Abstract
COVID-19 positive (194) and negative (212) pneumonia patients were selected to analyze bacterial pathogens coinfection. Results showed that 50% of COVID-19 patients were coinfected or carried bacterial pathogens. Bordetella pertussis infection rate was significantly higher in positive patients. Consequently, preventions should be taken to control bacterial pathogens coinfection in COVID-19 patients.
Keywords: COVID-19, Respiratory bacterial pathogen, Bordetella pertussis, Pneumonia
Up to 10 CEST, 24 August 2020, 23,311,719 COVID-19 infected patients were confirmed in the world according to the report of WHO (2019). It has attracted great attention from all over the world. COVID-19 is a respiratory pathogen, and its infection could result in severe pneumonia (Zhu et al., 2020). Coinfection with bacterial pathogens was one of the reasons why COVID-19 is fatal (Cox et al., 2020; Jose and Desai, 2020). Identification of coinfection pathogens plays a great important role in treatment of COVID-19 infected patients. Recently, COVID-19 infected pneumonia patients with coinfection of Mycoplasma pneumonia and Legionella pneumophila were found in Qingdao and Wuhan (Xing et al., 2020). However, the sample size was limited, and only several bacterial pathogens were analyzed. Moreover, COVID-19 infected samples were not compared with non–COVID-19 infected samples in the previous study. Hence, bacterial pathogens’ dynamic change in COVID-19 positive pneumonia patients could not be identified. In this study, 406 samples including COVID-19 positive and negative pneumonia patients were collected, and 14 respiratory bacterial pathogens were tested. The study aims to more accurately evaluate the coinfection of COVID-19 with common bacterial pathogens.
During February 8 to February 15, 2020, in Suizhou city, Hubei province, 2216 pneumonia patients were diagnosed based on chest X-ray or computed tomography (small slide shadows and interstitial change, obvious extrapulmonary bands, or even ground-glass opacity and infiltration shadows, lung consolidation, and pleural effusion), blood routine (reduction of lymphocytes), and blood biochemistry (increase of alanine aminotransferase, aspartate aminotransferase, lactate dehydrogenase, myozyme, myoglobin, and C-reactive protein, and normal procalcitonin) tests. These patients were admitted to hospital after being diagnosed as pneumonia patients. Then, the COVID-19 pneumonia patients were determined by throat swab nucleic acid test in Suizhou Center for Disease Control and Prevention. Total nucleic acids (DNA and RNA) were extracted from throat swab samples by Liferiver Nucleic acids extraction Kit (Shanghai ZJ Bio-Tech Co., Ltd.). The COVID-19 nucleic acid test was based on real-time polymerase chain reaction (PCR) according to the Chinese Center for Disease Control and Prevention's proposal (Chinese National Health Commission, 2020), and the used primers were shown in Table S1. Among them, there were 292 COVID-19 positive and 1924 COVID-19 negative pneumonia patients. In this study, 194 cases (47.78%) were selected from 292 positive cases randomly and 212 cases (52.22%) from 1924 negative cases randomly. Among COVID-19 positive patients, 99 (51.03%) were male and 94 (48.45%) were female, while the males and females in the COVID-19 negative patients were 119 (56.13%) and 93 (43.87%), respectively. The age range was 1–88 years old including 12 children, and the average age was 45 years old.
The residual total nucleic acids (DNA and RNA) after COVID-19 diagnosis were sent to Hubei Center for Disease Control and Prevention for bacterial pathogens analysis from Suizhou Center for Disease Control and Prevention. Chlamydia pneumonia, Streptococcus pneumonia, Bordetella pertussis, Streptococcus pyogenes, Staphylococcus aureus, Corynebacterium diphtheria, L. pneumophila, M. pneumonia, Mycobacterium tuberculosis, Neisseria meningitides, Haemophilus influenza, Streptococcus agalactiae, Pseudomonas aeruginosa, and Moraxella catarrhalis were detected by real-time PCR (Hirama et al., 2011; Schwartz et al., 2006; World Health Organization, 2011; Yang et al., 2015). All the primers and probes used in this study were shown in Table S1. The χ 2 test was carried out by SPSS version 16.0.
