Abstract
The discovery of activating mutations in EGFR and KRAS in a subset of lung adenocarcinomas was a major advance in our understanding of lung adenocarcinoma biology, and has led to groundbreaking studies that have demonstrated the efficacy of tyrosine kinase inhibitor therapy. Fine-needle aspirates and other cytologic procedures have become increasingly popular for obtaining diagnostic material in lung carcinomas. However, frequently the small amount of material or sparseness of tumor cells obtained from cytologic preparations limit the number of specialized studies, such as mutation analysis, that can be performed. In this study we used laser capture microdissection to isolate small numbers of tumor cells to assess for EGFR and KRAS mutations from cell block sections of 19 cytology samples from patients with known lung adenocarcinomas. We compared our results with previous molecular assays that had been performed on either surgical or cytology specimens as part of the patient’s initial clinical work-up. Not only were we able to detect the identical EGFR or KRAS mutation that was present in the patient’s prior molecular assay in every case, but we were also able to consistently detect the mutation from as few as 50 microdissected tumor cells. Furthermore, isolating a more pure population of tumor cells resulted in increased sensitivity of mutation detection as we were able to detect mutations from laser capture microdissection-enriched cases where the tumor load was low and traditional methods of whole slide scraping failed. Therefore, this method can not only significantly increase the number of lung adenocarcinoma patients that can be screened for EGFR and KRAS mutations, but can also facilitate the use of cytologic samples in the newly emerging field of molecular-based personalized therapies.
Keywords: cytology, EGFR, KRAS, laser capture microdissection, lung adenocarcinoma, mutation
The emergence of targeted therapeutics in lung adenocarcinoma has revolutionized the field of personalized medicine and established a prognostic and predictive role for molecular analysis in conjunction with morphologic diagnosis in determining clinical outcomes of patients with advanced-stage disease.1–3 The epidermal growth factor receptor (EGFR) is known to play a role in the development and progression of cancer, and somatic mutations within the tyrosine kinase domain of EGFR have been identified in a subset of lung adenocarcinomas. The most common mutations are a point mutation c.2573T>G (L858R) in exon 21 and small in-frame deletions in exon 19, which result in constitutive activation of tyrosine kinase.4 What has made targeted therapy an exciting and developing field is that several clinical trials using tyrosine kinase inhibitors, gefitinib and erlotinib, have shown that patients with advanced lung adenocarcinomas harboring an EGFR mutation have a longer progression-free survival and response to tyrosine kinase inhibitors as a first-line therapy, whereas patients without these mutations have better outcomes with chemotherapy.2,4–10 Approximately 15–30% of lung adenocarcinomas also harbor activating mutations in the downstream GTPase, KRAS, most frequently found in codons 12 and 13 of exon 2. Mutations in EGFR and KRAS are mutually exclusive and recent studies indicate that patients with mutant KRAS tumors do not respond to tyrosine kinase inhibitors.11,12
A large fraction of lung carcinoma patients are diagnosed by cytology on fine-needle aspirates, pleural fluids, bronchial washes/brushes and bronchoalveolar lavages,13 and often cytology samples may be the only available material for molecular analysis. The majority of these patients who present with advanced-stage disease due to unresectable tumor or metastatic disease are increasingly being managed by targeted therapy. Minimally invasive procedures like endobronchial ultrasound-guided transbronchial needle aspiration are gaining popularity in the staging of advanced lung carcinoma patients because of the multitude of information that can be obtained from the aspirated material, including pathologic diagnosis and molecular testing.14,15 These minimally invasive techniques also allow for serial sampling of a patient’s tumor to assess therapeutic response as well as identify additional molecular markers of resistance. With this recent paradigm shift in lung cancer diagnosis and management, cytology has moved away from being a screening modality or ancillary technique and has established itself as an independent diagnostic procedure that plays a predictive role in determining clinical management.13,16
Mutation analysis, however, has been discouraged in cytology specimens because of scant material,17 and cytology specimens are often underutilized for decisions regarding targeted cancer therapy.18 In a recent study from Smouse et al19 over a period of 2 years, from a total of 239 cases tested for EGFR mutations, only 12 were cytology material. A similar review by Clark18 reports only 13 cytology cases from a total of 59 specimens tested for EGFR mutations. Nonetheless, as shown in the retrospective study by Smouse et al,19 mutation analysis of cytology cell block material shows similar or higher sensitivity in comparison with surgical specimens and was likely dependent on the proportion of tumor cells present in a given specimen.
