Abstract
Therapeutic RNA molecules possess high potential for treating medical conditions if they can successfully reach the target cell upon administration. However, unmodified RNA molecules are rapidly degraded and cleared from the circulation. In addition, their large size and negative charge complicates their passing through the cell membrane. The difficulty of RNA therapy, therefore, lies in the efficient intracellular delivery of intact RNA molecules to the tissue of interest without inducing adverse effects. Here, we outline the recent developments in therapeutic RNA delivery and discuss the wide potential in manipulating the function of cells with RNAs. The focus is not only on the variety of delivery strategies but also on the versatile nature of RNA and its wide applicability. This wide applicability is especially interesting when considering the modular nature of nucleic acids. An optimal delivery vehicle, therefore, can facilitate numerous clinical applications of RNA.
Keywords: drug delivery, RNA therapy, nanoparticles, cell-specific targeting
Current Landscape of Biopharmaceutical Industry
For many years pharmaceutical companies have relied on small-molecule therapeutics to generate drugs. These small molecules bind to pockets of target proteins such as enzymes, receptors, or other proteins, mostly in an antagonistic way. By doing so they interfere with a certain biological process resulting in a therapeutic effect [1]. While small-molecule drugs possess certain favorable characteristics (such as ease of production, possibility of oral administration, favorable pharmacokinetics, and the ability to pass through the cell membrane), their potential is limited as they rely on the druggability of the target. Whether a biological target is druggable or not depends on a variety of factors such as the presence of suitable pockets in the protein structure in which small molecules can dock, a suitable size to accommodate binding, and the degree of polarity. Docking into a deep cavity is crucial in achieving sufficiently high binding affinities (generally with a KD in the nanomolar range or better) [1]. It is estimated that out of the ~20 000 human proteins, only ~3000 are druggable [2]. As of 2017, approved drugs targeted only 667 human proteins [3]. This not only indicates that more drugs can be developed to reach the 3000 druggable targets, but also highlights that the vast majority of 20 000 human proteins remain undruggable.
To tap into this unexplored potential, we need to look beyond small-molecule drugs. Over the years, more complex, biological macromolecules such as monoclonal antibodies (mAbs) have entered the pharmaceutical arena. Major benefits of their use, compared with small-molecule drugs, include long half-life, ability to target a broader group of proteins (due to the vast mAb repertoire), ability to be engineered to widen their applicability and increase their specificity, and lower toxicity. Disadvantages of the more complex biological macromolecules include more complicated pharmacological profiles, higher cost of production, and limits in route of administration (mostly intravenous) [4].While small molecules still dominate the pharmaceutical market, biologics have started to gain a higher share in the last few years. In 2018, 17 of the 59 newly approved drugs were biologics [5,6]. Altogether, this shows that besides traditional small-molecule drugs, larger biomolecules are becoming increasingly important to treat diseases.
In line with the trend of developing more specific and efficacious medicines, a new therapeutic avenue is gaining momentum: that of nucleic-acid-based therapy. Examples of such therapeutic agents include the use of oligos [7], plasmid DNA [8], mRNA [9], ribozymes [10,11] (see Glossary) and RNAi-related nucleic acids such as miRNA [12], siRNA [13], and short hairpin RNA (shRNA) [14]. While the use of mAbs are limited to cell surface receptors or secreted proteins, nucleic acids can interfere with protein expression itself and therefore circumvent the druggability issue during drug development.
While clinical development of RNA therapeutics has faced decades of significant challenges in terms of potency and immunogenicity, in recent years, the field has gained some momentum with the recent approval of two siRNA-based drugs patisiran and givosiran [15,16] within a short period of time. This, combined with a well-filled clinical pipeline of mRNA therapeutics [17], shows the potential of clinical development of RNA therapeutics in the coming years. Because of the rapid developments in this field, this review focuses specifically on RNA therapeutics, its delivery, challenges, and finally, the outlook on clinical applications.
Types of RNA Therapeutics
RNA therapeutics span from antisense oligonucleotides (ASOs), siRNA, miRNA, mRNA, RNA aptamers, short activating RNA (saRNA), to single guide RNA (sgRNA) for CRISPR/Cas9 systems. Below, we discuss each of these classes of RNA therapeutic molecules.
ASOs
ASOs are short, single-stranded oligonucleotides that are complementary to a target mRNA to which they hybridize and thereby modulate protein expression. ASOs can be based on both DNA and RNA [18].While classical ASOs are DNA based and act by formation of DNA–RNA hybrids that serve as a substrate for RNase H, other ASO functionalities also exist and open the door for RNA-based ASOs. For example, ASOs can hybridize close to the start codon and sterically interfere with translation or hybridize to untranslated regions and sterically interfere with RNA-binding proteins (Figure 1 ). Besides inhibiting translation, ASOs can also increase translation efficiency of the target protein by sterically inhibiting the translation of an upstream open reading frame and can alter splicing by binding to splice sites or to exonic/intronic inclusion signals [19]. An example of an advanced stage RNA-ASO is a splice-modulating oligonucleotide, QR-110, that is currently being investigated in clinical trials for use against a severe type of inherited retinal dystrophy (Table 1 ) [20]. Furthermore, other types of RNA-ASOs are anti-miRs which bind directly to the mature strand of the target miRNA and block its functionality [21]. Locked nucleic acids (LNAs) enhance the functionality of anti-miRs by increasing their stability [22].
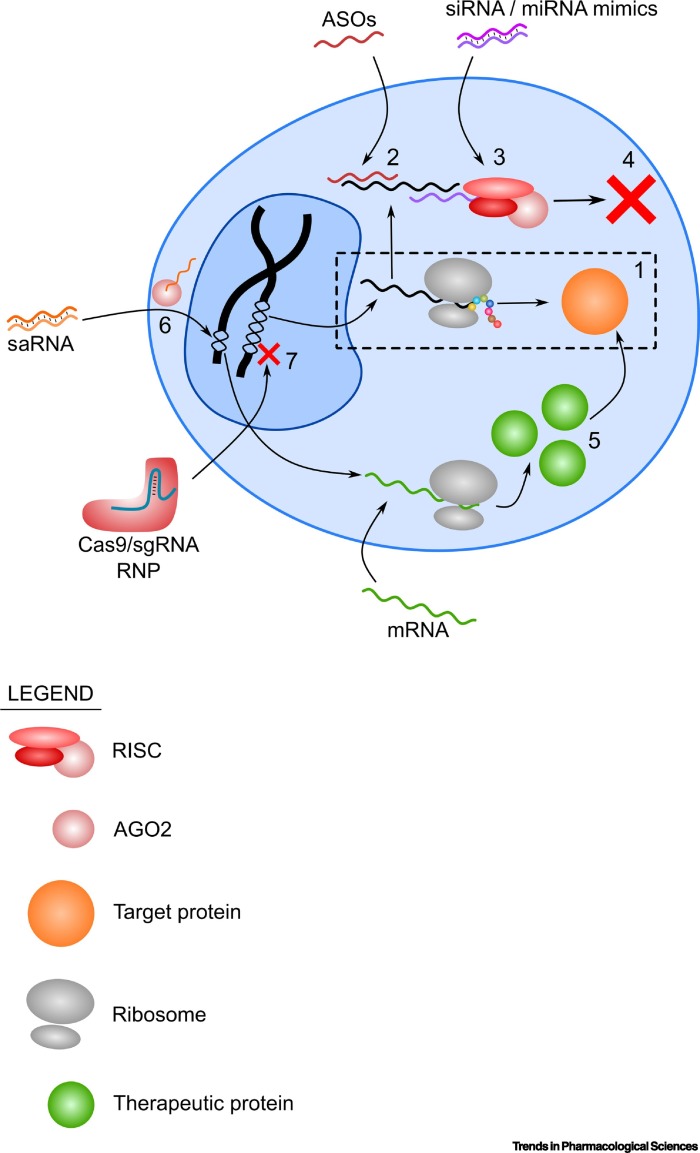
Figure 1.
Overview of Different Mechanisms of Action of Different RNA Therapeutics.
(1) Without therapeutic RNA molecules, the translation of a pathogenic protein proceeds without inhibition (shown in the broken line box). (2) ASOs hybridize to the target mRNA, while the (3) siRNA/miRNA mimics utilize the RISC in the RNAi pathway to (4) inhibit translation of target mRNA. (5) Overexpression of a therapeutic protein that counteracts the function of the pathogenic protein can be done by delivering the mRNA of the therapeutic protein. (6) saRNA can be delivered to the cell where it binds to AGO2, is imported to the nucleus, and in turn activates an endogenous gene. (7) A more permanent approach to remove the pathogenic protein is by gene knockout using Cas9 and sgRNA RNPs. Abbreviations: AGO2, argonaute 2; ASO, antisense oligonucleotide; RISC, RNA-induced silencing complex; RNP, ribonucleoprotein; saRNA, small activating RNA.
Table 1.