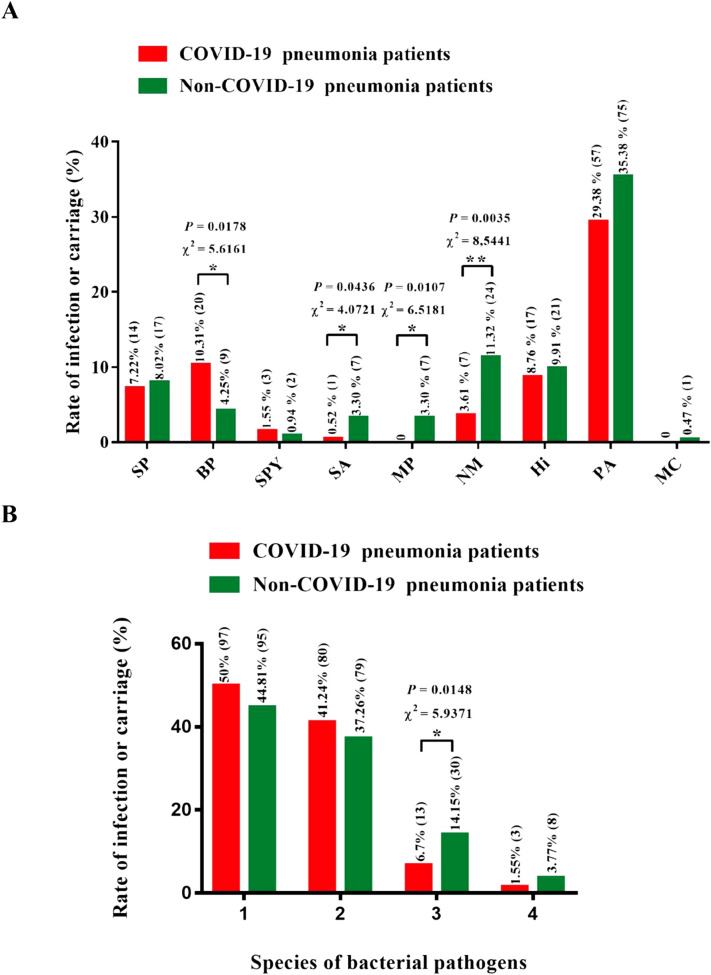
S. pneumonia, B. pertussis, S. pyogenes, S. aureus, N. meningitides, H. influenza, and P. aeruginosa were detected in both the COVID-19 positive and negative patients, while M. pneumonia and M. catarrhalis were only found in COVID-19 negative patients (Table 1 ). B. pertussis infection rate in COVID-19 positive patients was 10.31% (including 20 adult patients), which was significantly higher than 4.25% (including 2 children and 7 adult patients) in COVID-19 negative patients (χ 2 = 5.6161, P = 0.0178) (Fig. 1A). Pertussis vaccination status was not available among the cohort. Furthermore, there were 5 deaths in the COVID-19 positive patients. In these 5 patients, 2 were coinfected with B. pertussis and 3 patients were detected with P. aeruginosa (Table S2). One of the 2 patients was first infected by COVID-19 and then infected by B. pertussis and P. aeruginosa (Table S3). The bacterial coinfection might be one of the reasons why disease worsened among these 3 patients who died. B. pertussis causes cough to spread the pathogens in the air, which can further enhance the pathogen’s transmission (Bisgard et al., 2004). It also has the ability to inhibit the function of the host's immune system and cause various respiratory disorders (Carbonetti, 2007). Accordingly, B. pertussis should be regarded with more care in the treatment of COVID-19. The infection or carriage rates of M. pneumonia and N. meningitides in COVID-19 positive patients were significantly lower comparing to the COVID-19 negative patients (Fig. 1A). This might be explained by the fact that moxifloxacin and some Chinese traditional medicines (Huoxiang Zhengqi capsules, Jinhua Qinggan granule, Lianhua Qingwen capsule, Shufeng Jiedu capsules, and Fangfeng Tongsheng Wan) were used for treatment in all pneumonia patients during the period (Chinese National Health Commission, 2020; Medical expert group of Tongji Hospital Affiliated to Tongji Medical College of Huazhong University of Science and Technology, 2020). Among the positive patients, some of them had been given this treatment for 5–9 days already (the second time for COVID-19 test), and the others had taken this treatment for 1–2 days (the first time for COVID-19 test). However, all the negative patients accepted this treatment for only 1–2 days (the first time for COVID-19 test). M. pneumonia and N. meningitides were both sensitive to moxifloxacin, and these Chinese traditional medicines are capable of antibacterial functions according to the reports (Grayson, 2010; Zhang et al., 2019). Hence, the longer time treatment might reduce the infection of M. pneumonia and N. meningitides according to this result. The rate of P. aeruginosa carriage was quite high in both COVID-19 positive (29.38%) and negative (35.38%) patients compared to other pathogens (Fig. 1A). This suggested that the P. aeruginosa carriage in this area was very high and might not be reduced by moxifloxacin and these Chinese traditional medicines. It is necessary to prevent P. aeruginosa infection in the treatment of COVID-19 because it is an opportunistic pathogen and its infection is common in some pulmonary diseases (Faure et al., 2018). In addition, none of C. pneumonia, C. diphtheria, L. pneumophila, M. tuberculosis, and S. agalactiae was detected in both COVID-19 positive and negative patients. L. pneumophila coinfection with COVID-19 in Wuhan and Qingdao (Xing et al., 2020) might be due to high-level contamination of L. pneumophila compared to Suizhou.