Molecular analytical techniques have rapidly gained pace in recent years and high-throughput methods have changed the study of molecular events associated with pathological processes.20–22 However, molecular diagnosis is limited by the amount of tissue available for analysis and the number of tumor cells present within the sample. Tumor cell heterogeneity presents a challenge for molecular assays, where it is often necessary to isolate subpopulations of cells within a neoplasm to obtain a pure sample of tumor cells for DNA isolation and amplification. Laser capture microdissection provides a simple technique for rapid and accurate selection of pure populations of cells under direct microscopic visualization.23–26
In this study, we assessed the feasibility of using laser capture microdissection to enable and facilitate EGFR and KRAS mutation detection from cytologic samples (formalin-fixed, paraffin-embedded cell blocks), and compared the performance of this technique with results from standard mutational assays that had been previously performed on surgical or cytology materials as part of the routine clinical work-up. Our data indicate that laser capture microdissection-enabled mutation detection on cytologic material provides highly accurate and reproducible data comparable or superior to standard methods, and could be invaluable particularly when the tumor sample is limited.
Materials and methods
Clinical Samples
A total of 19 cytology specimens of either primary or metastatic lung adenocarcinomas from 14 patients were examined, including 9 fine-needle aspirates of the lung, 3 fine-needle aspirates of regional lymph nodes, 1 fine-needle aspirate of a distant metastasis (iliac crest), 1 bronchoalveolar lavage and 5 pleural fluids. All cases had cytomorphological evaluation of direct smears and/or cytospins as well as immunohistochemical studies (TTF-1 and Napsin A positivity) confirming the diagnosis. All specimens also had formalin-fixed, paraffin-embedded cell blocks that were cut into 4 μm sections and stained with hematoxylin and eosin (H&E) without a coverslip. Eight specimens were analyzed for EGFR mutations, seven specimens were analyzed for KRAS mutations and four specimens were analyzed for both EGFR and KRAS mutation status.
Laser Capture Microdissection
Laser capture microdissection was performed using an Arcturus XT (Life Technologies, Carlsbad, CA, USA). To improve visualization of the cells at the microscope, a drop of xylene was applied to the section. The tumor cells were identified by morphology, captured using a near-infrared laser pulse and transferred onto a cap (Capsuret™ Macro LCM Caps, Life Technologies, catalog number LCM0211). The DNA was extracted from the cap after overnight incubation with proteinase K buffer at 56 °C, using the QIAamp DNA micro kit (Qiagen, Valencia, CA, USA, catalog number 56304). Following proteinase K digestion, the buffer was incubated at 90 °C for 1 h and DNA was isolated and eluted in 20 μl of buffer following the kit manufacturer’s instructions.
PCR Amplification and Mutation Detection
Targeted analysis for EGFR mutation hot spots at codons 858, 861 and 863 within exon 21 was performed using pyrosequencing on a PyroMark Q24 instrument (Qiagen), whereas exon 19 deletions were assessed by capillary electrophoresis using a Genetic Analyzer 3130xl (Applied Biosystems). The pyrosequencing assay was designed using PyroMark Assay Design v2.0 (Qiagen). PCR amplification primers for exon 21 are as follows: EGFR-Ex21-FW 5′-biotin-GAGGACCGTCGCTTGGTG-3′ and EGFR-Ex21-REV 5′-TGCCTCCTTCTGCATGGTATTC-3′. Sequencing primer EGFR-Ex21-SEQ 5′-TGCATGGTATTCTTTCTC-3′ was used with nucleotide dispensation order 5′-CTCGCGAGTACATGCATGTAGCATGC-3′ to interrogate the sequence 5′-TTCCGCAYCCAGCNGTTTGGCCNGCCCA-3′. The primers for the exon 19 deletion assay were modified from the design by Pan et al27 to generate a shorter amplicon (154 bps). The primer sequences are as follows: EGFR-Ex19-FW 5′-ACTCTGGATCCCAGAAGGTGAGA-3′ and EGFR-Ex19-REV 5′-fam-AAAGGTGGGCCTGAGGTTCA.
Targeted analysis for KRAS mutation hot spots in codons 12 and 13 within exon 2 was performed using the PyroMark Q24 KRAS v2.0 kit (Qiagen), as originally described by Ogino et al.28
PCR reactions were conducted in a total volume of 25 μl containing genomic DNA template, 200 nM of each forward and reverse primers, 12.5 μl 2× HotStarTaq Master Mix (Qiagen). PCR cycling conditions for EGFR assays were 95 °C, 15 min; 40× (95 °C, 30 s; 60 °C, 1 min; 72 °C, 1 min), 72 °C, 10 min; 8 °C, hold. COLD-PCR conditions for KRAS were 95 °C 15 min, 10× (95 °C 20 s, 53 °C 30 s, 72 °C 20 s), 72 °C 5 min, 95 °C 2 min, 35× (95 °C 20 s, 70 °C 8 min, 80 °C 3 s, 53 °C 30 s, 72 °C 20 s), 72 °C 5 min, 8 °C hold. For EGFR deletion detection, 1 μl of the 100-fold diluted PCR product was analyzed by capillary electrophoresis on an ABI 3130xl Genetic Analyzer. For the pyrosequencing assays, 10 μl of PCR product was immobilized on streptavidin-coated Sepharose beads (GE Healthcare) and processed according to the manufacturer’s instructions.
Results
Patient and Tumor Characteristics
The clinical data for all cases analyzed are summarized in Table 1. Eight patients were male, ranging in age from 32 to 76 years, of which four had adenocarcinomas with gene mutations in either EGFR or KRAS that had been previously established by molecular assays performed on either cytology or surgical biopsies (Table 3). Six patients were female, ranging in age from 51 to 68 years, and all six cases had previously documented mutations in either EGFR or KRAS on cytology or surgical pathology material (Table 3). Most patients had advanced-stage lung carcinomas and received multiple modalities of treatment. The clinical management available at the time of our study is also summarized in Table 1.
Table 1.
Clinical characteristics of lung adenocarcinoma patients analyzed for EGFR and KRAS mutations
| Gender | Age (years) | Stage | Smoking history | Management | |
|---|---|---|---|---|---|
| 1 | M | 55 | IB | Yes | Chemo/Rad/Surgery/Erlotinib |
| 2 | M | 75 | IV | Yes | Chemo |
| 3 | M | 56 | IV | No | Surgery/Chemo/Erlotinib |
| 4 | F | 67 | IIIA | Yes | Surgery/Rad/Chemo/Erlotinib |
| 5 | F | 61 | IV | No | Erlotinib |
| 6 | F | 62 | IV | No | Chemo/Erlotinib |
| 7 | F | 68 | IV | Yes | Chemo |
| 8 | F | 51 | IIA | Yes | Chemo/Rad |
| 9 | M | 63 | IV | Yes | Chemo |
| 10 | F | 58 | IV | Yes | Chemo |
| 11 | M | 37 | I | Yes | NA |
| 12 | M | 76 | III | No | NA |
| 13 | M | 32 | IV | No | Chemo/Rad/Erlotinib |
| 14 | M | 62 | IV | Yes | Surgery/Chemo |
Abbreviations: M, male; F, female; Chemo, chemotherapy; Rad, radiation; NA, not available.
Table 3.
Summary of EGFR and KRAS mutational analysis of laser capture microdissection-assisted cytology specimens in comparison with previously performed assays on surgical or cytology specimens
| Patient | Case | LCM source (C) | No. of cells | LCM mutation | Original source | Original mutation |
|---|---|---|---|---|---|---|
| 1 | 1 | Lung | 300 | EGFR c.2573T>G | Lung (S) | EGFR c.2573T>G |
| 2 | Lung | 300 | EGFR c.2573T>G | |||
| 2 | 9 | Lung | 300 | EGFR WT | Lung (C) | EGFR WT |
| 13 | Lung | 50 | KRAS 12 c.34G> A | KRAS 12 c.34G> A | ||
| 3 | 3 | Lung | 300 | EGFR Ex19del21 | Lung (C) | EGFR Ex19del21 |
| 4 | 4 | PLFL | 300 | EGFR c.2573T>G | Lung (S) | EGFR c.2573T>G |
| 5 | 5 | PLFL | 300 | EGFR c.2573T>G | PLFL (C) | EGFR c.2573T>G |
| 6 | 6 | Lung | 300 | EGFR c.2573T>G | LN (S) | EGFR c.2573T>G |
| 7 | LN | 50 | EGFR c.2573T>G | |||
| 8 | BAL | 300 | EGFR c.2573T>G | |||
| 7 | 14 | LN | 50 | KRAS12 c.34_35GG>TT | LN (S) | KRAS12 c.34_35GG>TT |
| 8 | 15 | Lung | 50 | KRAS12 c.34G> T | Lung (S) | KRAS12 c.34G>T |
| 9 | 16 | LN | 50 | KRAS 12 c.35G> A | Lung (S) | KRAS 12 c.35G>A |
| 10 | 12 | Bone | 50 | EGFR WT | Bone (S) | EGFR WT |
| 17 | Bone | 50 | KRAS 12 c.35G>A | KRAS 12 c.35G>A | ||
| 11 | 10 | PLFL | 50 | EGFR WT | Lung (S) | EGFR WT |
| 20 | PLFL | 50 | KRAS WT | KRAS WT | ||
| 12 | 11 | Lung | 50 | EGFR WT | Lung (S) | EGFR WT |
| 21 | Lung | 50 | KRAS WT | KRAS WT | ||
| 13 | 18 | Lung | 50 | KRAS WT | Lung (S) | KRAS WT |
| 19 | PLFL | 50 | KRAS WT | |||
| 22 | PLFL | 50 | KRAS WT | PLFL (C) | KRAS WT | |
| 14 | 23 | Lung | 50 | KRAS WT | PLFL (C) | KRAS WT |
Abbreviations: LCM, laser capture microdissection; C, cytology specimen; S, surgical biopsy; PLFL, pleural fluid; LN, lymph node; BAL, bronchoalveolar lavage; WT, wild type.
Estimating the Minimal Number of Cells Required for Mutational Analyses
To assess the minimal number of cells required to successfully run mutation analysis, we microdissected decreasing numbers of tumor cells from cell block sections of four cytology cases and compared the results with the data from the original clinical work-up (Figure 1). For one case we had 300, 250, 200, 150, 100 and 50 cells, and for the remaining three cases we used 300, 100 and 50 cells (Table 2). Two of the cases had EGFR mutations in exon 21 (c.2573T>G) and two had KRAS mutations in codon 12. The mutations were consistently detected in all the samples tested, including samples containing only 50 cells, and the results were comparable to the original mutation analyses (Table 2). This suggests that mutation analyses for both EGFR and KRAS could be consistently performed with as few as 50 tumor cells. Below 50 cells there was significant reduction in signal strength; therefore, we did not perform mutation analysis for samples with <50 cells. We subsequently performed our remaining mutation analyses, 3 cases for EGFR and 9 cases for KRAS, using ~50 tumor cells and successfully amplified DNA (either wild type or mutant), comparable to previous data (Table 3). These results demonstrate that as few as 50 tumor cells, either in groups or as individual cells dispersed within a cell block, are sufficient to detect EGFR or KRAS mutations.
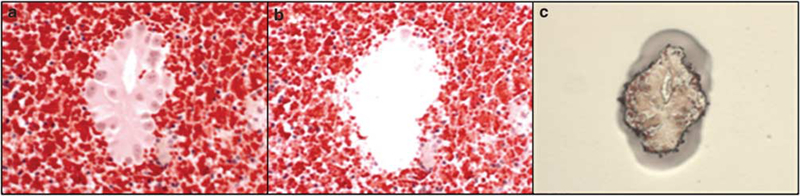
Figure 1.
Laser capture microdissection of tumor cells. (a) H&E-stained cell block section showing lung adenocarcinoma cells before microdissection. (b) Same section after microdissection. (c) Tumor cells following microdissection on the cap.
Table 2.
Quantitative assay for estimating the minimal number of cells required for mutational analysis
| Patient | LCM source | Number of cells | Mutation detected |
|---|---|---|---|
| 1 | FNA lung | 300 | EGFR c.2573T>G |
| 250 | EGFR c.2573T>G | ||
| 200 | EGFR c.2573T>G | ||
| 150 | EGFR c.2573T>G | ||
| 100 | EGFR c.2573T>G | ||
| 50 | EGFR c.2573T>G | ||
| 6 | FNA lymph node | 300 | EGFR c.2573T>G |
| 100 | EGFR c.2573T>G | ||
| 50 | EGFR c.2573T>G | ||
| 7 | FNA lymph node | 300 | KRAS12 c.34_35GG>TT |
| 100 | KRAS12 c.34_35GG>TT | ||
| 50 | KRAS12 c.34_35GG>TT | ||
| 8 | FNA lung | 300 | KRAS12 c.34G> T |
| 100 | KRAS12 c.34G> T | ||
| 50 | KRAS12 c.34G> T | ||
Abbreviations: LCM, laser capture microdissection; FNA, fine-needle aspiration.