Overview of Clinical Trials of Various RNA Therapeutics Discussed in This Reviewa
| Candidate | Type of RNA | Vehicle | Route of administration | Biological target | Condition or disease | Clinical stage | Year (first posted) | Clinical trial number |
|---|---|---|---|---|---|---|---|---|
| Antisense oligonucleotides | ||||||||
| QR-010 | ASO | NA | Inhalation | Cystic fibrosis transmembrane conductance regulator (ΔF508 mutation) | Cystic fibrosis | Phase Ib | 2015 | NCT02532764 |
| QR-421a | ASO | NA | Intravitreal injection | Usherin (exon 13 of USH2A) | Retinitis pigmentosa | Phase I/II | 2018 | NCT03780257 |
| QR-110 | ASO | NA | Intravitreal injection | Centrosomal protein 290 (p.Cys998X mutation) | Leber’s Congenital amaurosis | Phase II/III | 2019 | NCT03913143 |
| QR-1123 | ASO | NA | Intravitreal injection | Mutant P23H of the rhodopsin gene | Retinitis pigmentosa | Phase I/II | 2019 | NCT04123626 |
| Candidate | Type of RNA | Vehicle | Route of administration | Biological target | Condition or disease | Clinical stage | Year (first posted) | CT identifier |
|---|---|---|---|---|---|---|---|---|
| siRNA and miRNA | ||||||||
| TD101 | siRNA | NA | Intralesional injection | Keratin 6A | Pachyonychia congenita | Phase I | 2008 | NCT00716014 |
| PF-04523655 | siRNA | NA | Intravitreal injection | RTP801 | Age-related macular degeneration | Phase II | 2008 | NCT00713518 |
| Choroidal neovascularization Diabetic retinopathy Diabetic macular edema |
Phase II | 2011 | NCT01445899 | |||||
| ALN-RSV01 | siRNA | NA | Inhalation of nebulized solution | Nucleocapsid protein of respiratory syncytial virus | Lung transplant patients infected with respiratory syncytial virus | Phase IIb | 2010 | NCT01065935 |
| ALN-VSP02 | siRNA | siRNA-LNPb (2 siRNAs) | i.v. infusion | Kinesin spindle protein and vascular endothelial growth factor | Solid tumor | Phase I | 2009/2010 |
NCT01158079 NCT00882180 |
| TKM-080301 | siRNA | Stable nucleic acid–lipid particles (SNALPs) | i.v. infusion | Polo-like kinase 1 | Neuroendocrine tumors Adrenocortical carcinoma |
Phase I/II | 2010 | NCT01262235 |
| Primary or secondary liver cancer | Phase I | 2011 | NCT01437007 | |||||
| Advanced hepatocellular carcinoma | Phase I/II | 2014 | NCT02191878 | |||||
| QPI-1007 | siRNA | NA | Intravitreal injection | Caspase 2 | Nonarteritic anterior ischemic optic neuropathy | Phase II/III | 2015 | NCT02341560 |
| Acute primary angle closure glaucoma | Phase II | 2013 | NCT01965106 | |||||
| ALN-PCS02 | siRNA | siRNA–LNP | i.v. infusion | Proprotein convertase subtilisin/kexin type 9 | Elevated LDL-cholesterol | Phase I | 2011 | NCT01437059 |
| siG12D-LODER | siRNA | Miniature biodegradable polymeric matrix | Intratumoral using endoscopic ultrasound biopsy needle | KRAS | Pancreatic cancer | Phase II | 2012 | NCT01676259 |
| DOPC-encapsulated siRNA | siRNA | Neutral liposome | i.v. infusion | EphA2 | Advanced malignant solid neoplasm | Phase I | 2012 | NCT01591356 |
| Atu027 | siRNA | Liposomal siRNA | i.v. infusion | Protein kinase N3 | Pancreatic cancer | Phase Ib/IIa | 2013 | NCT01808638 |
| SYL040012 | siRNA | NA | Ophthalmic administration | β2-Adrenergic receptor | Open angle glaucoma Ocular hypertension |
Phase II | 2014 | NCT02250612 |
| ND-L02-s0201 | siRNA | LNP | i.v. infusion | Heat shock protein 47 | Hepatic fibrosis | Phase I | 2014 | NCT02227459 |
| Idiopathic pulmonary fibrosis | Phase II | 2018 | NCT03538301 | |||||
| QPI-1002 | siRNA | NA | i.v. injection | P53 | Delayed graft function | Phase III | 2015 | NCT02610296 |
| Risk for AKI following cardiac surgery | Phase III | 2018 | NCT03510897 | |||||
| ALN-PCSSC (Inclisiran) | siRNA | GalNAc-siRNA | s.c. injection | Proprotein convertase subtilisin/kexin type 9 | Elevated LDL-cholesterol | Phase I | 2014 | NCT02314442 |
| Atherosclerotic cardiovascular disease (or risk equivalents) | Phase II | 2015 | NCT02597127 | |||||
| Renal impairment | Phase I | 2017 | NCT03159416 | |||||
| High cardiovascular risk and elevated LDL-cholesterol | Phase II | 2017 | NCT03060577 | |||||
| Atherosclerotic cardiovascular disease Elevated LDL-cholesterol |
Phase III | 2018 |
NCT03399370 NCT03400800 |
|||||
| Homozygous Familial Hypercholesterolemia | Phase III | 2019 | NCT03851705 | |||||
| Atherosclerotic cardiovascular disease (or risk equivalents) Hypercholesterolemia Elevated LDL cholesterol |
Phase III | 2019 | NCT03814187 | |||||
| SYL1001 | siRNA | NA | Ophthalmic administration | Transient receptor potential cation channel subfamily V Member 1 | Dry eye disease | Phase III | 2017 | NCT03108664 |
| ALN-GO1 (Lumasiran) | siRNA | GalNAc-siRNA | s.c. injection | hydroxyacid oxidase 1 | Primary hyperoxaluria type 1 | Phase III | 2018 |
NCT04152200 NCT03905694 NCT03681184 |
| ALN-AT3 (Fitusiran) | siRNA | GalNAc-siRNA | s.c. injection | Antithrombin | Hemophilia | Phase IIIα | 2018 |
NCT03549871 NCT03974113 NCT03754790 NCT03417102 NCT03417245 |
| ALN-AAT02 | siRNA | GalNAc-siRNA | s.c. injection | Mutant of α-1 antitrypsin | ZZ Type α-1 Antitrypsin deficiency liver disease | Phase I/II | 2018 | NCT03767829 |
| Patisiran (label expansion studies) | siRNA | LNP | i.v. infusion | transthyretin | hATTR with cardiomyopathy hATTR progression post liver transplant Polyneuropathy |
Phase III | 2019 |
NCT03997383 NCT03862807 NCT02510261 NCT04201418 (Phase IV) |
| ALN-CC5 (Cemdisiran) | siRNA | GalNAc-siRNA | s.c. injection | Complement component C5 | IgA nephropathy Berger disease |
Phase II | 2019 | NCT03841448 |
| ALN-AGT01 | siRNA | GalNAc-siRNA | s.c. injection | Angiotensinogen | Hypertension | Phase I | 2019 | NCT03934307 |
| ALN-TTRsc02 (Vutrisiran) | siRNA | GalNAc-siRNA | s.c. injection | Transthyretin | Amyloidosis, hereditary transthyretin amyloidosis | Phase III | 2018 | NCT03759379 |
| Transthyretin amyloidosis (ATTR) with cardiomyopathy | Phase III | 2019 | NCT04153149 | |||||
| ARO-APOC3 | siRNA | TRiM™ formulation | s.c. injection | Apolipoprotein C3 | Hypertriglyceridemia Familial chylomicronemia |
Phase I | 2018 | NCT03783377 |
| ARO-ANG3 | siRNA | TRiM™ formulation | s.c. injection | Angiopoietin like protein 3 | Dyslipidemia | Phase I | 2018 | NCT03747224 |
| RG6346 (DCR-HBVS) | siRNA | GalNAc-siRNA | s.c. injection | Hepatitis B surface antigen | Chronic hepatitis B viral infection | Phase I | 2018 | NCT03772249 |
| KrasG12D siRNA (iExosomes) | siRNA | Mesenchymal Stromal cell derived Exosomes |
i.v. infusion | KRAS | Pancreatic cancer patients with KrasG12D mutation | Phase I | 2018 | NCT03608631 |
| OLX10010 | siRNA | cp-asiRNA | i.d./s.c. injection | Connective tissue growth factor | Hypertrophic scar | Phase I | 2018 | NCT03569267 |
| ARO-AAT | siRNA | TRiM™ formulation | s.c. injection | Mutant of α1-antitrypsin (Z-AAT) | α1-Antitrypsin deficiency | Phase II | 2019 | NCT03946449 |
| ARO-HSD | siRNA | TRiM™ formulation | s.c. injection | Hydroxysteroid 17β-dehydrogenase 13 | Nonalcoholic steatohepatitis | Phase I | 2019 | NCT04202354 |
| ARO-HIF2 | siRNA | TRiM™ formulation | i.v. infusion | Hypoxia-inducible factor 2α | Clear cell renal cell carcinoma | Phase I | 2019 | NCT04169711 |
| DCR-PHXC | siRNA | GalXC-siRNA | s.c. injection | Lactate dehydrogenase A | Primary hyperoxaluria type 1 and type 2 | Phase II | 2019 |
NCT03847909 NCT04042402 (Phase 3) |
| JNJ-3989 | siRNA | TRiM™ formulation | s.c. injection | Hepatitis B surface antigen | Chronic hepatitis B viral infection | Phase I | 2019 | NCT04208386 |
| Phase II | 2020 | NCT04439539 | ||||||
| BMT101 | siRNA | cp-asiRNA | i.d. injection | Connective tissue growth factor | Hypertrophic scar | Phase II | 2019 | NCT04012099 |
| DCR-A1AT | siRNA | GalXC-siRNA | s.c. injection | α1- Antitrypsin | α1-Antitrypsin deficiency | Phase I/II | 2019 | NCT04174118 |
| ARO-ENaC | siRNA | NA | Inhalation of nebulized solution | Epithelial sodium channel α subunit | Cystic fibrosis | Phase I/IIa | 2020 | NCT04375514 |
| AMG 890 | siRNA | GalNAc-siRNA | s.c. injection | Lipoprotein(a) | Elevated plasma lipoprotein(a) | Phase II | 2020 | NCT04270760 |
| STP705 | siRNA | Histidine–lysine copolymer peptide | Intralesional injection | TGF-β, Cox-2 | Cutaneous squamous cell carcinoma in situ | Phase I/II | 2020 | NCT04293679 |
| Miravirsen | LNA-modified anti-miR | NA | s.c. injection | miR-122 | Hepatitis C | Phase II | 2010/2015 |
NCT01200420 NCT02508090 NCT02452814 |
| TargomiRs | miR-mimic | EDV (nonliving bacterial minicells) | i.v. infusion | miR-16 | Malignant pleural mesothelioma Non-small cell lung cancer |
Phase I | 2015 | NCT02369198 |
| RG-125 (AZD4076) |
Anti-miR | GalNac | s.c. injection | miR-103/107 | Nonalcoholic steatohepatitis | Phase I | 2015 | NCT02612662 |
| SAR339375 | Anti-miR | NA | s.c. injection | miR-21 | Alport’s syndrome | Phase II | 2016 | NCT02855268 |
| MRG-106 (Cobomarsen) | LNA-modified anti-miR | NA | i.v. infusion | miR-155 | Cutaneous T cell lymphoma Mycosis fungoides |
Phase II | 2018 | NCT03713320 |
| s.c. and intratumoral injection i.v. infusion |
miR-155 | Lymphoma/leukemia | Phase I | 2015 | NCT02580552 | |||
| MRG-110 | Anti-miR | NA | i.d. injection | miR-92a | Wound healing | Phase I | 2018 | NCT03603431 |
| MRG-201 | miR-mimic | Cholesterol | i.d. injection | miR-29 | Keloid | Phase II | 2018 | NCT03601052 |
| mRNA | ||||||||
| Melanoma mRNA vaccine | mRNA | NA | i.d. injection | Melan-A Mage-A1 Mage-A3 Survivin GP100 Tyrosinase |
Melanoma | Phase I/II | 2005 | NCT00204516 |
| Melanoma mRNA vaccine | mRNA | Protamine-complexed mRNA | i.d. injection | Melan-A Mage-A1 Mage-A3 Survivin GP100 Tyrosinase |
Melanoma | Phase I/II | 2005 | NCT00204607 |
| CV9201 | mRNA | RNActive® technology | i.d. injection | Tumor-associated antigens | Non-small cell lung cancer | Phase I/II | 2009 | NCT00923312 |
| RBL001/RBL002 | mRNA | NA | i.nod. injection | Melanoma antigens | Melanoma | Phase I | 2012 | NCT01684241 |
| BNT114 | mRNA | Proprietary size- and charge-based RNA-LPX | i.v. infusion | Shared TAA for TNBC and neoantigens identified by NGS | TNBC | Phase I | 2014 | NCT02316457 |
| CV7201 | mRNA | RNActive® technology | i.m. injection i.d. injection |
Rabies virus glycoprotein | Rabies | Phase I | 2014 | NCT02241135 |
| Lipo-MERIT | mRNA | Liposome-complexed mRNA | i.v. infusion | RBL001.1 RBL002.2 RBL003.1 RBL004.1 |
Melanoma | Phase I | 2015 | NCT02410733 |
| mRNA-4157 | mRNA | Lipid encapsulated mRNA | i.m. injection | Tumor-associated antigens (personalized) | Solid tumors | Phase I | 2017 | NCT03313778 |
| Melanoma | Phase II | 2019 | NCT03897881 | |||||
| BI 1361849 | mRNA | RNActive® technology | i.d. injection | Tumor-associated antigens | Non-small cell lung cancer | Phase I/II | 2017 | NCT03164772 |
| mRNA-1647 mRNA-1443 |
mRNA | Undisclosed | i.m. injection | Viral antigens | Cytomegalovirus infection | Phase I | 2017 | NCT03382405 |
| mRNA1325 | mRNA | LNP | i.d. injection | Viral antigen | Zika virus infection | Phase I | 2017 | NCT03014089 |
| VAL506440 | mRNA | LNP | i.d. injection | H10N8 antigen influenza | Influenza | Phase I | 2017 | NCT03076385 |
| VAL339851 | mRNA | LNP | i.d. injection | H7N9 Antigen influenza | Influenza | Phase I | 2017 | NCT03345043 |
| mRNA-2416 | mRNA | LNP | Intratumoral injection | OX40 ligand | Advanced malignancies | Phase I/II | 2017 | NCT03323398 |
| CV9202 (BI 1361849) | mRNA | RNActive® technology | i.d. injection | Six non-small cell lung cancer antigens (NY-ESO-1, MAGE-C1, MAGE-C2, survivin, 5T4, and MUC-1) | Non-small cell lung cancer | Phase I/II | 2017 | NCT03164772 |
| Neo-antigen mRNA |
mRNA | NA | s.c. injection | Tumor antigen | Digestive tract adenocarcinomas | NA | 2018 | NCT03468244 |
| CV7202 | mRNA | LNP | i.m. injection | Rabies virus glycoprotein | Rabies | Phase I | 2018 | NCT03713086 |
| mRNA1653 | mRNA | LNP | i.d. injection | Viral antigen | Human metapneumovirus and human parainfluenza infection | Phase I | 2018 | NCT03392389 |
| mRNA-2752 | mRNA | LNP | Intratumoral injection | OX40L IL-23 IL-36γ |
Advanced malignancies | Phase I | 2018 | NCT03739931 |
| mRNA-1944 | mRNA | LNP | i.v. infusion | Anti-Chikungunya virus mAb | Chikungunya virus infection | Phase I | 2019 | NCT03829384 |
| BNT131 (SAR441000) | mRNA | Proprietary size- and charge-based RNA-LPX | Intratumoral injection | IL-12sc IL-15sushi IFNα GM-CSF |
Metastatic neoplasm | Phase I | 2019 | NCT03871348 |
| mRNA-1893 | mRNA | LNP | i.m. injection | Undisclosed | Zika virus | Phase I | 2019 | NCT04064905 |
| mRNA-3704 | mRNA | LNP | i.v. infusion | Methylmalonyl-coenzyme A mutase | Methylmalonic acidemia | Phase I/II | 2019 | NCT03810690 |
| mRNA-1273 | mRNA | LNP | i.m. injection | SARS-CoV-2 glycoproteins | COVID-19 | Phase III | 2020 | NCT04470427 |
| BNT162 | mRNA | LNP | i.m. injection | Undisclosed SARS-CoV-2 proteins | COVID-19 | Phase I/II Phase II/III |
2020 |
NCT04380701 NCT04368728 |
| BNT122 (RO7198457) | mRNA | Proprietary size- and charge-based RNA-LPX | i.v. infusion | Tumor-associated antigens | Colorectal cancer stage II/III Advanced melanoma |
Phase II | 2020 |
NCT04486378 NCT03815058 |
| Locally advanced or metastatic solid tumors | Phase I | 2017 | NCT03289962 | |||||
| CVnCoV | mRNA | LNP | i.m. injection | SARS-CoV-2 spike protein | COVID-19 | Phase I | 2020 | NCT04449276 |
| RNA Aptamers | ||||||||
| NOX-E36 | Aptamer | L-RNA, PEGylated |
i.v. injection s.c. injection |
Chemokine (cysteine–cysteine motif) ligand 2 |
Chronic inflammatory diseases Type 2 diabetes mellitus Systemic lupus erythematosus |
Phase I | 2009 | NCT00976729 |
| Renal impairment | Phase I | 2011 | NCT01372124 | |||||
| Type 2 diabetes mellitus albuminuria | Phase II | 2012 | NCT01547897 | |||||
| NOX-A12 | Aptamer | L-RNA, PEGylated |
i.v. injection | Chemokine (C-X-C motif) ligand 12 Stromal-derived factor 1 |
Autologous stem cell transplantation | Phase I | 2009 | NCT00976378 |
| Chronic lymphocytic leukemia | Phase II | 2011 | NCT01486797 | |||||
| Multiple myeloma | Phase II | 2012 | NCT01521533 | |||||
| Colorectal cancer Pancreatic cancer |
Phase I/II | 2017 | NCT03168139 | |||||
| Glioblastoma | Phase I/II | 2019 | NCT04121455 | |||||
| NOX-H94 | Aptamer | L-RNA, PEGylated |
Undisclosed | Hepcidin | Anemia of chronic disease | Phase II | 2012 | NCT01691040 |
| Anemia End-stage renal disease |
Phase I/II | 2014 | NCT02079896 | |||||
| Zimura (ARC1905) |
Aptamer | NA | Intravitreal injection | Complement component 5 | Age-related macular degeneration | Phase II | 2017 | NCT03362190 |
| Stargardt disease 1 | Phase II | 2017 | NCT03364153 | |||||
| saRNA | ||||||||
| MTL-CEBPA | saRNA | SMARTICLES® liposomal nanoparticle | i.v. infusion | CCAAT enhancer binding protein | Hepatocellular carcinoma Liver cancer |
Phase I | 2016 | NCT02716012 |
| Other | ||||||||
| CV8102 | Noncoding ssRNA | NA | Intratumoral injection | Activation of TLR7/8/RIG-1 | Melanoma Squamous cell carcinoma of the skin or head/neck Adenoid cystic carcinoma |
Phase I | 2017 | NCT03291002 |
Only the most advanced clinical stage for each RNA candidate is mentioned. Table was compiled using data from i–ix, [25], and [81].