Table 1.
Respiratory bacterial pathogen spectrum analysis.
| Sample source | Age (frequency) | Gender (frequency) | Frequency of pathogen carriage |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CP | SP | BP | SPY | SA | CD | LP | MP | MT | NM | Hi | SAG | PA | MC | |||
| COVID 19 positive pneumonia patients | 0–10 (1) | Male (0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Female (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 10–18 (1) | Male (0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Female (1) | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 18–45 (78) | Male (40) | 0 | 3 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 14 | 0 | |
| Female (38) | 0 | 6 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 8 | 0 | ||
| 45–65 (94) | Male (52) | 0 | 2 | 4 | 1 | 1 | 0 | 0 | 0 | 0 | 4 | 6 | 0 | 16 | 0 | |
| Female (42) | 0 | 1 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 14 | 0 | ||
| 65– (20) | Male (7) | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 1 | 0 | |
| Female (13) | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 0 | 4 | 0 | ||
| Total (194) | Male (99) | 0 | 6 | 12 | 2 | 1 | 0 | 0 | 0 | 0 | 4 | 11 | 0 | 31 | 0 | |
| Female (95) | 0 | 8 | 8 | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 6 | 0 | 26 | 0 | ||
| COVID 19 negative pneumonia patients | 0–10 (11) | Male (7) | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 | 0 | 5 | 1 |
| Female (4) | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 2 | 0 | ||
| 10–18 (4) | Male (2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | |
| Female (2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | ||
| 18–45 (96) | Male (60) | 0 | 6 | 3 | 1 | 3 | 0 | 0 | 2 | 0 | 7 | 5 | 0 | 19 | 0 | |
| Female (36) | 0 | 1 | 1 | 1 | 2 | 0 | 0 | 2 | 0 | 4 | 0 | 0 | 12 | 0 | ||
| 45–65 (80) | Male (40) | 0 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 5 | 0 | 17 | 0 | |
| Female (40) | 0 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 4 | 5 | 0 | 11 | 0 | ||
| 65– (21) | Male (10) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | |
| Female (11) | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | ||
| Total (212) | Male (119) | 0 | 12 | 5 | 1 | 3 | 0 | 0 | 3 | 0 | 15 | 13 | 0 | 45 | 1 | |
| Female (93) | 0 | 5 | 4 | 1 | 4 | 0 | 0 | 4 | 0 | 9 | 8 | 0 | 30 | 0 | ||
CP = C. pneumonia; SP = S. pneumonia; BP = B. pertussis; SPY = S. pyogenes; SA = S. aureus; CD = C. diphtheria; LP = L. pneumophila; MP = M. pneumonia; MT = M. tuberculosis; NM = N. meningitides; Hi = H. influenza; SAG = S. agalactiae; PA = P.aeruginosa; MC = M. catarrhalis.
Fig. 1.
Infection or carriage rate of specific bacterial pathogens (A) and multiple bacterial pathogens in COVID-19 and non–COVID-19 infected pneumonia patients (B). SP, BP, SPY, SA, MP, NM, Hi, PA, and MC stand for S. pneumonia, B. pertussis, S. pyogenes, S. aureus, M. pneumonia, N. meningitides, H. influenza, P. aeruginosa, and M. catarrhalis, respectively.
In total, 50% of COVID-19 positive patients were coinfected or carried common bacterial pathogens (Fig. 1B). Among them, 41.24% were coinfected or carried 1 species of bacterial pathogen, while 8.25% were coinfected or carried 2 or 3 species of bacterial pathogens (Fig. 1B). The rate of coinfection or carrying 2 species of bacterial pathogens in COVID-19 negative patients was significantly higher than that in COVID-19 positive patients (Fig. 1B). Meanwhile, 1- and 3-species bacterial pathogens co-infection or carriage rate showed no difference between COVID-19 positive and negative patients.
Although there are some limitations in this study including 1) bacterial culture and susceptibility studies were not performed on these samples to corroborate PCR findings, 2) lower respiratory tract samples for PCR or culture were not obtained to further determine whether PCR findings were reflective of actual infection, and 3) viral coinfections were not studied, it can provide some insights for COVID-19 prevention and treatment. In summary, 50% of COVID-19 positive patients were coinfected or carried the tested common bacterial pathogens according to this study. Usually, when a single or multiple pathogens invade the upper respiratory tract and damage the respiratory system, the other pathogens can easily infect the patient at that moment and further lead to the clinical diagnosis and treatment being more complicated (Bosch et al., 2013). The increased risk of B. pertussis infection in COVID-19 positive patients and high carriage of P. aeruginosa in this area might worsen the pathogenic condition. Consequently, some preventions should be taken to treat the common bacterial pathogens coinfection, especially for B. pertussis. The reason why B. pertussis increased in COVID-19 positive patients needs to be clarified and further confirmed in further study.