Gene Sequencing and Mutation Status
A total of 12 samples were tested for EGFR mutation status. Eight cases showed EGFR mutations, with seven cases containing a point mutation c.2573T>G (L858R) in exon 21 and one case with an in-frame deletion in exon 19. Four cases were wild type for EGFR. All 12 cases had mutation status analyses that had been previously documented on surgical biopsies or cytology material that were in concordance with the current results (Table 3).
For KRAS mutation, 11 samples were analyzed. Five cases showed mutations in codon 12, with two cases showing c.35G>A, one case showing c.34G>A, one case showing c.34_35GG>TT and one case showing c.34G>T. Six cases were wild type for KRAS. All the cases had mutation status analyses that had been previously documented on surgical biopsies or cytology material that were identical to the current results. Table 3 shows a summary of the mutation analyses performed for EGFR and KRAS along with the original molecular test data that had been previously performed.
Laser Capture Microdissection Increased Sensitivity of Mutation Detection
To evaluate the effectiveness of selectively using a pure population of tumor cells by laser capture microdissection, three cases (two pleural fluids: cases 4 and 5, and one bronchoalveolar lavage: case 8) with approximately ≤20% tumor cells were selected for mutation analysis (Figure 2a). We compared mutation assays following whole slide scraping of cell block sections (4 slides) with laser capture microdissection-assisted analysis from a single slide (~300 cells). We failed to detect the mutation from the standard method of whole slide scraping; however, the laser capture microdissection-assisted analysis was able to identify the appropriate mutation for all three cases (Figure 2b and c). This suggests that in cases where the tumor load is low and tumor cells are sparse and dispersed, isolating a more pure population of tumor cells can yield a more sensitive method for mutation detection in comparison with standard methods.
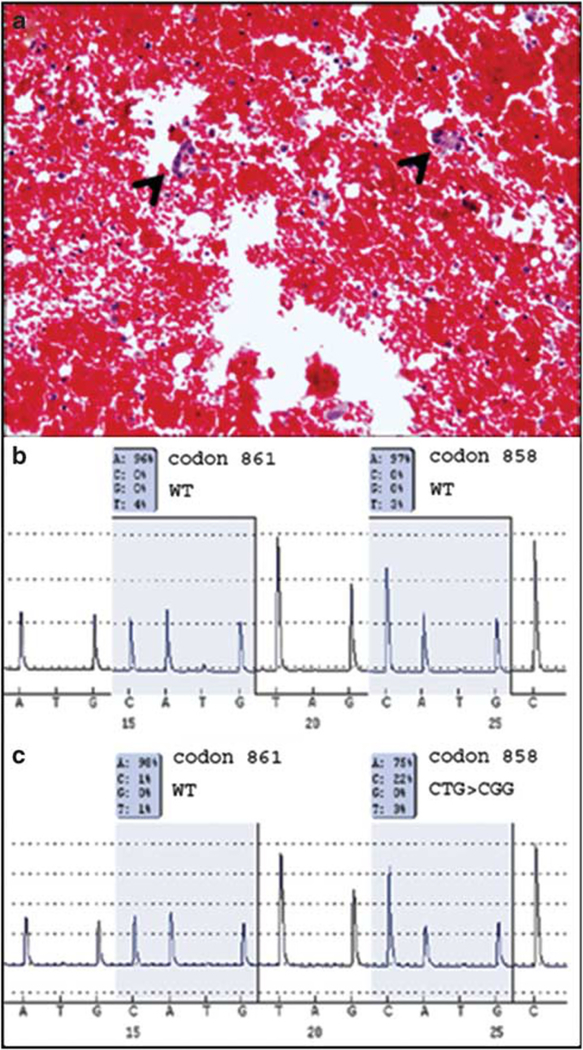
Figure 2.