Abbreviations: AKI, acute kidney injury; cp-asiRNA, proprietary cell-penetrating asymmetric interfering RNA by OliX Pharmaceuticals; GalXCL, proprietary siRNA formulation by Dicerna Pharmaceuticals; GM-CSF, granulocyte-macrophage colony-stimulating factor; i.d., intradermal; IFN, interferon; Il, interleukin; i.m., intramuscular; i.nod., intranodal; i.v., intravenous; LNA, locked nucleic acid; LNP, lipid nanoparticle; NA, not applicable; NGS, next-generation sequencing; s.c., subcutaneous; TRiM™, targeted RNAi molecule, platform for ligand-mediated targeted RNA delivery by Arrowhead Pharmaceuticals; TAA, tumor-associated antigen; TNBC, triple-negative breast cancer.
siRNA and miRNA
siRNAs are short (20–25 nucleotides), double-stranded RNA molecules that use the RNAi pathway to degrade a target mRNA in a sequence-specific manner. Upon delivery into the cytoplasm, argonaute (AGO)2 cleaves the passenger (sense) strand and the guide (antisense) strand of the siRNA is loaded into the RNA-induced silencing complex (RISC). The guide strand then guides the RISC to the target mRNA which is recognized and cleaved (Figure 1). The RISC and guide strand can be recycled and therefore one siRNA molecule can drive the cleavage of multiple mRNA molecules resulting in highly efficient gene silencing [23].
Another type of RNA that uses the RNAi pathway are miRNA mimics (Figure 1). miRNA mimics are synthetic, double-stranded RNAs that mimic a naturally occurring miRNA. These miRNA mimics can replenish altered miRNA functionality. For instance, in certain types of cancer, miRNAs that silence oncogenes are downregulated and miRNA mimics can give a therapeutic effect in such cases [22]. For examples of siRNA and miRNA therapeutics that are currently in advanced clinical trials for various disease indications, see Table 1.
mRNA
The concept of transfecting mRNA directly into cells instead of transfecting DNA has been around for a few decades [24,25]. mRNA is seen as a safer alternative to DNA [26] for therapeutic purposes such as protein replacement therapy, as it degrades quickly leading to no concerns about potential adverse effects from long-lasting expression or genomic integration. However, mRNA expression is more difficult to regulate than DNA expression, which remains a concern in mRNA therapy, as possible toxicity can arise from mRNA expression at off-target sites, leading to unwanted protein expression. For instance, unspecific expression of factor VIII, factor IX, and interleukin-12 has been shown to be dangerous [27]. Strategies that are commonly used for spatial control of DNA expression (such as use of transcription factors, inclusion of tissue specific promoters [28]) and for temporal control [such as the use of transcriptional on/off switches, e.g., tetracycline-responsive repressor (TetR) [29]] are not available for mRNA control. This, along with the unstable nature of mRNA, has delayed its potential for in vivo applications, and the field of mRNA therapeutics received a major boost only as advances in chemistry created more stable modified nucleotides (see later) and sophisticated regulation systems for mRNA expression were generated. mRNA therapeutics have found a role in protein replacement therapy [e.g., vascular endothelial growth factor (VEGF)-A delivery after myocardial infarction] [30], vaccines for infectious diseases (e.g., expression of viral antigens in dendritic cells) [31,32], or for in vivo production of mAbs [33]. For examples of mRNA therapeutics that are currently in advanced clinical trials for various disease indications, see Table 1.
RNA Aptamers
Aptamers are short single-stranded oligonucleotides that can consist of both DNA and RNA. Aptamers were first generated in 1990 using the Systematic Evolution of Ligands by Exponential Enrichment (SELEX) selection method. Using SELEX, aptamers that selectively bind small molecular ligands or proteins with high affinity and high specificity are selected from a library [34,35]. To date, only one RNA aptamer has received FDA approval: pegabtanib, which is used for treatment of age-related macular degeneration (mechanism of action is the binding to the VEGF isoform 165) [36]. Several other aptamers are currently being investigated in clinical trials (Table 1). Besides the therapeutic potential of RNA aptamers, aptamers are also used solely as targeting moieties to aid delivery of other RNA payloads such as siRNA (see more discussion in the section ‘Delivery of RNA Therapeutics’).
saRNA
saRNAs are 21-nucleotide, double-stranded, noncoding RNA that possess two nucleotide overhangs on both ends [37] (Figure 1). saRNAs are initially loaded on the AGO2 protein where the passenger strand is cleaved. The saRNA–AGO2 complex then enters the nucleus and binds to promoter regions of genes to enhance transcription [38]. In a study by Zhao et al., a combination of saRNA and siRNA was used to modulate the balance between the transcription factors CCAAT/enhancer-binding protein α and β (CEBPA, a tumor suppressor and CEBPB, an oncogene, respectively), where saRNA-driven activation of CEBPA decreased proliferation and migration in the differentiated hepatocellular carcinoma (HCC) cell lines [39]. saRNAs have recently progressed to the clinical setting as well, and the first saRNA-based clinical trial is currently ongoing (clinical trial numberi: NCT02716012; Table 1). This clinical trial uses liposomal nanoparticles, termed SMARTICLES, encapsulated with saRNA that activates the CEBPA gene for treatment of patients with HCC. CEBPA is considered a master regulator in normal liver function and its expression is decreased in HCC. Lowered CEBPA expression is seen in many liver abnormalities and increasing CEBPA expression in HCC might result in improved therapeutic outcome [40].
gRNA for CRISPR/Cas9-Directed Knockout
The development of CRISPR-Cas9 gene editing technology (Box 1 ) has further pushed the development of RNA therapeutics to the forefront. The codelivery of Cas9 mRNA and sgRNA against a certain genomic target has promising applications for gene knock out strategies (Figure 1). While in a laboratory setting, many options are available for the introduction of the Cas9 protein and sgRNA to cells, including plasmid DNA, viral transfection, or electroporation, but this does not easily translate to an in vivo clinical setting. [43]. Therefore, the most realistic method right now involves ex vivo manipulation of cells with re-introduction of edited cells into the body [44].
Box 1. The CRISPR/Cas System.
The CRISPR/Cas system, a form of acquired immunity in bacteria and archaea, has been harnessed as a genome-editing tool and has also revolutionized the field of RNA therapeutics. The CRISPR system consists of two distinct classes (1 and 2). Class 2 is the most frequently utilized for genome editing applications, in particular, CRISPR/Cas9. CRISPR/Cas9 requires the CRISPR-associated nuclease Cas9 along with a gRNA. The gRNA consists of two RNA molecules: the CRISPR/RNA (crRNA) and the transactivating RNA (tracrRNA). To simplify the tool, these two RNAs are combined on a single guide RNA chimera (sgRNA) [41]. While the gRNA guides the Cas9 nuclease to a specific genomic location, the Cas9 cuts the DNA, resulting in a double-strand break, which in eukaryotes can be repaired by two mechanisms: nonhomologous end joining (NHEJ) and homology-directed repair (HDR). The more prominent of the two DNA repair pathways, NHEJ, is prone to introducing indel errors during the repair causing frameshift mutations resulting in premature termination of translation, generating a knockout of the gene of interest. When an HDR (donor) template is introduced, HDR-directed repair can be utilized, which enables correction of mutated genes, insertion of genes, or replacement of genes [42].
Alt-text: Box 1
Chemical Modifications to Increase RNA Stability and Decrease Immunogenicity
While the field has seen significant progress, some of the major obstacles in RNA therapeutics are the unstable nature (due to the high stability and activity of RNases) and high immunogenicity of the RNA molecules [45]. Both single-stranded and double-stranded RNA molecules induce the production of type I interferons and various other proinflammatory cytokines through multiple signaling pathways, involving Toll-like receptor (TLR) 3, 7, or 8, or retinoic-acid inducible gene (RIG)/melanoma differentiation-associated (MDA)5 [46,47]. The high immunogenicity combined with low RNA stability necessitates chemical modifications of the RNA molecule to make advancement to the clinic more realistic. Such modifications can involve alterations of the ribose group, the phosphate backbone, the RNA termini, or modification of the nucleobases themselves [45]. For example, modifying the ribose on the 2'-O position dramatically increased the in vivo potency of siRNA. At least 13 ribose modifications have been reported previously and especially 2'-OMe, 2'-F, and 2′-O-methoxyethyl modifications turned out to be highly successful for increasing siRNA serum stability [45]. Effective phosphate modifications include phosphorothioates (PSs) and borine-modified phosphorus (boranophosphate). Such modifications replace nonbridging phosphate oxygen atoms with either sulfur or borane, leading to nuclease- resistant nucleotide linkages [45,48]. Another popular modification is the phosphorodithioate linkage (PS2) that replaces both nonbridging phosphate oxygens with sulfur [48]. Furthermore, as the RNA termini are vulnerable to exonucleases, protecting them is equally important. This has been done by including inverted thymidine residues at the 3' end [45]. Other strategies towards stabilizing RNA termini have included addition of palmitic acid [49] and the covalent attachment of aromatic compounds (such as phenyl, hydroxylphenyl, pyrenyl, and naphthyl derivatives) to the 5' sense strand of siRNAs [50].
Delivery of RNA Therapeutics
The major challenge in RNA therapeutics is delivery and difficulties in intracellular uptake of RNA molecules due to the large molecular weight and negatively charged phosphate backbone that hinders internalization [51]. Previous studies on systemic administration of naked RNA has revealed that their pharmacokinetic profile is generally poor [52., 53., 54.]. While chemical modifications of the RNA molecules dramatically increase the stability and decrease the immunogenicity, intravenous injection of naked RNA remains challenging where multiple barriers hamper RNA uptake. Cellular barriers include difficulties in both passing of the cell membrane as well as the subsequent endosomal escape. While other RNA molecules might be hampered with these cellular barriers, naked mRNA delivery somewhat circumvents these barriers as dendritic cells actively internalize naked mRNA by micropinocytosis [55,56]. Another barrier is the mononuclear phagocyte system (MPS), which effectively clears administered particulates through phagocytosis [57]. Although the smaller RNA molecules such as siRNA are not taken up by the MPS, a disadvantage of the small size is the increased clearance rate by the kidneys as the glomerular filtration barrier limit is about 8 nm [57]. Kidney clearance is therefore the major obstacle for intravenous administration of naked RNA. Fortunately, including PS modifications decreases the rate of kidney clearance of RNA molecules by increasing binding to plasma proteins, thereby increasing the serum half-life of the administered RNA [58].
When naked siRNA is injected intravenously, generally high doses are required. A study from 2012 reported that when ≤200 mg/kg siRNA was administered, the plasma concentration 30 min after injection was ~90% lower than 5 min after injection. At 2 h after injection, there was virtually no siRNA left in the plasma (<2%) due to rapid clearance by the kidneys. Even in partially nephrectomized rats, the clearance of siRNA was rapid. Higher dosages like 800 and 1200 mg/kg, however, yielded better results, which was mirrored in monkeys [52]. The fact that the clearance rate is slower with the higher doses indicates that the high dose, at least partially, saturates the kidney clearance system. However, the clearance is still fast and such high doses are clinically irrelevant both from a financial and safety point of view. Furthermore, the transient nature of siRNA will require multiple doses and dosing multiple times at such high concentrations seems unrealistic.
Local administration seems to be the more viable way of delivering naked RNA. Naked siRNA has been successfully delivered to eye, brain, and tumor tissues when injected locally [59]. Likewise, naked mRNA has also been successfully delivered locally to the heart. In 2018, Carlsson and colleagues reported efficient uptake of naked mRNA encoding VEGF by cardiomyocytes when injected directly into the heart. This resulted in an improved cardiac function in an animal model of myocardial infarction [60]. Especially for vaccination purposes, naked mRNA delivery is an option as dendritic cells actively internalize naked mRNA by micropinocytosis [55,56]. Other strategies that have been successful for naked mRNA delivery include intradermal injection of mRNA using hollow microneedles for expression in the skin [61], and injection of self-amplifying mRNA resulting in prolonged protein expression [62]. Below, we discuss several delivery strategies that have been developed to enable safe and efficient delivery of RNA therapeutics.
RNA Conjugated with a Targeting Moiety
The attachment of an active targeting moiety such as an antibody to the RNA can aid in both tissue/cell targeting and in internalization into target cells. Although it does not directly protect the RNA against degradation, it facilitates accumulation into the cells of interest [63]. Important characteristics of the targeting moiety are presence of active groups for conjugation purposes, good binding affinity, and reduced immunogenicity [64]. A recent notable success story of RNA conjugated to a targeting moiety is givosiran, which has been recently approved by the FDA. Givosiran is composed of siRNA conjugated to three N-acetylgalactosamine (GalNAc) molecules. GalNAc binds the asialoglycoprotein receptor (ASGR) with high affinity and internalizes siRNA into the hepatocytes rapidly, although endosomal escape remains an issue, like in other delivery strategies [65]. This liver-specific uptake makes GalNAc suitable for siRNA delivery to the liver for hepatic diseases.
Antibodies are a popular choice as targeting moieties due to their specificity, high affinity, well-defined structure, and long in vivo circulation time [66]. Multiple studies have used antibody–siRNA conjugates (ARCs) with different methods of linking the antibody to the siRNA molecule. These conjugation methods can include either a covalent attachment (e.g., Sugo et al. used maleimide/thiol chemistry [67]) or a noncovalent attachment (e.g., Ibtehaj et al. used protamine as a linker using its electrostatic interactions with siRNA [68]) (Figure 2A). Moreover, linkers that allow for attachment between the siRNA and antibody can be made cleavable. A feature of cleavable linkers is that once the conjugate enters the target cell, the siRNA and antibody separate, which can alter the intracellular tracking including endosomal release and RISC loading [69., 70., 71.]. pH sensitivity and photosensitivity are examples of strategies used in cleavable linker design [63].
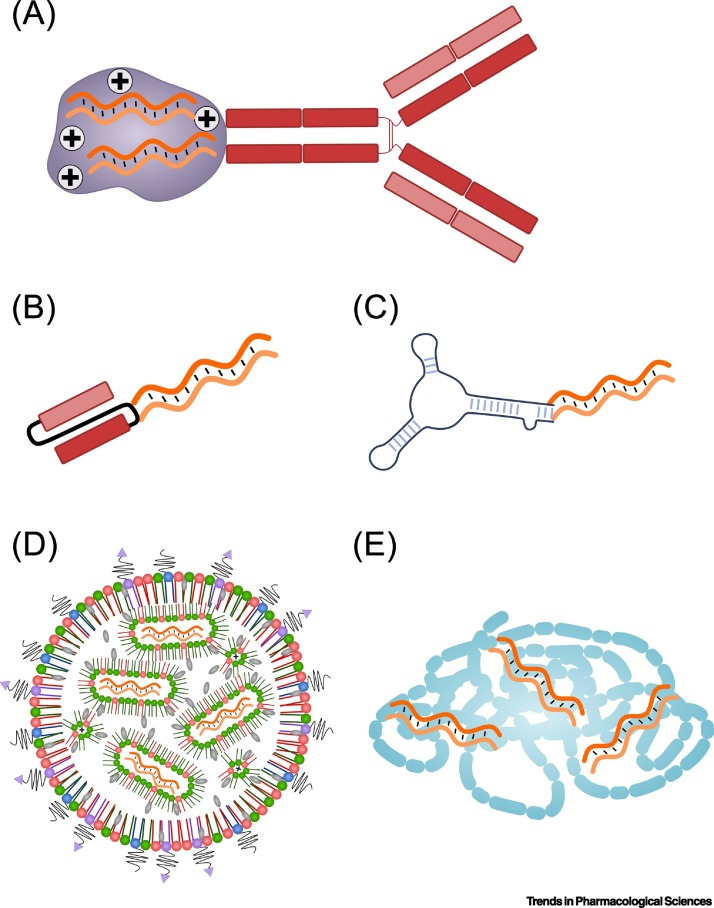
Figure 2.
Examples of Delivery Vehicles for Different RNA Payloads.
(A) Antibody conjugated to RNA molecules which can be mediated by using for instance positively charged protamine (shown as plus signs). (B) Conjugate of RNA with a single-chain variable fragment (scFv). (C) RNA–aptamer conjugates. (D) RNA encapsulated in lipid nanoparticles (LNPs). Cationic or ionizable lipids (shown in green) aid in encapsulating the RNA payload through electrostatic interactions. This way, the RNA is encapsulated in inverted micelles. Cholesterol (shown in grey) provides stability to the LNPs. The surface of the LNPs are generally coated with PEG (black lines). Reactive groups such as maleimide (purple triangles) can be linked to the PEG and are used to functionalize the LNPs with targeting moieties (chemical conjugation of targeting moieties). (E) Cationic polymers can encapsulate RNA therapeutics by electrostatic interactions.
An alternative to the use of full-length mAbs is the use of smaller antibody derivatives such as single-chain variable fragment (scFv ) [72] (Figure 2B). Examples of such applications include the delivery of CXC chemokine receptor (CXCR)4–siRNA for HER2+ breast cancer therapy [73] and CD44–siRNA targeted by a scFv against epidermal growth factor receptor (EGFR) in triple-negative breast cancer [74].
Aptamers
Aptamers have been used to direct therapeutic RNA molecules to target sites [75] (Figure 2C). In terms of specificity and affinity, they are highly comparable to antibodies but are smaller, have a higher stability, and are easier to generate [76]. In 2018, Zhou et al. used an RNA aptamer against gp120 (the exterior envelope glycoprotein in HIV that drives its entry into the host CD4+ T cells) to deliver anti-HIV siRNA to infected T cells. The siRNA induces transcriptional gene silencing by targeting specific sites within the viral promoter, resulting in suppression of HIV infection and protection of CD4+ T cells in mice [77]. Another study used an RNA aptamer against prostate-specific membrane antigen (PSMA), a cell-surface receptor that is highly upregulated in certain prostate cancers, to target prostate cancer cells in a mouse xenograft model engrafted with PSMA+ human cancer cells. The RNA therapeutic delivered in this study was the CRISPR/Cas9 system against polo-like kinase (PLK)1. The results demonstrated high cell-type-specific delivery that translated into antitumor activity in vivo [78].