Authors' contributions
Jing Lv and Faxian Zhan designed this study. Danwen Nie and Fang Guo collected the samples. Fei He, Hongmei Yang, Bin Fang, and Bing Hu performed the nucleic acids extraction and RT-PCR tests. Xian Xia, Honglin Jiang, Yongzhong Jiang, and Xixiang Huo analyzed the data and prepared the manuscript.
Conflict of interest
All authors declare no conflict of interest.
Acknowledgments
We are grateful to Dr. Bingqing Zhu and Min Yuan for their suggestions in manuscript preparation and revision. This work was supported by Medical Scientific Research Foundation of Hubei Province, China (no. JX6B23); Innovation Team Project (T201907) and Open Foundation of the Hubei Key Laboratory of Edible Wild Plants Conservation and Utilization (EWPL201901) at Hubei Normal University; and the “Thirteenth Five-Year” National Key Program for Infectious Disease of China “Routine Surveillance of Five Major Syndromes in Hubei Province” (2017ZX10103005003).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.diagmicrobio.2020.115199.
Appendix A. Supplementary data
Supplementary tables
References
- Bisgard K.M., Pascual F.B., Ehresmann K.R. Infant pertussis: who was the source? Pediatr Infect Dis J. 2004;23(11):985–989. doi: 10.1097/01.inf.0000145263.37198.2b. [DOI] [PubMed] [Google Scholar]
- Bosch A.A., Biesbroek G., Trzcinski K. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog. 2013;9(1) doi: 10.1371/journal.ppat.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonetti NH: Immunomodulation in the pathogenesis of Bordetella pertussis infection and disease. Curr Opin Pharmacol 2007; 7(3):272–278. DOI: 10.1016/j.coph.2006.12.004. [DOI] [PubMed]
- Chinese National Health Commission . 3nd ed. 2020. Prevention and control plan for novel coronavirus pneumonia. [Google Scholar]
- Cox M.J., Loman N., Bogaert D. Co-infections: potentially lethal and unexplored in COVID-19. Lancet Microbe. 2020;1(1) doi: 10.1016/S2666-5247(20)30009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure E., Kwong K., Nguyen D. Pseudomonas aeruginosa in chronic lung infections: how to adapt within the host? Front Immunol. 2018;9:2416. doi: 10.3389/fimmu.2018.02416. [eCollection 2018] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson M.L. 6th ed. Hodder Arnold; Melbourne, Australia: 2010. Kucers' the use of antibiotics. [Google Scholar]
- Hirama T., Yamaguchi T., Miyazawa H. Prediction of the pathogens that are the cause of pneumonia by the battlefield hypothesis. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0024474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose M., Desai K. Fatal superimposed bacterial sepsis in a healthy coronavirus (COVID-19) patient. Cureus. 2020;12(5) doi: 10.7759/cureus.8350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medical expert group of Tongji Hospital Affiliated to Tongji Medical College of Huazhong University of Science and Technology. A quick guide to the diagnosis and treatment of pneumonia infected by new coronavirus (3rd Edition). Her Med. 2020; 1–9. [Chinese].
- Schwartz T., Volkmann H., Kirchen S. Real-time PCR detection of Pseudomonas aeruginosa in clinical and municipal wastewater and genotyping of the ciprofloxacin-resistant isolates. FEMS Microbiol Eco. 2006;57(1):158–167. doi: 10.1111/j.1574-6941.2006.00100.x. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO). Laboratory methods for the diagnosis of meningitis caused by Neisseria meningitidis, Streptococcus pneumoniae, and Haemophilus influenza. 2nd Ed. WHO Meningitis Manual 2011.
- World Health Organization (WHO). Coronavirus disease 2019 (COVID-19) Situation Report.
- Xing Q., Li G., Xing Y. Precautions are needed for COVID-19 patients with coinfection of common respiratory pathogens. medRxiv. 2020 doi: 10.1101/2020.02.29.20027698. [DOI] [Google Scholar]
- Yang H., Zhan J., Fang B. Research on acute non-viral respiratory tract infection pathogens spectrum of four hundred influenza-like cases. Chin J Prev Med. 2015;49(6):567–570. [PubMed] [Google Scholar]
- Zhang L., Yu F., Sang W. Research progress of Lianhua Qingwen in the treatment of influenza. Pharm Clin Chin Materia Medica. 2019;10(1):54–58. [Google Scholar]
- Zhu N., Zhang D., Wang W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables