) Deconvolved instant TIRF-SIM image of immunolabeled microtubules in a fixed U2OS cell. Inset shows power spectrum of data, on a logarithmic intensity scale. Circles indicating the diffraction limit (blue) and 125 nm spatial resolution (red) are also shown. b) Higher magnification views of the green rectangular region in a) showing diffraction-limited TIRFM (obtained using only the excitation microlenses, left), instant TIRF-SIM (raw data after employing pinholes and emission microlenses, middle), and deconvolved instant TIRF-SIM (right). c) Higher magnification views of diffraction-limited TIRFM (top), raw instant TIRF-SIM (middle) and deconvolved instant TIRF-SIM images, corresponding to blue, yellow, and red rectangular regions in b). Comparative line profiles (d, dashed lines in c; e, solid lines in c) are also shown. f) Examples of 120 nm DNA nanoruler resolution targets, imaged in instant TIRF-SIM. Red arrows indicate lobes on each dumbbell. All data are delined (Supplementary Fig. 5) except left and middle columns in b) and top and middle rows in c). Scale bars: 5 μm in a, 2 μm in b, 0.5 μm in c, 250 nm in f. Experiments were repeated at least 4 times with similar results; representative data are shown.
We next used instant TIRF-SIM to examine the dynamics of protein distributions in living cells (Fig. 2). First, we recorded microtubule dynamics over 500 time-points by imaging the fluorescence microtubule binding probe, EMTB-3xEGFP15,16, in Jurkat T cells after they settled on anti-CD3 coated coverslips (Fig. 2a, Supplementary Video 1). Our imaging rate of 20 Hz was sufficient to easily follow buckling, shortening, and sliding of microtubule bundles at the base of the cell within the evanescent field (Fig. 2b). As a second example, we recorded the dynamics of the small GTPase HRas, which is lipidated and then targeted to the plasma membrane17. Images were acquired every 0.75 s over 60 timepoints in U2OS cells (Fig. 2c, Supplementary Video 2). Intriguingly, GFP-HRas localized in highly dynamic microdomains at the plasma membrane (Fig. 2c, d, Supplementary Video 3). The high spatiotemporal resolution of our technique revealed rich dynamics of this reticulated pattern, as we observed reorganization of domains on the second timescale, including transient ‘filling in’ of the void areas between microdomains (Fig. 2d), and coordinated, ‘wave-like’ motion between microdomains (Supplementary Video 3). To our knowledge, neither the distribution nor the dynamics of Ras has been reported at this length scale in living cells, perhaps due to the lack of spatial resolution or optical sectioning (we found that in diffraction-limited TIRFM, microdomains were poorly resolved, Supplementary Fig. 10a,b).
Figure 2, Instant TIRF-SIM enables high speed super-resolution imaging at the plasma membrane over hundreds of time points.
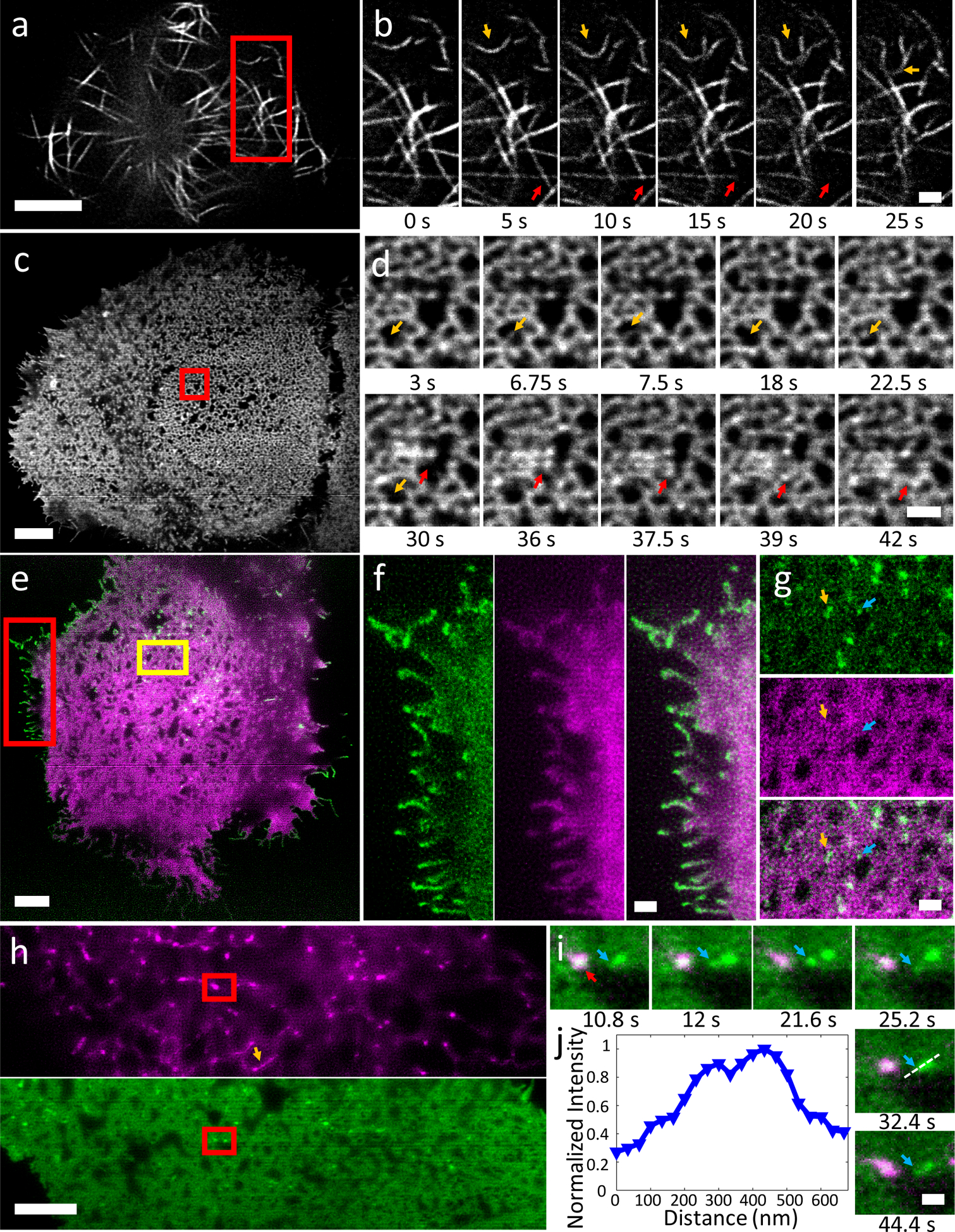
a) Image of EMTB-3xEGFP expressed in Jurkat T cells, taken from 500 frame series (images recorded every 50 ms). Higher magnification series b) of red rectangular region in a highlights microtubule buckling (orange arrows) and movement (red arrows mark microtubule bundle that moves left and up during image series). See also Supplementary Video 1. c) Image of EGFP-HRAS expressed in U2OS cell, taken from series spanning 60 time points, images recorded every 0.75 s. d) Higher magnification view of red rectangular region in c) emphasizing dynamics, including transient filling in (orange arrows) and reorganization (red arrows) of microdomains. See also Supplementary Videos 2, 3. e) Two-color image showing EGFP-VSVG (green) and Halotag-Ras (labeled with Janelia Fluor 546, magenta), derived from series spanning 100 time points, dual-color images recorded every 2.3 s. f) Higher magnification view of red rectangular region in e), showing EGFP-VSVG (left), Halotag-Ras (middle) and merged (right) distributions, highlighting concentrated VSVG at cell periphery. g) Higher magnification view of orange rectangular region in e), showing EGFP-VSVG (top), Halotag-Ras (middle) and merge (bottom). Arrows mark VSVG puncta located near Ras microdomains. See also Supplementary Video 4. h) Two-color image showing pDsRed2 ER (magenta, top) and EGFP-HRas (green, bottom), derived from image series spanning 100 time points, images recorded every 1.2 s. Orange arrow highlights ER tubule. i) Higher magnification series of red rectangular region in h, highlighting dynamics of Ras puncta (blue arrow) in vicinity of ER contact site (red arrow). j) profile of dashed line in i indicating peak-to-peak separation of 134 nm in HRas channel. See also Supplementary Video 5, 6. Scale bars: 5 μm in a, c, e, h; 1 μm in b, d, f, g; 500 nm in i. Experiments were repeated at least 4 times with similar results; representative data are shown.
We also imaged Halotag-HRas (labeled with Janelia Fluor 54918) with GFP-tagged vesicular stomatitis virus G protein (VSVG19, Fig. 2e, Supplementary Video 4), highlighting the ability of instant TIRF-SIM for dual-color imaging. Despite similar targeting to the plasma membrane20, GFP-VSVG and Halotag-HRas displayed distinct localization within living cells (Fig. 2g). VSVG showed some localization around Ras microdomains within the cell interior (Fig. 2g) and preferential enrichment at the cell boundary, particularly at cell filopodia and filamentous structures. In another example, we imaged GFP-HRas with pDsRed2-ER (Fig. 2h), which marks endoplasmic reticulum (ER). The ER mostly appeared as a set of bright punctate spots and occasional tubules near the plasma membrane, while the rest of the ER appeared as a network structure presumably further from the coverslip. Although punctate ER structures occasionally colocalized with Ras, the protein distributions were mostly distinct and exhibited different dynamics (Supplementary Video 5), consistent with their differential localization and function within the cell. The spatial resolution of our technique proved key in resolving apparent fission and fusion of Ras microclusters adjacent to more stable ER contacts (Fig. 2i), a phenomenon otherwise obscured by diffraction (Fig. 2j, Supplementary Fig. 10c, d, 11, Supplementary Video 6). Additional recordings of intracellular calcium flux, actin, and myosin IIB dynamics (Supplementary Videos 7–10) underscored our ability to perfom super-resolution imaging well matched to the dynamics of interest, either matching or surpassing the image acquisition rate offered by more traditional TIRF-SIM systems (Supplementary Table 2).
A key advantage in iSIM is the ability to image at much faster frame rates. To illustrate this capability, we imaged GFP tagged Rab11, a recycling-endosome specific GTPase that drives constant turnover of endosomes from the plasma membrane to the cytosol and modulates extracellular release of vesicles21, in U2OS cells at 37°C at 100 Hz (Supplementary Video 11). This imaging rate was sufficient to visualize and track22 the rapid motion of 1713 Rab11-decorated particles (Fig. 3a). An analysis of track motion revealed that most particles underwent < 1 μm displacement over our 6 s imaging period, yet we also observed tens of particles that showed greater displacements, with mean speed greater than 1 μm/s (Fig. 3b, c, Supplementary Fig. 12) and with instantaneous speed in some cases exceeding 10 μm/s (Fig. 3d, f). A closer analysis at the single particle level (Fig. 3b, c) also revealed qualitative differences in particle motion, with some particles undergoing diffusive motion, as revealed by a linear mean square displacement (MSD) vs. time and others showing supralinear MSD vs. time (Fig. 3e) with bouts of directed motion (Fig. 3d, Supplementary Video 12, 13).
Fig. 3, Rapid dynamics of Rab11 are resolved at 100 Hz with instant TIRF-SIM.