Lipid Nanoparticles
Initially developed as carriers for in vivo siRNA delivery [79], lipid nanoparticles (LNPs) are complex and large structures (~100 nm) that also have been used to deliver large RNA molecules such as mRNA in vivo [80]. LNPs can encapsulate large amounts of RNA [81] and protect them against RNase degradation and renal clearance (Figure 2D) [82]. Addition of polyethylene glycol (PEG) lipids has been proven to enhance LNP circulation time due to the steric barrier it creates around the LNP surface that protects the LNPs from interactions with plasma proteins, typically opsonins, that would target the LNPs for degradation by the MPS [83]. In fact, the use of PEG-lipids has become the standard since novel microfluidic mixing systems require the steric barrier provided by PEG to facilitate self-assembly. However, the use of PEG-lipids is a double-edged sword as the downside is that the same steric barrier inhibits interactions with the cell membrane and subsequent endosomal escape of therapeutic RNA [84]. Therefore, fine tuning of the amount of PEG-lipid and the length of the PEG chain is pivotal.
The interaction of PEG-lipid with the plasma proteins upon injection results in release of PEG from the LNP, which further complicates optimization of the LNP formulation. An important study in 2013 reported on the desorption rate of PEG-lipids from the LNPs into the aqueous phase. This desorption involves a transition state that requires a large amount of free energy. The length of the hydrophobic chain influences the amount of energy required to enter this activation state; the longer the hydrophobic chain, the more energy is required and hence the lower the in vivo PEG-lipid desorption rate. The results demonstrated that PEG-C18 desorption is low (0.2%/h), while initial desorption rates of PEG-C14 is ~2%/min [85]. Due to the positive and negative effects of PEG-lipids on in vivo intracellular RNA delivery, an optimal balance is required and, in most cases, including ~1.5% PEG-lipid (as percentage of the total lipid composition) is ideal. An increased concentration would result in a decrease of RNA delivery and can only be compensated by including shorter chain PEG-lipids (e.g. PEG-C14) that will enhance RNA desorption rates [85].
Polymers
Cationic polymers are well suited for the delivery of RNA due to their positive charge, chemical diversity, and compatibility with various targeting moieties (Figure 2E). Polymers can be linear or branched polymers, or consist of many branched repeats such as in the case of dendrimers [86]. Examples of the use of polymers for RNA delivery include polyethyleneimine (PEI) for the delivery of siRNA [87] and self-immolative polycarbonate-block-poly(α-amino)esters (dynamic materials that function as polycations that can lose their cationic charge to enhance endosomal escape) for mRNA delivery [9,88]. Dendrimers are popular for RNA delivery due to their tunable structure and monodispersity (all dendrimer molecules are as similar as possible and of a well-defined size upon synthesis). These hyperbranched macromolecules can be functionalized with many functional groups for use in, for instance, ligand attachment when targeted delivery is required [89,90]. Dong et al. described an example of targeted dendrimers for RNA delivery in 2018 where dendrimers encapsulated with siRNA were targeted with the RDGK peptide, which can target both the tumor endothelium (through ανβ3 integrin) and the tumor cells themselves (through neuropilin-1 receptor targeting). Using that system, the researchers silenced the cancer survival gene Hsp27 in vivo and achieved significant anticancer activity [91].
Moving to the Clinic
Since the FDA approval of the RNA therapeutic, pegaptanib (Macugen) in 2004 [92], the field of RNA has not progressed far. Besides the general difficulties in intracellular delivery of oligonucleotides to the cell/tissue of interest, RNA molecules are notoriously unstable due to the presence of ribonucleases. As discussed here, advances in nucleic acid chemistry combined with suitable delivery vehicles boosted the field, and these efforts eventually crystalized into the approval of the first siRNA-based drug, patisiran in 2018 [15]. Since then, the field of RNA therapeutics has received a boost, and this is evident when looking at the clinical pipeline that contains a wide range of RNA therapeutics at all stages of clinical development for a variety of medical conditions [34,93] (Table 1).
During the clinical development of patisiran, the LNP delivery vehicle seemed by far the most advanced and suitable carrier for siRNA delivery in vivo. The reason LNPs were so successful in hepatic gene silencing was due to the ability of ionizable LNPs to adsorb apoliporotein E (ApoE) in the circulation, giving rise to a natural targeting ligand that binds with high affinity to the low-density lipoprotein (LDL) receptor that is widely expressed on hepatocytes [94]. In late 2019, a second siRNA drug, givosiran, was approved [16]. Instead of LNPs, givosiran was delivered as a GalNAc-conjugated siRNA. Both currently approved siRNA-based drugs thus have a strong preferential uptake in the liver but use distinct mechanisms (ApoE vs GalNAc) [16,94].
Based on studies that led to givosiran, it seems that GalNAc-based siRNA delivery strategies have some important benefits over LNPs. In terms of toxicity, ease of production and the required dose frequency, GalNAc is superior [95]. Furthermore, GalNAc-based strategies can be administered subcutaneously [96] as opposed to intravenously for LNPs. LNPs also require an extensive premedication regime to deal with infusion-related reactions [97]. Because of this, many siRNA candidates currently in the clinical pipeline are based on GalNAc or similar conjugates (see the siRNA section in Table 1). Alnylam currently has three candidate drugs in Phase III studies (fitusiran, lumasiran, and vutrisiran)ii. A fourth one, inclisiran, showed positive results in Phase III studies and is currently under review for approval [98]. Arrowhead Pharmaceuticals has two candidates in Phase II studies (ARO-AAT and JNJ-3989)iii and Dicerna is building a clinical pipeline with three programs (Phyox, RG6346, and Shine)iv.
Multiple mRNA therapeutics are also being developed: Moderna has two candidate drugs in Phase II and an impressive early stage pipelinev; CureVac has several candidates in Phase Ivi; and BioNtech has one candidate against melanoma in Phase II and many candidates in developmentvii. Moreover, the recent outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in accelerated development of several mRNA vaccines: mRNA-1273 (Phase III) by Moderna; BNT162 (Phase II/III) by BioNTech; and CVnCoV Vaccine (Phase I) by CureVac (Table 1).
While siRNA and mRNA candidates are the most abundant, dozens of other RNA therapeutic candidates have also entered clinical trials (Table 1). Indeed, four RNA-based ASOs are currently under clinical investigation: QR-010, QR-421a, QR-1123, and QR110, all developed by ProQR. Furthermore, anti-miR and miRNA mimics, including miravirsen, SAR339375, cobomarsen, and MRG-201 are all currently in Phase II clinical trials (Table 1).
While the first clinical trial with the in vivo delivery of CRISPR/Cas9 components was launched recently (clinical trial number: NCT03872479), the delivery is via viral vectors [35]. This might soon change as several studies have reported on the in vivo delivery of CRISPR/Cas9 components using RNA-LNP in animal models. Notable examples include: delivery of Cas9 mRNA with LNPs (although viral vectors were used to deliver the sgRNA and repair template) to correct a fumarylacetoacetate hydrolase splicing mutation in a mouse model of hereditary tyrosinemia [36], and delivery of both Cas9 mRNA and sgRNA with LNPs to knockout the mouse transthyretin gene, resulting in >97% reduction of serum proteins levels that lasted for at least 12 months [37].
Concluding Remarks and Future Perspectives
This review covered the recent advances in delivery of RNA therapeutics. Various RNA payloads were described with different roles, ranging from overexpression to silencing and knockout of the target gene. The versatility of RNA molecules makes them popular therapeutic agents for a wide range of medical conditions. However, issues with delivery have slowed down the field of RNA therapeutics for many years. Poor cellular uptake, fast clearance from the circulation, and the induction of immune responses have necessitated optimization of both the RNA molecules as well as the delivery vehicles. Chemical modifications of RNA molecules truly boosted the field of RNA therapeutics and facilitated the shift from completely encapsulated RNA nanoparticles to the use of less complex RNA conjugates (e.g., GalNAc). The ability to protect RNA molecules from degradation in vivo turned out to be crucial as early attempts with RNA-based therapy resulted in direct enzymatic degradation eliminating any therapeutic potential [95]. Additionally, the discovery of circular RNA (circRNA) provided the scientific community with even more possibilities to protect and deliver therapeutic RNA. As circRNA is stable against exonucleolytic decay, it might become an important agent for therapeutic applications for longer-lasting effects [99].
There is, however, still plenty of work to be done to advance the dream of RNA therapy. For example, most RNA-based therapeutics are chemically modified RNAs and it is important to pay attention to potential adverse effects of these modifications. Not only does modified RNA differ structurally from unmodified RNA but it can also affect the functionality. Therefore, some caution is necessary when focusing on an improved pharmacokinetic profile of synthetic RNA molecules with disregard to altered biological functionalities that might not be directly visible. To address this issue, a new class of RNAs has been explored. These are the bioengineered RNA agents (BERAs), which are made and folded in living cells and demonstrate favorable stability in human cells. They could represent a more natural alternative to the extensively chemical modified RNAs currently used [100].