Laser capture microdissection increases sensitivity of mutation detection. (a) H&E-stained cell block section showing scattered tumor cells (arrow head) in a background of red blood cells, lymphocytes and mesothelial cells. (b) Pyrogram from EGFR mutation assay for exon 21 from a whole slide scrape of the same case showing wild type. (c) Pyrogram from EGFR mutation assay for exon 21 following laser capture microdissection showing detection of c.2573 T>G (L858R) mutation.
Discussion
It is becoming routine clinical practice to perform mutational analyses for prognostic and predictive markers in tumor samples of lung adenocarcinoma patients. The majority of lung cancer patients are diagnosed at later stages that preclude surgical resection, and in advanced-stage lung cancer, platinum-based combined chemotherapy is effective in ~30% of cases.29 Detecting activating mutations in EGFR plays an essential role in determining responsiveness to tyrosine kinase inhibitors for targeted therapeutics. Clinical decisions regarding the use of different agents in patients with lung adenocarcinomas might be improved by pretreatment mutational profiling of both EGFR and KRAS.
The diagnosis of lung cancer is often based solely on cytology specimens.16,30 As often the number of tumor cells is limited on cytology specimens, it is critical to develop sensitive assays for mutation detection from small samples. Previous studies have shown that direct sequencing cannot consistently detect mutant DNA in specimens with <50% tumor cells and can rarely detect mutations in specimens with <25% tumor cells.19 For mutation analysis, enrichment of tumor cells is important to avoid diluting tumor DNA with the nonmutated DNA of benign cells, which in most cases represents the larger proportion of the cell population. Manual microdissection has been previously used on cytology specimens to extract DNA for EGFR mutation analysis;31 however, samples with lower numbers of tumor cells were rejected or reported as inconclusive because of technical difficulties of manually isolating a pure population of malignant cells. Laser microdissection has been successfully employed by other groups using a laser pressure catapulting system32,33 on Papanicolaou-stained cytology material or paraffin-embedded samples with very small groups of tumor cells. In our hands, using direct lysates of tumor cells interfered with the consistent sensitivity of the mutation assay, and incorporating the DNA extraction step resulted in better DNA quality (data not shown), suggesting that the DNA extraction step is important in removing potential PCR inhibitors.
In this study we validated a method for mutation analysis in cytology cell block samples containing as few as 50 tumor cells using laser capture microdissection. Laser capture microdissection can be successfully used to selectively isolate tumor cells, even in cases where the stochastic distribution of tumor and benign cells precludes more traditional methods of enrichment such as scraping whole samples or manual microdissection with a needle. There have been arguments against using extremely small amounts of DNA for PCR-based mutational analysis as artifactual mutations have been described especially in DNA extracted from paraffin-embedded tissue.34–37 However, improved methods using DNA extraction kits specially adapted for laser capture microdissected specimens from paraffin-embedded tissues, and more sensitive and reliable detection methods such as pyrosequencing and using small PCR amplicons (150–200 bp) have improved downstream analysis for samples with small amounts of DNA. To ensure we did not interpret false-positive results as true mutations, all our assays were performed in duplicate. Our method not only detected mutations consistently from 50 tumor cells, but also detected the exact same mutation that had been previously identified in each of these patients. It is important to note that number of tumor cells does not necessarily equate to number of copies of the gene target, particularly for EGFR for which mutations often occur in an amplified allele.38 In our experience, selecting and microdissecting a tumor population of only 50 tumor cells is fast, reproducible, specific and quite feasible in the clinical set-up. Laser capture microdissection increased the sensitivity of mutation detection in specimens with limited number of tumor cells, and therefore could significantly increase the number of patients who can be screened for both EGFR and KRAS mutations.
New diagnostic modalities like endobronchial ultrasound-guided transbronchial needle aspiration are increasingly being used to stage lung cancer through evaluation of mediastinal and hilar lymph nodes for metastasis.39 These new minimally invasive sampling procedures provide opportunities, but to fully take advantage of their promise, more sensitive analytic approaches capable of interrogating very small amounts of tissue must be developed. As the field moves from simple mutational analysis of one or a few genes to more challenging assays like whole pathway DNA mutation testing and complex expression-based assays using proteomic or transcriptomic measurements, the need for simple and reliable methods of isolating relatively pure cell populations will become essential,40 and the use of microdissection is likely to be an integral component of such approaches.
In conclusion, our study demonstrates that laser capture microdissection-assisted EGFR and KRAS mutation analysis from cytology cell block samples provides results that match those obtained from whole histology/cytology slide scrapes typically used for clinical molecular diagnostic testing. We have shown that EGFR and KRAS mutation analysis can be performed consistently and reproducibly with as few as 50 tumor cells for both assays, and that tumor cell enrichment attainable through the use of laser capture microdissection allows for more sensitive mutation detection. Highly sensitive and reproducible approaches to mutation analysis, such as the method described here, should facilitate the use of cytologic materials for the molecular testing that underpins the newly emerging molecular-based personalized therapies.
Acknowledgement
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute.
Disclosure/conflict of interest
Michael Emmert-Buck is an inventor on all NIH-held patents covering laser capture microdissection technology and receives royalty payments through the NIH technology transfer program.
References
- 1.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol 2005;23:5900–5909. [DOI] [PubMed] [Google Scholar]
- 2.Jackman DM, Miller VA, Cioffredi LA, et al. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated nonsmall cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res 2009;15:5267–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.West H, Lilenbaum R, Harpole D, et al. Molecular analysis-based treatment strategies for the management of non-small cell lung cancer. J Thorac Oncol 2009;4:S1029–S1039; quiz S41–2. [DOI] [PubMed] [Google Scholar]
- 4.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA 2004;101:13306–13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129–2139. [DOI] [PubMed] [Google Scholar]
- 6.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497–1500. [DOI] [PubMed] [Google Scholar]
- 7.Janne PA, Engelman JA, Johnson BE. Epidermal growth factor receptor mutations in non-small-cell lung cancer: implications for treatment and tumor biology. J Clin Oncol 2005;23:3227–3234. [DOI] [PubMed] [Google Scholar]
- 8.Han SW, Kim TY, Hwang PG, et al. Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small-cell lung cancer patients treated with gefitinib. J Clin Oncol 2005;23:2493–2501. [DOI] [PubMed] [Google Scholar]
- 9.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947–957. [DOI] [PubMed] [Google Scholar]
- 10.Rosell R, Taron M, Reguart N, et al. Epidermal growth factor receptor activation: how exon 19 and 21 mutations changed our understanding of the pathway. Clin Cancer Res 2006;12:7222–7231. [DOI] [PubMed] [Google Scholar]
- 11.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med 2005;2:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Massarelli E, Varella-Garcia M, Tang X, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res 2007;13:2890–2896. [DOI] [PubMed] [Google Scholar]
- 13.Malapelle U, Bellevicine C, Zeppa P, et al. Cytology-based gene mutation tests to predict response to anti-epidermal growth factor receptor therapy: a review. Diagn Cytopathol 2011;39:703–710. [DOI] [PubMed] [Google Scholar]
- 14.Nakajima T, Yasufuku K. How I do it–optimal methodology for multidirectional analysis of endobronchial ultrasound-guided transbronchial needle aspiration samples. J Thorac Oncol 2011;6:203–206. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Olive I, Monso E, Andreo F, et al. Endobronchial ultrasound-guided transbronchial needle aspiration for identifying EGFR mutations. Eur Respir J 2010;35:391–395. [DOI] [PubMed] [Google Scholar]
- 16.Travis WD, Rekhtman N, Riley GJ, et al. Pathologic diagnosis of advanced lung cancer based on small biopsies and cytology: a paradigm shift. J Thorac Oncol 2010;5:411–414. [DOI] [PubMed] [Google Scholar]
- 17.Sequist LV, Bell DW, Lynch TJ, et al. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol 2007;25:587–595. [DOI] [PubMed] [Google Scholar]
- 18.Clark DP. Seize the opportunity: underutilization of fine-needle aspiration biopsy to inform targeted cancer therapy decisions. Cancer Cytopathol 2009;117: 289–297. [DOI] [PubMed] [Google Scholar]
- 19.Smouse JH, Cibas ES, Janne PA, et al. EGFR mutations are detected comparably in cytologic and surgical pathology specimens of nonsmall cell lung cancer. Cancer Cytopathol 2009;117:67–72. [DOI] [PubMed] [Google Scholar]
- 20.Asano H, Toyooka S, Tokumo M, et al. Detection of EGFR gene mutation in lung cancer by mutant-enriched polymerase chain reaction assay. Clin Cancer Res 2006;12:43–48. [DOI] [PubMed] [Google Scholar]
- 21.Janne PA, Borras AM, Kuang Y, et al. A rapid and sensitive enzymatic method for epidermal growth factor receptor mutation screening. Clin Cancer Res 2006;12:751–758. [DOI] [PubMed] [Google Scholar]
- 22.Smith GD, Chadwick BE, Willmore-Payne C, et al. Detection of epidermal growth factor receptor gene mutations in cytology specimens from patients with non-small cell lung cancer utilising high-resolution melting amplicon analysis. J Clin Pathol 2008;61: 487–493. [DOI] [PubMed] [Google Scholar]
- 23.Harrell JC, Dye WW, Harvell DM, et al. Contaminating cells alter gene signatures in whole organ versus laser capture microdissected tumors: a comparison of experimental breast cancers and their lymph node metastases. Clin Exp Metastasis 2008;25:81–88. [DOI] [PubMed] [Google Scholar]
- 24.Emmert-Buck MR, Bonner RF, Smith PD, et al. Laser capture microdissection. Science 1996;274:998–1001. [DOI] [PubMed] [Google Scholar]
- 25.Fend F, Raffeld M. Laser capture microdissection in pathology. J Clin Pathol 2000;53:666–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Serag HB, Nurgalieva ZZ, Mistretta TA, et al. Gene expression in Barrett’s esophagus: laser capture versus whole tissue. Scand J Gastroenterol 2009;44:787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Q, Pao W, Ladanyi M. Rapid polymerase chain reaction-based detection of epidermal growth factor receptor gene mutations in lung adenocarcinomas. J Mol Diagn 2005;7:396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogino S, Kawasaki T, Brahmandam M, et al. Sensitive sequencing method for KRAS mutation detection by pyrosequencing. J Mol Diagn 2005;7:413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gandara DR, Lara PN Jr, Mack P, et al. Individualizing therapy for non-small-cell lung cancer: a paradigm shift from empiric to integrated decision-making. Clin Lung Cancer 2009;10:148–150. [DOI] [PubMed] [Google Scholar]
- 30.Rivera MP, Detterbeck F, Mehta AC. Diagnosis of lung cancer: the guidelines. Chest 2003;123:129S–136S. [DOI] [PubMed] [Google Scholar]
- 31.Boldrini L, Gisfredi S, Ursino S, et al. Mutational analysis in cytological specimens of advanced lung adenocarcinoma: a sensitive method for molecular diagnosis. J Thorac Oncol 2007;2:1086–1090. [DOI] [PubMed] [Google Scholar]
- 32.Savic S, Tapia C, Grilli B, et al. Comprehensive epidermal growth factor receptor gene analysis from cytological specimens of non-small-cell lung cancers. Br J Cancer 2008;98:154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molina-Vila MA, Bertran-Alamillo J, Reguart N, et al. A sensitive method for detecting EGFR mutations in nonsmall cell lung cancer samples with few tumor cells. J Thorac Oncol 2008;3:1224–1235. [DOI] [PubMed] [Google Scholar]
- 34.Marchetti A, Felicioni L, Buttitta F. Assessing EGFR mutations. N Engl J Med 2006;354:526–528; author reply −8. [DOI] [PubMed] [Google Scholar]
- 35.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med 2005;353:133–144. [DOI] [PubMed] [Google Scholar]
- 36.Quach N, Goodman MF, Shibata D. In vitro mutation artifacts after formalin fixation and error prone translesion synthesis during PCR. BMC Clin Pathol 2004;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams C, Ponten F, Moberg C, et al. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol 1999; 155:1467–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li AR, Chitale D, Riely GJ, et al. EGFR mutations in lung adenocarcinomas: clinical testing experience and relationship to EGFR gene copy number and immunohistochemical expression. J Mol Diagn 2008;10:242–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yasufuku K, Chiyo M, Koh E, et al. Endobronchial ultrasound guided transbronchial needle aspiration for staging of lung cancer. Lung Cancer 2005;50: 347–354. [DOI] [PubMed] [Google Scholar]
- 40.Silvestri A, Colombatti A, Calvert VS, et al. Protein pathway biomarker analysis of human cancer reveals requirement for upfront cellular-enrichment processing. Lab Invest 2010;90:787–796. [DOI] [PubMed] [Google Scholar]