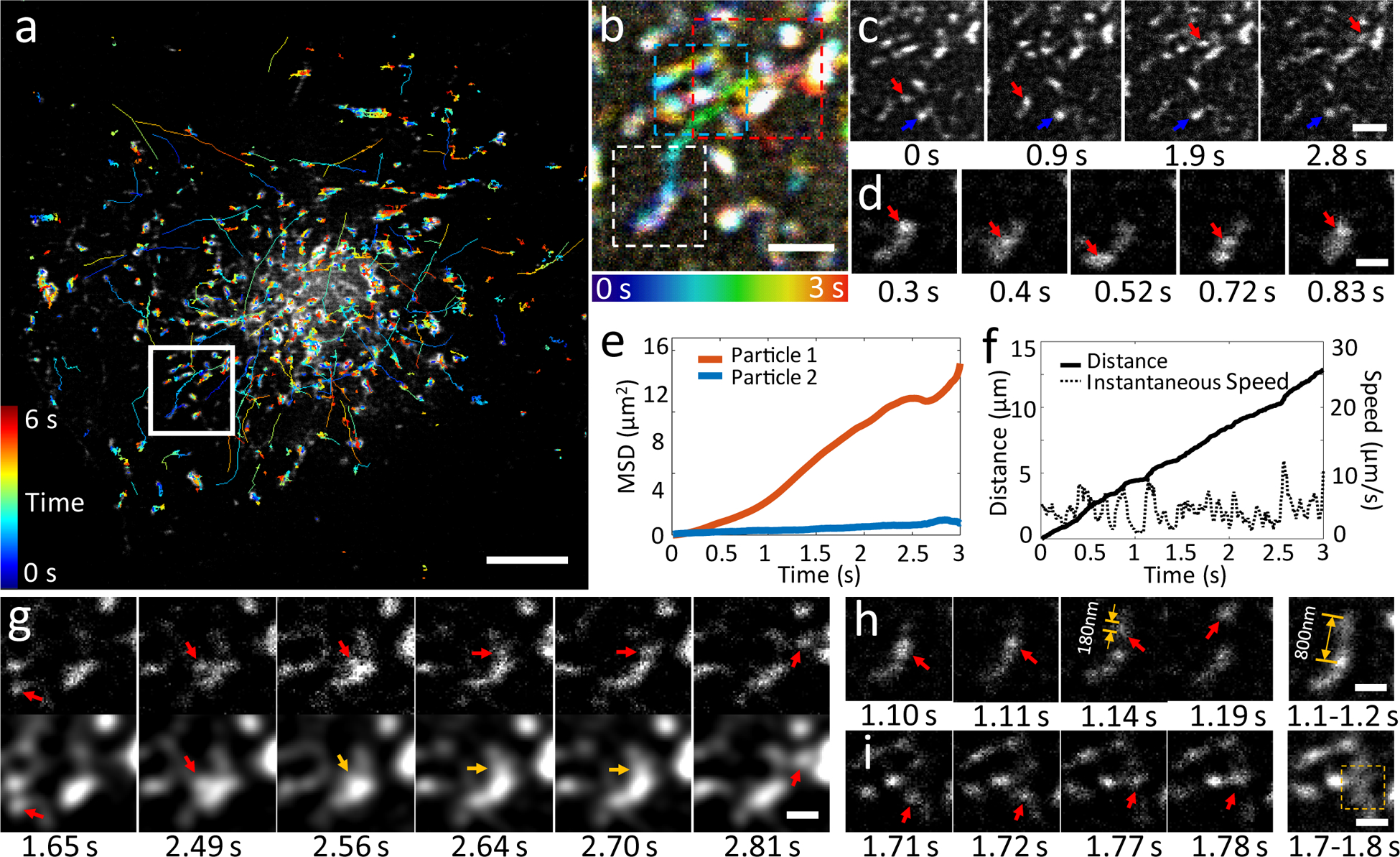
EGFP-Rab11 was transfected into U2OS cells and imaged at 37°C at 100 Hz. a) First frame from image series, with overlaid tracks (lines colored to indicate time, color bar indicated at left). b) Higher magnification view of white rectangular region in a), over the first 3 seconds of acquisition. Time evolution indicated in color bar at bottom. c) Selected images corresponding to region in b), emphasizing motile (red arrow) and more stationary (blue arrow) particle. d) Magnified view of more motile particle indicated with red arrow in c) (white dashed rectangular region in b) highlighting bidirectional motion. Mean square displacements of both particles (e) and distance and instantaneous speed (f) of the motile particle are also quantified. g) Selected images corresponding to red dashed rectangular region in b). Particle, highlighted by arrows, is resolved in instant TIRF-SIM (top row) but poorly or not (yellow arrows) resolved in spatially blurred images (bottom row) that simulate diffraction-limited TIRFM. h) Selected images corresponding to white dashed rectangular region in b), demonstrating that recording at fast 5 ms exposures resolves particles (red arrows), but combining frames into 100 ms bins does not (particle of size ~180 nm is blurred into extended structure of length ~800 nm) i) Same analysis as in h (compare particle indicated by red arrows in left images to yellow dashed region in right-hand image), but particle and region correspond to cyan dashed region in b). Scale bars: 5 μm in a; 1 μm in b, c; 0.5 μm in d, g, h, i. See also Supplementary Fig. 12, 13 and Supplementary Videos 11–14. Experiments were repeated at least 4 times with similar results; representative data are shown.
Imaging at slower frame rates (as with previous implementations of TIRF-SIM) distorts track lengths because long tracks are broken into shorter tracks, short tracks are discarded, and multiple independent short tracks may be classified falsely as longer tracks (Supplementary Fig. 13a, b). Imaging at lower spatial resolution (as in diffraction-limited TIRFM) failed to resolve particles in densely packed regions of the cell (Fig. 3g, Supplementary Video 14) or missed hundreds of tracks (Supplementary Fig. 13c, d).
If the sample moves on a lengthscale comparable to or greater than the resolution limit in a single exposure, motion blur degrades spatial resolution and introduces artifacts (Supplementary Fig. 14, Supplementary Videos 15,16). For example, when binning frames together to simulate the slower exposures afforded by traditional TIRF-SIM, Rab11 particles that were otherwise resolved formed artificial structures larger than the diffraction limit (Fig. 3h, i). To further illustrate this point, we imaged Rab11 on a state-of-the-art TIRF-SIM. While this instrument could not match the 100 Hz frame rate of our system, we were able to acquire raw images at an exposure time identical to our instrument (5 ms), resulting in a frame rate of 12.9 Hz. Rab11 particles were sharply defined in raw diffraction-limited images, but the time delay introduced by the need to acquire nine such images for a single reconstructed frame resulted in obvious artifacts (Supplementary Videos 17, 18, Supplementary Fig. 15), worsening spatial resolution to the extent that images were no longer even diffraction-limited.
Such artifacts were absent in instant TIRF-SIM, which is fundamentally faster than traditional TIRF-SIM, as only one image needs to be acquired, instead of the standard nine3. Related advantages of our method over previous approaches include less read noise and less computational processing. Although our ~115 nm spatial resolution is worse than claimed in state-of-the-art linear TIRF-SIM6 (84 nm), our existing implementation of instant TIRF-SIM is ~50 fold faster.
Room for technical improvement remains. The excitation efficiency of our setup is low, as ~60% of the illumination is blocked by the annular mask. Using a spatial light modulator (SLM) to generate the pattern might direct the illumination through the annular mask more effectively, facilitating lower power illumination. Controlling the phase of the illumination might also reduce the sidelobes in each focus, improving contrast in the focal plane and perhaps even removing the need for pinholes (although pinholes are still useful in reducing scattered light that continues to contaminate objective-based TIRF23). Using optics that allow rapid adjustment of the annulus dimensions (such as an SLM or a digital micromirror device) could facilitate adjustment of the evanescent field depth, providing additional axial information within the TIRF zone24. Finally, we did not exploit the narrower central maximum in each excitation focus for (marginally) higher spatial resolution, due to the coupling between inter-focus distance and focus size (Supplementary Note 1). Combining TIRFM with single-point rescanning SIM9,25 would address this issue, albeit at the cost of temporal resolution.
Online Methods
Instant TIRF-SIM
The instant TIRF-SIM is built directly upon our previously reported instant SIM system7, but with two important modifications in the excitation path. First, we used a 1.7 NA objective (Olympus, APON100XHOTIRF) for excitation and detection. When imaging into aqueous samples with refractive index 1.33, 1-(1.33/1.7) = 0.22 of the objective back focal plane diameter (dBFP) is available for TIRF, implying that sub-critical illumination rays within a diameter 0.78 * dBFP = 0.78 * 2 * NAOBJ * fOBJ = 0.78 * 2 * 1.7 * 1.8 mm = 4.77 mm must be blocked. Second, we inserted a relay system into the excitation arm of the instant SIM to block these rays. Excitation from 488 nm and 561 nm lasers was combined and beam expanded as before, and directed to a microlens array (Amus, f = 6 mm, 222 μm spacing between microlenses, 1 mm thick, 25 mm diameter, antireflection coated over 400–650 nm, APO-Q-P222-F6(633)+CHR) to produce an array of excitation foci. We used a matched pair of scan lenses (Scan lens 1 and 2, f=190 mm, Special Optics, 55-S190–60-VIS) placed in 4f configuration to relay these excitation foci to the rest of the optical system, inserting an opaque circular mask (Photosciences, 2.68 mm diameter chrome circle with optical density 5 on 4” × 4” × 0.090” quartz wafer) at the focal point between scan lenses (and the Fourier plane of the excitation foci produced by the microlens array) to filter subcritical rays. Given the 350 mm/ 190 mm = 1.84x magnification between the mask and the back focal plane of the objective, we designed the mask to block the central 2.68 mm * 1.84 = 4.93 mm diameter of the illumination. An iris placed just after the mask ensured that the outer diameter of the beam was ~3.33 mm, a diameter that magnified to 3.33 * 1.84 = 6.13 mm, or ~dBFP, thereby reducing stray light that would otherwise fall outside the objective back focal plane. Alignment of the opaque mask and microlens array, which is critical, was greatly aided by placing the former on a 3-axis translation stage (Thorlabs, LT3, used for correct positioning of the mask image at the back focal plane) and the latter on a uniaxial translation stage (Thorlabs, LNR50M, used to position excitation foci precisely at the focal plane of the objective lens). We also used an alignment reticle (Leica) that screwed into our objective turret to further check that the annular illumination pattern was properly positioned (concentric with the optical axis of the objective) and focused at the back focal plane of our objective.
In the emission path, optics were identical to our previous design, except that we used a pinhole array with larger pinholes (Photosciences, Chrome on 0.090″ thick quartz, 222 μm pinhole spacing, 50 μm pinhole diameter) and an emission-side microlens array with longer focal length (f = 1.86 mm, Amus, APO-Q- P222-F1.86(633)). The total magnification between sample and our scientific grade complementary metal-oxide semiconductor camera (PCO-TECH, pco.edge 4.2) detector was 350 mm / 1.8 mm = 194.4, resulting in an image pixel size of 33.4 nm. These elements are shown in Supplementary Fig. 1.
The excitation laser power was measured immediately prior to the objective. Depending on the sample, the average power ranged from 0.2 – 2 mW, implying an intensity range from ~7 – 70 W/cm2 (given our 58 μm × 52 μm field of view).
Samples were deposited on 20 mm diameter high index coverslips (Olympus, 9-U992) designed for use with the 1.7 NA lens. Coverslips were mounted in a magnetic chamber (Live Cell Instrument, CM-B20–1) that attached to the microscope stage. For temperature maintenance at 37 °C, the magnetic chamber was mounted within an incubation chamber (Okolab, H301-MINI).
Estimating the evanescent field depth
We used two methods to estimate evanescent field depth. First, we used an analytical method26. For excitation of wavelength λ impinging at angle θ1 upon an interface with indices n1 and n2, n1 > n2, the intensity I of an evanescent field decays along the optical axis with decay constant d according to I(z) = Ioexp(−z/d), with d = λ/(4π) (n12sin2(θ1) - n22)−0.5. The term n12sin2(θ1) is equivalent to the square of an “effective” NA, in our case ≤ 1.7. If considering the smallest angles in our annular excitation (corresponding to the inner radius used in the mask, producing evanescent waves with the longest decay length), this effective NA is 4.93/6.12 * NAOBJ = 1.37. Assuming n2 = 1.33 and λ = 488 nm leads to d = 118 nm. If considering the largest angles (corresponding to the outer annulus radius, producing evanescent waves with the shortest decay length), the effective NA is NAOBJ = 1.7, leading to d = 37 nm. By these simple calculations, the “average” decay thus lies between 37 nm – 118 nm, weighted by the distribution of intensity in the annular excitation.
Since such an intensity distribution is difficult to measure accurately, we instead opted to measure the average evanescent decay length more directly using silica beads (diameter 7.27 μm, refractive index, 1.42, Bangs Laboratories) placed in a solution of fluorescein dye (Fluka, Cat #32615) (Supplementary Fig. 3a). In this method, the known diameter of the bead is used to convert the apparent radii observed with TIRF to an axial depth26, z (Supplementary Fig. 3b). Following previous work27, we integrate the intensity I(z) from the coverslip surface to some depth z, as this corresponds to the observed signal F(z) at each depth. First, we assume the fluorescence is well modeled by a sum of two exponentials. The first term corresponds to signal derived from “pure” TIRF (with decay d) and the second term models scattering that is known to contaminate objective-type TIRF (with decay D):
where A and B are constants that account for incident beam intensity, concentration, and the relative weight of the scattering term. Integrating this expression yields
Fitting the measured fluorescence intensity at each depth (derived at each bead radius) to this expression (Supplementary Fig. 3c) with the MATLAB curve fitting toolbox gave d = 123 nm with 95% confidence interval (117nm, 129 nm). The scattering amplitude B represented ~24 % of the signal.
Diffraction-limited TIRF comparisons
For some comparisons (Fig. 1b, c, Supplementary Fig. 8, 10a, b, 13c, d), the sample was imaged with instant TIRF-SIM and then again after removing pinholes and emission microlenses, yielding a diffraction-limited image. In other cases (Fig. 3g, Supplementary Fig. 10c, d, 14), we simulated this effect (Supplementary Fig. 11) by blurring the deconvolved instant-SIM data with a 2D Gaussian function of σ = (σ2DIFFRACTION-LIMITED -σ2INSTANT TIRF-SIM)0.5 where σINSTANT TIRF-SIM is derived from the apparent width of beads measured in the instant TIRF-SIM after deconvolution (=115 nm/2.355) and σDIFFRACTION-LIMITED is the corresponding diffraction-limited value (=249 nm/2.355).
Comparisons with commercial TIRF-SIM
Data were collected using a DeltaVision OMX SR microscope (GE Healthcare Life Sciences) equipped with a 60x NA1.42 objective (Olympus). Samples were maintained at 37C with the environmental chamber housed within the microscope. The exposure time for each raw diffraction-limited frame was set to 5 ms, resulting in a total acquisition time of 77.8 ms for each 9-frame grouping. Data were processed with OMX software to yield SIM reconstructions.
Data Processing
Flat fielding
Due to the spatially nonuniform profile of the excitation laser beam, the excitation intensity is not distributed uniformly even when the excitation is scanned. The scanned excitation distribution has highest intensity in the center of the field of view and diminishes at increasing distances perpendicular to the scanning direction. To normalize for this variation in excitation intensity (‘flat fielding’), we averaged 100 images of a thin fluorescein layer, smoothed the average perpendicular to the scan direction, and divided the raw data by this smoothed average prior to deconvolution. All datasets shown in the paper were flat-fielded before display (Supplementary Software).
Deconvolution
Unless otherwise indicated, data presented in this paper were deconvolved to further enhance spatial resolution. Before deconvolution, background was subtracted from the raw images. Background was estimated by averaging 100 “dark” images acquired without illumination. For deconvolution, we used the Richardson-Lucy algorithm28,29, blurring with a 2D PSF:
For k = 1,2,…N
where ⊗ denotes convolution operation, ImageMeasured is the measured image (after background subtraction) and is the flipped PSF:
with m, n the PSF dimensions.
The PSF was experimentally derived by registering and then averaging the images of 20 100 nm yellow-green beads. Deconvolution was implemented in MATLAB 2017a with the number of iterations N set to 10 (Supplementary Software).
Bleach correction
For several time-lapse datasets (Fig. 2c–i, 3, Supplementary Fig. 10c, d, 12–15, Supplementary Video 1–6, 11–18, we performed standard bleaching correction using an ImageJ Plugin (Bleach Correction30, https://imagej.net/Bleach_Correction) with the “simple ratio” method.
Delining data
Close inspection of the data acquired in our instant TIRF-SIM revealed a horizontal line artifact that arises because the instant SIM performs a 1D scan of a 2D space with a 2D grid of illumination points7. This problem is slightly exacerbated under TIRF illumination due to the smaller foci size (Supplementary Note 1, Supplementary Software). However, we found that the lines showed repeatable signatures in Fourier space and that by applying notch filters at these locations we could suppress line artifacts in our images (Supplementary Fig. 5–7, Methods).
More specifically, Fourier transforming the image data and displaying the power spectrum revealed high intensity puncta along the vertical axis. These puncta correspond to horizontal line artifacts in our data, as discussed in the main text. Replacing these high-intensity pixels with zero valued pixels and taking the inverse Fourier transform suppressed the lines without compromising spatial resolution (Supplementary Fig. 5). As 10 rectangular regions, each 20×20 pixels, were zeroed in each 2048×2048 Fourier transform, delining affected less than < 0.1% of the total power spectrum in each image.
A similar strategy was proposed to reduce artifacts that continue to plague more traditional SIM implementations31. All data shown in this paper, unless otherwise noted, was ‘delined’ using this procedure.
Drift Correction
Some datasets (Supplementary Video 1, 3) were corrected for drift using the ‘StackReg’ plugin in ImageJ32 (https://imagej.net/StackReg), using the ‘Rigid Body’ condition.
Image display
All images are displayed in grayscale, except images in Fig. 2 e–i, displayed in green and/or magenta colormaps from ImageJ, and Supplementary Fig. 14, displayed using the ‘hot’ colormap in ImageJ.
Tracking
For the Rab11 dataset (Fig. 3), we performed semi-automated tracking using the TrackMate ImageJ Plugin33 (https://imagej.net/TrackMate). For particle detection, the Difference of Gaussian (DoG) detector was used with estimated spot diameter of 0.25 μm, and an initial quality threshold of 150. The particles were further filtered with a quality threshold of 200 and linked with a simple Linear Assignment Problem (LAP) linker. Linking maximum distance, the gap-closing maximum distance, and the maximum frame gap were set to 0.3 μm, 0.4 μm and 8, respectively. Manual inspection of the data revealed that the vast majority of tracks < 10 frames were spurious, so these were excluded from further analysis. For tracking on the whole image (Fig. 3), the linking filters were manually adjusted to filter out obviously spurious tracks. Then manual editing was performed within the plugin interface to improve tracking results. Within the cropped region used for downsampling analysis (Supplemental Fig. 13a, b, Supplemental Video 12), images were downsampled 5- and 10 times in the time domain. Then, automated tracking was performed independently for the cropped images (100Hz) and the downsampled images (20 Hz and 10 Hz) without manually adjusting either linking filters or links. Automated tracking parameters were also used in additional Rab11 datasets (Supplemental Fig. 13c, d) except that the initial quality threshold and filtered quality threshold for the spots detection were replaced by 100 and 120, respectively, due to the slightly weaker image intensity.
From the particle tracks (i.e., the sequences of coordinates denoting the position of each tracked particle at each time point), we computed several quantitative metrics including displacement, distance, instantaneous speed, mean speed and mean squared displacement (MSD).
Given a trajectory consisting of N time points and the particle coordinates at ith time point pi=(xi,yi), we define the distance between any two points pi and pj as the Euclidean norm
The total distance traversed at the jth time point is calculated from the starting point (the 1st time point) and defined as
and the displacement (magnitude), also known as net distance
Then the total distance for the whole trajectory is DN and the total displacement for the whole trajectory is FN.
The instantaneous speed is defined as
where Δt is the time interval between two successive time points. The instantaneous speed is also the derivative of the traveled distance Dj.
Then the mean speed is calculated as the average of the instantaneous speed:
The mean squared displacement is calculated as
Sample preparation
Bead Samples
High index coverslips were cleaned with 100% ethanol and coated with 0.1% Poly-L-Lysine (PLL, Sigma-Aldrich) for 10 min. Then 100 nm yellow-green beads (Thermo Fisher Scientific, F8803) were diluted ~105-fold and 20 μL added to the coverslip. After 10 min, the coverslip was washed four times with clean water before imaging (Supplementary Fig. 8).
DNA Nanorulers
High index coverslips were incubated with BSA-biotin solution (1 mg/ml in PBS) for 5 min, washed three times with PBS, incubated with neutravidin solution (1 mg/ml) for 5 min in PBS, and washed three times with PBS containing 10 mM magnesium chloride (IB buffer). 1–2 μL 120 nm DNA nanorulers labeled with Alexa Fluor 488 (GATTAquant DNA Nanotechnologies, GATTA-SIM 120B) diluted into 200 μL IB were incubated with the coverslip for 5 minutes immediately prior to imaging (Fig. 1f).
Fixed Samples
For imaging microtubules within fixed samples (Fig. 1a–c, Supplementary Fig. 4, 5a), high index coverslips were first immersed in 70% ethanol for ~ 1 min and allowed to air dry in a sterile cell culture hood. U2OS cells were grown on uncoated high index coverslips until ~50% confluency. The entire coverslip was submerged for 3 minutes in methanol pre-chilled to −20 °C to fix the cells. Coverslips were then washed in PBS at room temperature extensively before blocking in antibody dilution buffer (Abdil; 1%BSA, 0.3% Triton-X 100 in PBS) for 1 hour at room temperature. The primary antibody stain was performed overnight at 4 °C using 1/500 mg/ml of mouse anti α-Tubulin (Thermo Fisher Scientific #62204) in Abdil. The secondary antibody stain was performed for 1–2 hours at room temperature using 1/200 mg/ml of goat anti-mouse Alexa 488 (Invitrogen A11001) in Abdil.
Live Jurkat T cells
High index coverslips were rinsed with 70% ethanol and dried with filtered air. The coverslips were then incubated in Poly-L-Lysine (PLL) at 0.01% W/V (Sigma Aldrich, St. Louis, MO) for 10 min. PLL solution was aspirated and the coverslip was left to dry for 1 hour at 37 °C. Coverslips were next incubated with streptavidin (Invitrogen) at 2 μg/ml for 1 hour at 37 °C and excess streptavidin was washed with PBS. Antibody coating for T cell activation was performed by incubating the coverslips in a 10 μg/ml solution of biotin labeled anti-CD3 antibody (OKt3, eBiosciences, San Diego, CA) for 2 hours at 37 °C. Excess antibody was removed by washing with L-15 imaging media immediately prior to the experiment. E6–1 Jurkat T-cells were transiently transfected with EMTB-3xEGFP (Fig. 2a, b, Supplementary Video 1) or F-tractin EGFP (Supplementary Video 8) plasmid using the Neon (Thermo Fisher Scientific) electroporation system two days before the experiment. Transfected cells were centrifuged and resuspended in L-15 imaging media prior to pipetting them onto the coverslip. Imaging was performed 10 minutes after the cells settled on the substrate. EMTB-3xEGFP was a gift from William Bement (Addgene plasmid # 26741) and pEGFP-C1 F-tractin EGFP was a gift from Dyche Mullins (Addgene plasmid # 58473).
Live U2OS cells
Ras, Rab, VSVG, ER imaging
Human osteosarcoma U2OS cells were routinely passaged in DMEM (Life technologies) plus 10% FBS (Hyclone) at 37 °C, with 5% CO2. For cleaning prior to live cell imaging, high index coverslips were boiled for 5 minutes with distilled water, thoroughly rinsed with distilled water and stored in 90% ethanol for at least 2 hours. To facilitate cell adherence, the coverslips were coated with FBS for 2 hours at 37°C. 24 – 48 hours prior to transfection, cells were plated on cleaned coverslips, at a density of ~60%. Cells were transfected with the appropriate plasmid using Turbofect (Life Technologies) at a ratio of 3:1 (Liposomes:DNA). The next day, the medium was replaced with fresh DMEM plus 10% FBS without phenol red, which was also used as the imaging medium. To monitor wild type Ras dynamics, we used EGFP-HRas (Fig. 2c, d, Supplementary Fig. 5b, 10a, b, Supplementary Video 2, 3), or if imaged with VSVG-GFP (Addgene #11912) (Fig. 2e–g, Supplementary Video 4), we used a HaloTag chimera of HRas (gift of Dominic Esposito, NCI). Halotag proteins were labeled using Janelia Fluor549 (gift of Luke Lavis, Janelia Research Campus) at a final concentration of 100 nM for 15 minutes. Following labelling, the cells were rinsed twice with plain DMEM, incubated with fresh medium plus 10% FBS for 20 minutes, and finally the medium replaced with fresh, phenol red free DMEM plus 10% FBS. The dynamics of Rab GTPase were followed for GFP tagged Rab1134 (Addgene #12674)(Fig. 3, Supplementary Fig. 12–13, Supplementary Video 11–13). For dual labelling of Ras and the endoplasmic reticulum, we co-transfected the EGFP-HRas construct with pDsRed2-ER (Clontech, cat #632409) (Fig. 2h, i, Supplementary Fig. 10c, d, Supplementary Video 5, 6), which carries a KDEL ER retention signal.
For OMX SR imaging of GFP-tagged Rab11 (Supplementary Fig. 15, Supplementary Video 17, 18), U2OS cells were seeded onto 35 mm glass bottom MatTek dishes, immersed in DMEM supplemented with 10% FBS and penicillin/streptomycin, and maintained at 37°C, with 5% CO2. Cells were transfected with GFP-Rab11 plasmid using X-tremeGENE™ (Sigma Aldrich) at a ratio of 2:1 (transfection agent:DNA). 24 hours post transfection, cells were transferred to the microscope for imaging.
Membrane imaging
U2OS were plated on high index coverslips as described above. CellTracker CM-DiI (Thermo Fisher Scientific) was diluted to 1 mg/mL in DMSO, and diluted 1:1000 in PBS. The cells were incubated with this solution for 5 minutes at 37°C, 15 minutes at 4°C, and the solution replaced with media. Cells were then incubated at 37°C, 5% CO2 for 48 hours and imaged (Supplementary Fig. 14, Supplementary Video 15, 16).
Myosin imaging
For imaging moysin IIA bipolar filaments (Supplementary Fig. 9a, Supplementary Video 10), high index coverslips were plasma cleaned (PDC-001, Harrick Plasma) for 5 minutes, and then coated with 10 μg/ml human plasma fibronectin (Millipore, cat. # FC010) in PBS (Thermo Fisher Scientific). U2OS cells were cultured in McCoys media (Invitrogen) supplemented with 10% fetal calf serum (Thermo Fisher Scientific), at 37 °C in 5% CO2. Cells were transfected with GFP-myosin IIA expression and mApple-F-tractin plasmids as previously described35 and cultured for 12 hours prior to plating on fibronectin coated coverslips.
Actin imaging
For actin imaging (Supplementary Video 9), U2OS cells were cultured at 37°C in the presence of 5% CO2 in high glucose DMEM medium (Thermo Fisher Scientific) with 10% fetal bovine serum, 1% Pen/Strep and GlutaMAXTM (Thermo Fisher Scientific). Cells were seeded on high index coverslips and transfected with F-tractin EGFP by X-tremeGENE HP (Sigma-Aldrich) 24 hours prior to imaging.
Live INS-1 cells
For calcium imaging (Supplementary Video 7), INS-1 cells were cultured at 37°C with 5% CO2 in modified RPMI media (10% fetal bovine serum, 1% pen/strep, 11.1 mM glucose, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], 2 mM glutamine, 1 mM pyruvate, and 50 μM β-mercaptoethanol). Cells were seeded on high index coverslips that were cleaned by successive washing in detergent and bleach and then thoroughly rinsed with PBS. After PBS rinsing, coverslips with dipped in ethanol to sterilize and allowed to dry. Coverslips were treated with 0.1% poly-L-lysine (Sigma-Aldrich) for (5 to 10 minutes) followed by thorough rinsing with media. Cells were transfected with Lipofectamine 2000 (Thermo Fisher Scientific) using 1 μg of DNA per coverslip. For calcium imaging, cells were transfected with GCamp6S-CAAX and imaged one day after transfection.
Live SK-MEL cells
For FCHO (Supplementary Fig. 9b) imaging, SK-MEL cells were cultured in standard DMEM without phenol red (10% fetal bovine serum, 1% pen/strep, 1% GlutaMAX) at 37°C with 5% CO2. Cells were seeded and transfected (with FCHO2-GFP in this case) as described above for calcium imaging.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
We thank Steve Lee and Abhishek Kumar for useful discussion on the instant TIRF-SIM, Henry Eden and Patrick La Riviere for providing feedback on the manuscript, Chris Combs for loaning us the Leica alignment reticle, Ethan Tyler for help with the illustrations, William Bement for the gift of EMTB-3xEGFP, Dyche Mullins for the gift of pEGFP-C1 F-tractin-EGFP, Dominic Esposito for the gift of HaloTag chimera of HRas, and Luke Lavis for the gift of Janelia Fluor549. Support for this work was provided by the Intramural Research Programs of the National Institute of Biomedical Imaging and Bioengineering; the National Heart, Lung, and Blood Institute; A.U. and I.R. were supported by NSF grant 1607645. JT and HS acknowledge funding from the NIH Director’s Challenge Innovation Award Program.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Poulter NS, Pitkeathly WTE, Smith PJ & Rappoport JZ in Advanced Fluorescence Microscopy: Methods and Protocols Vol. 1251 Methods in Molecular Biology (ed Verveer Peter J) (Springer, 2015). [Google Scholar]
- 2.Fiolka R, Beck M & Stemmer A Structured illumination in total internal reflection fluorescence microscopy using a spatial light modulator. Optics Letters 33, 1629–1631 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Kner P, Chhun BB, Griffis ER, Winoto L & Gustafsson MGL Super-resolution video microscopy of live cells by structured illumination. Nat. Methods 6, 339–342 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung E, Kim D, Cui Y, Kim YH & So PT Two-dimensional standing wave total internal reflection fluorescence microscopy: superresolution imaging of single molecular and biological specimens. Biophys. J 93, 1747–1757 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gliko O, Reddy GD, Anvari B, Brownell WE & Saggau P Standing wave total internal reflection fluorescence microscopy to measure the size of nanostructures in living cells. J Biomed Opt 11, 064013 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Li D et al. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science 349, aab3500 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.York AG et al. Instant super-resolution imaging in live cells and embryos via analog image processing. Nat Methods 10, 1122–1126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roth S, Sheppard CJR, Wicker K & Heintzmann R Optical photon reassigment microscopy (OPRA). Optical Nanoscopy 2, 1–6 (2013). [Google Scholar]
- 9.De Luca GM et al. Re-scan confocal microscopy: scanning twice for better resolution. Biomed Opt Express 4, 2644–2656 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curd A et al. Construction of an instant structured illumination microscope.. Methods 15, 30029–30023 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stout AL & Axelrod D Evanescent field excitation of fluorescence by epi-illumination microscopy. Applied Optics 28, 5237–5242 (1989). [DOI] [PubMed] [Google Scholar]
- 12.Gould TJ, Myers JR & Bewersdorf J Total internal reflection STED microscopy. Opt Express 19, 13351–13357 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Burnette DT et al. A contractile and counterbalancing adhesion system controls the 3D shape of crawling cells. J Cell Biol. 205, 83–96 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sochacki KA, Dickey AM, Strub MP & Taraska JW Endocytic proteins are partitioned at the edge of the clathrin lattice in mammalian cells. Nat Cell Biol 19, 352–361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faire K et al. E-MAP-115 (ensconsin) associates dynamically with microtubules in vivo and is not a physiological modulator of microtubule dynamics.. J Cell Sci 112, 4243–4255 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Miller AL & Bement WM Regulation of cytokinesis by Rho GTPase flux Nat Cell Biol 11, 71–77 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apolloni A, Prior IA, Lindsay M, Parton RG & Hancock JF H-ras but Not K-ras Traffics to the Plasma Membrane through the Exocytic Pathway. Mol Cell Biol. 20, 2475–2487 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimm JB et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods 12, 244–250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Presley JF et al. ER-to-Golgi transport visualized in living cells. Nature 389, 81–85 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Choy E et al. Endomembrane Trafficking of Ras: The CAAX Motif Targets Proteins to the ER and Golgi. Cell 98, 69–80 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Takahashi S et al. Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J Cell Sci 125, 4049–4057 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Shen H et al. Single Particle Tracking: From Theory to Biophysical Applications. Chemical Reviews 117, 7331–7376 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Fiolka R Clearer view for TIRF and oblique illumination microscopy. Optics Express 24, 29556–29567 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Olveczky BP, Periasamy N & Verkman AS Mapping Fluorophore Distributions in Three Dimensions by Quantitative Multiple Angle-Total Internal Reflection Fluorescence Microscopy. Biophys. J 73, 2836–2847 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winter PW et al. Two-photon instant structured illumination microscopy improves the depth penetration of super-resolution imaging in thick scattering samples. Optica 1, 181–191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattheyses AL & Axelrod D Direct measurement of the evanescent field profile produced by objective-based total internal reflection fluorescence. Journal of Biomedical Optics 11, 014006 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Fu Y et al. Axial superresolution via multiangle TIRF microscopy with sequential imaging and photobleaching. Proc Natl Acad Sci U S A 113, 4368–4373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson WH Bayesian-Based Iterative Method of Image Restoration. JOSA 62, 55–59 (1972). [Google Scholar]
- 29.Lucy LB An iterative technique for the rectification of observed distributions. Astronomical Journal 79, 745–754 (1974). [Google Scholar]
- 30.Miura K, Rueden C, Hiner M, Schindelin J & Rietdorf J ImageJ Plugin CorrectBleach V2.0.2. Zenodo (2014). [Google Scholar]
- 31.Li D & Betzig E Response to Comment on “Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics”. Science 352, 527 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Thevenaz P et al. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process 7, 27–41 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Tinevez JY et al. TrackMate: An open and extensible platform for single-particle tracking.. Methods 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Choudhury A et al. Rab proteins mediate Golgi transport of caveola-internalized glycosphingolipids and correct lipid trafficking in Niemann-Pick C cells. J Clin Invest 109, 1541–1550 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baird MA et al. Local pulsatile contractions are an intrinsic property of the myosin 2A motor in the cortical cytoskeleton of adherent cells.. Mol Biol Cell 28, 240–251 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