Furthermore, in terms of delivery of RNA therapeutics, most strategies have focused on the ‘low-hanging fruit’ by delivering the RNA to the liver and most drug candidates have hence focused on liver- and kidney-related diseases (with some upcoming potential in the central nervous system as well) [101]. Future ambitions should focus on sites in the body that are harder to reach such as leukocytes that are dispersed over the body and intrinsically hard to transfect. Currently, it seems that this necessitates the inclusion of targeting moieties such as peptides, antibodies, other proteins, polysaccharides, and more. Besides cell targeting, such targeting moieties might also aid in enhancing internalization of the payload.
Internalization and endosomal escape is a real bottleneck in RNA delivery. While a small fraction of siRNA release in the cytosol is sufficient for knockdown, other RNA-based approaches still lack potency due to endosomal entrapment, and we wonder how delivery systems can enhance this (see Outstanding Questions). This is not trivial, as can be seen from the use of PEG, which on the one hand is a critical component in many delivery systems, while on the other hand, it inhibits endosomal escape.
While most delivery efforts for small RNAs such as siRNA are now invested in GalNAc-based or similar conjugates, larger delivery vehicles such as polymers or LNPs should not yet be disregarded. This is because other vehicles might aid in the improvement of extrahepatic delivery. We and others have demonstrated the feasibility of accurately targeting specific cell populations in vivo in animal models by functionalizing the surface of the delivery vehicle with targeting moieties [102., 103., 104., 105., 106., 107., 108., 109., 110.]. Therefore, for leukocyte-implicated diseases such as chronic lymphocytic leukemia or autoimmunity, tumors with insufficient enhanced permeability and retention (EPR) effect or any other condition where specific targeting to the cell of interest is critical, a variety of delivery vehicles such as LNPs and dendrimers that can be functionalized with targeting moieties, are in high demand [111]. Perhaps, including targeting moieties can also bring down costs by maximizing accumulation of the therapeutic agents at the cell/tissue of interest. This is especially important for mRNA, which is expensive to prepare and might therefore be universally less accessible (see Outstanding Questions).
In conclusion, the field of RNA therapeutics has seen major developments at multiple levels (targeting, RNA modifications, delivery vehicles, etc.) and a wide variety of different RNA molecules are currently at different stages of (pre-)clinical development (Table 1). Owing to the invested efforts, RNA therapeutics moved from unrealistic dreams to genuine realities and many actors in the field are determined to drive the RNA revolution to the next level.
Outstanding Questions.
What options can to be explored to enhance endosomal escape for RNA therapeutics?
As mRNA is expensive and difficult to deliver intracellularly, could saRNA replace mRNA to boost expression of endogenous genes, leaving mRNA dedicated only for the introduction of exogenous proteins (such as for vaccines or Cas9)?
How can we overcome the problem of saturation of certain intracellular components (such as AGO2) in situations where multiple RNAs are used to maximize efficacy for simultaneous silencing/knockout of one gene and overexpression of another gene?
Alt-text: Outstanding Questions
Acknowledgments
This work was supported in part by an European Research Council (ERC) grant LeukoTheranostics (Award # 647410), by EXPERT EU Horizon 2020, Israel Science Foundation (ISF) grants (1178/16; 3150/19 and 2012/20), Israel Cancer Research Fund (ICRF) (Grant # 16-1285-PG) and by the Lewis Trust grant awarded to D.P.
Glossary
- Asialoglycoprotein receptor (ASGR)
a C-type lectin that is highly expressed on hepatocytes and removes desialylated glycoproteins from the circulation.
- Endosomal escape
when a payload is internalized through endocytosis, it enters endocytic vesicles called endosomes. The payload needs to escape from the endosome in order to reach the cytosol, failure to escape results in transport to late endosomes and eventually lysosomes resulting in degradation of the payload.
- Enhanced permeability and retention (EPR) effect
a theory by which high-molecular-weight nontargeted drugs accumulate in cancer tissues or tissues with inflammation due to hypervascularization and leaky vascular walls.
- Locked nucleic acid (LNA)
nucleic acid analog that possesses a 2'-O, 4'-C methylene bridge that reduces the flexibility of the pentose ring. LNAs have higher resistance against nucleases and altered hybridization properties.
- Macropinocytosis
ingestion of extracellular liquid and dissolved molecules by cells.
- Ribozymes
RNA enzymes; RNA molecules with catalytic activity.
Resources
ihttps://clinicaltrials.gov/iiwww.alnylam.com/alnylam-rnai-pipeline/iiihttps://arrowheadpharma.com/pipelineivhttps://dicerna.com/pipelinevwww.modernatx.com/pipelineviwww.curevac.com/our-pipelineviihttps://biontech.de/science/pipelineviii www.cancer.gov/
ixwww.olixpharma.com/References
- 1.Gurevich E.V., Gurevich V.V. Therapeutic potential of small molecules and engineered proteins. Handb. Exp. Pharmacol. 2014;219:1–12. doi: 10.1007/978-3-642-41199-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodgers G., et al. Glimmers in illuminating the druggable genome. Nat. Rev. Drug Discov. 2018;17:301–302. doi: 10.1038/nrd.2017.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santos R., et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2016;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sierra J.R., et al. Molecular mechanisms of acquired resistance to tyrosine kinase targeted therapy. Mol. Cancer. 2010;9:75. doi: 10.1186/1476-4598-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mullard A. 2018 FDA drug approvals. Nature reviews. Drug discovery. 2019;18:85–89. doi: 10.1038/d41573-019-00014-x. [DOI] [PubMed] [Google Scholar]
- 6.De La Torre B.G., Albericio F. The pharmaceutical industry in 2018. An analysis of FDA drug approvals from the perspective of molecules. Molecules. 2019;24:809. doi: 10.3390/molecules24040809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett C.F. Therapeutic antisense oligonucleotides are coming of age. Annu. Rev. Med. 2019;70:307–321. doi: 10.1146/annurev-med-041217-010829. [DOI] [PubMed] [Google Scholar]
- 8.Hobernik D., Bros M. DNA vaccines—how far from clinical use? Int. J. Mol. Sci. 2018;19:3605. doi: 10.3390/ijms19113605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kowalski P.S., et al. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewin A.S., Hauswirth W.W. Ribozyme gene therapy: applications for molecular medicine. Trends in Molecular Medicine. 2001;7:221–228. doi: 10.1016/s1471-4914(01)01965-7. [DOI] [PubMed] [Google Scholar]
- 11.Trujillo A.J., et al. A discovery with potential to revitalize hammerhead ribozyme therapeutics for treatment of inherited retinal degenerations. Adv. Exp. Med. Biol. 2019;1185:119–124. doi: 10.1007/978-3-030-27378-1_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanna J., et al. The potential for microRNA therapeutics and clinical research. Front. Genet. 2019;10:478. doi: 10.3389/fgene.2019.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ledford H. Gene-silencing technology gets first drug approval after 20-year wait. Nature. 2018;560:291–292. doi: 10.1038/d41586-018-05867-7. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L., et al. In vivo targeted delivery of CD40 shRNA to mouse intestinal dendritic cells by oral administration of recombinant Saccharomyces cerevisiae. Gene Ther. 2014;21:709–714. doi: 10.1038/gt.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Titze-de-Almeida S.S., et al. Leading RNA interference therapeutics part 1: silencing hereditary transthyretin amyloidosis, with a focus on patisiran. Mol. Diagn. Ther. 2020;24:49–59. doi: 10.1007/s40291-019-00434-w. [DOI] [PubMed] [Google Scholar]
- 16.de Paula Brandão P.R., et al. Leading RNA interference therapeutics part 2: silencing delta-aminolevulinic acid synthase 1, with a focus on givosiran. Mol. Diagn. Ther. 2019;24:61–68. doi: 10.1007/s40291-019-00438-6. [DOI] [PubMed] [Google Scholar]
- 17.Dolgin E. Unlocking the potential of vaccines built on messenger RNA. Nature. 2019;574:S10–S12. doi: 10.1038/d41586-019-03072-8. [DOI] [PubMed] [Google Scholar]
- 18.Bajan S., Hutvagner G. RNA-based therapeutics: from antisense oligonucleotides to miRNAs. Cells. 2020;9:137. doi: 10.3390/cells9010137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rinaldi C., Wood M.J.A. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat. Rev. Neurol. 2018;14:9–22. doi: 10.1038/nrneurol.2017.148. [DOI] [PubMed] [Google Scholar]
- 20.Dulla K., et al. Splice-modulating oligonucleotide QR-110 restores CEP290 mRNA and function in human c.2991+1655A>G LCA10 Models. Mol. Ther. Nucleic Acids. 2018;12:730–740. doi: 10.1016/j.omtn.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stenvang J., et al. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3:1. doi: 10.1186/1758-907X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–221. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 23.Dykxhoorn D.M., Lieberman J. Running interference: prospects and obstacles to using small interfering RNAs as small molecule drugs. Annu. Rev. Biomed. Eng. 2006;8:377–402. doi: 10.1146/annurev.bioeng.8.061505.095848. [DOI] [PubMed] [Google Scholar]
- 24.Ni H., et al. Extracellular mRNA induces dendritic cell activation by stimulating tumor necrosis factor-α secretion and signaling through a nucleotide receptor. J. Biol. Chem. 2002;277:12689–12696. doi: 10.1074/jbc.M110729200. [DOI] [PubMed] [Google Scholar]
- 25.Wolff J.A., et al. Direct gene transfer into mouse muscle in vivo. Science (80- ) 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 26.Youn H., Chung J.K. Modified mRNA as an alternative to plasmid DNA (pDNA) for transcript replacement and vaccination therapy. Expert Opin. Biol. Ther. 2015;15:1337–1348. doi: 10.1517/14712598.2015.1057563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner T.E., et al. Small-molecule-based regulation of RNA-delivered circuits in mammalian cells. Nat. Chem. Biol. 2018;14:1043–1050. doi: 10.1038/s41589-018-0146-9. [DOI] [PubMed] [Google Scholar]
- 28.Zheng C., Baum B.J. Evaluation of promoters for use in tissue-specific gene delivery. Methods Mol. Biol. 2008;434:205–219. doi: 10.1007/978-1-60327-248-3_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roney I.J., et al. Improvement of the reverse tetracycline transactivator by single amino acid substitutions that reduce leaky target gene expression to undetectable levels. Sci. Rep. 2016;6:1–8. doi: 10.1038/srep27697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magadum A., et al. mRNA-based protein replacement therapy for the heart. Mol. Ther. 2019;27:785–793. doi: 10.1016/j.ymthe.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang C., et al. Advances in mRNA vaccines for infectious diseases. Front. Immunol. 2019;10:594. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Versteeg L., et al. Enlisting the mRNA vaccine platform to combat parasitic infections. Vaccines. 2019;7 doi: 10.3390/vaccines7040122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Hoecke L., Roose K. How mRNA therapeutics are entering the monoclonal antibody field. J. Transl. Med. 2019;17:54. doi: 10.1186/s12967-019-1804-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou L.-Y., et al. Current RNA-based therapeutics in clinical trials. Curr. Gene Ther. 2019;19:172–196. doi: 10.2174/1566523219666190719100526. [DOI] [PubMed] [Google Scholar]
- 35.Kaur H., et al. Aptamers in the therapeutics and diagnostics pipelines. Theranostics. 2018;8:4016–4032. doi: 10.7150/thno.25958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim Y.-K. RNA therapy: current status and future potential. Chonnam Med. J. 2020;56:87. doi: 10.4068/cmj.2020.56.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn J.D., et al. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018;22:2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 38.Kwok A., et al. Developing small activating RNA as a therapeutic: current challenges and promises. Ther. Deliv. 2019;10:151–164. doi: 10.4155/tde-2018-0061. [DOI] [PubMed] [Google Scholar]
- 39.Zhao X., et al. Mechanisms involved in the activation of C/EBPα by small activating RNA in hepatocellular carcinoma. Oncogene. 2019;38:3446–3457. doi: 10.1038/s41388-018-0665-6. [DOI] [PubMed] [Google Scholar]
- 40.Reebye V., et al. Gene activation of CEBPA using saRNA: preclinical studies of the first in human saRNA drug candidate for liver cancer. Oncogene. 2018;37:3216–3228. doi: 10.1038/s41388-018-0126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jinek M., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (80-. ) 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenblum D., et al. Progress and challenges towards CRISPR/Cas clinical translation. Adv. Drug Deliv. Rev. 2020 doi: 10.1016/j.addr.2020.07.004. Published online July10, 2020. [DOI] [PubMed] [Google Scholar]
- 43.Glass Z., et al. Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol. 2018;36:173–185. doi: 10.1016/j.tibtech.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y., et al. Ex vivo cell-based CRISPR/Cas9 genome editing for therapeutic applications. Biomaterials. 2020;234 doi: 10.1016/j.biomaterials.2019.119711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selvam C., et al. Therapeutic potential of chemically modified siRNA: recent trends. Chem. Biol. Drug Des. 2017;90:665–678. doi: 10.1111/cbdd.12993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freund I., et al. RNA modifications modulate activation of innate toll-like receptors. Genes. 2019;10 doi: 10.3390/genes10020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brisse M., Ly H. Comparative structure and function analysis of the RIG-I-like receptors: RIG-I and MDA5. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang X., et al. Gene silencing activity of siRNA molecules containing phosphorodithioate substitutions. ACS Chem. Biol. 2012;7:1214–1220. doi: 10.1021/cb300078e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kubo T., et al. Palmitic acid-conjugated 21-nucleotide siRNA enhances gene-silencing activity. Mol. Pharm. 2011;8:2193–2203. doi: 10.1021/mp200250f. [DOI] [PubMed] [Google Scholar]
- 50.Kubo T., et al. SiRNAs conjugated with aromatic compounds induce RISC-mediated antisense strand selection and strong gene-silencing activity. Biochem. Biophys. Res. Commun. 2012;426:571–577. doi: 10.1016/j.bbrc.2012.08.128. [DOI] [PubMed] [Google Scholar]
- 51.Kaczmarek J.C., et al. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med. 2017;9 doi: 10.1186/s13073-017-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson J.D., et al. Toxicological and pharmacokinetic properties of chemically modified siRNAs targeting p53 RNA following intravenous administration. Nucleic Acid Ther. 2012;22:255–264. doi: 10.1089/nat.2012.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park J., et al. Pharmacokinetics and biodistribution of recently-developed siRNA nanomedicines. Adv. Drug Deliv. Rev. 2016;104:93–109. doi: 10.1016/j.addr.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh A., et al. Advances in siRNA delivery in cancer therapy. Artif. Cells Nanomed. Biotechnol. 2018;46:274–283. doi: 10.1080/21691401.2017.1307210. [DOI] [PubMed] [Google Scholar]
- 55.Kramps T., Probst J. Messenger RNA-based vaccines: progress, challenges, applications. Wiley Interdiscip. Rev. RNA. 2013;4:737–749. doi: 10.1002/wrna.1189. [DOI] [PubMed] [Google Scholar]
- 56.Diken M., et al. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther. 2011;18:702–708. doi: 10.1038/gt.2011.17. [DOI] [PubMed] [Google Scholar]
- 57.Tatiparti K., et al. siRNA delivery strategies: a comprehensive review of recent developments. Nanomaterials. 2017;7 doi: 10.3390/nano7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.C. Stanley T., et al. Phosphorothioate modified oligonucleotide–protein interactions. Nucleic Acids Res. 2020;48:5235–5253. doi: 10.1093/nar/gkaa299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan M., et al. New paradigms on siRNA local application. BMB Rep. 2015;48:147–152. doi: 10.5483/BMBRep.2015.48.3.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carlsson L., et al. Biocompatible, purified VEGF-A mRNA improves cardiac function after intracardiac injection 1 week post-myocardial infarction in swine. Mol. Ther. Methods Clin. Dev. 2018;9:330–346. doi: 10.1016/j.omtm.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Golombek S., et al. Intradermal delivery of synthetic mRNA using hollow microneedles for efficient and rapid production of exogenous proteins in skin. Mol. Ther. Nucleic Acids. 2018;11:382–392. doi: 10.1016/j.omtn.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vogel A.B., et al. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol. Ther. 2018;26:446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chernikov I.V., et al. Current development of siRNA bioconjugates: from research to the clinic. Front. Pharmacol. 2019;10 doi: 10.3389/fphar.2019.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steichen S.D., et al. A review of current nanoparticle and targeting moieties for the delivery of cancer therapeutics. Eur. J. Pharm. Sci. 2013;48:416–427. doi: 10.1016/j.ejps.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prakash T.P., et al. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N -acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 2014;42:8796–8807. doi: 10.1093/nar/gku531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dammes N., Peer D. Monoclonal antibody-based molecular imaging strategies and theranostic opportunities. Theranostics. 2020;10:938–955. doi: 10.7150/thno.37443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sugo T., et al. Development of antibody-siRNA conjugate targeted to cardiac and skeletal muscles. J. Control. Release. 2016;237:1–13. doi: 10.1016/j.jconrel.2016.06.036. [DOI] [PubMed] [Google Scholar]
- 68.Ibtehaj N., Huda R. High-dose BAFF receptor specific mAb-siRNA conjugate generates Fas-expressing B cells in lymph nodes and high-affinity serum autoantibody in a myasthenia mouse model. Clin. Immunol. 2017;176:122–130. doi: 10.1016/j.clim.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 69.Benizri S., et al. Bioconjugated oligonucleotides: recent developments and therapeutic applications. Bioconjug. Chem. 2019;30:366–383. doi: 10.1021/acs.bioconjchem.8b00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lorenzer C., et al. Vol. 141. 2019. Targeted delivery and endosomal cellular uptake of DARPin-siRNA bioconjugates: influence of linker stability on gene silencing; pp. 37–50. [DOI] [PubMed] [Google Scholar]
- 71.Winkler J. Oligonucleotide conjugates for therapeutic applications. Ther. Deliv. 2013;4:791–809. doi: 10.4155/tde.13.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Safdari Y., et al. Use of single-chain antibody derivatives for targeted drug delivery. Mol. Med. 2016;22:258–270. doi: 10.2119/molmed.2016.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang K., et al. Targeted delivery of CXCR4-siRNA by scFv for HER2+ breast cancer therapy. Biomaterials. 2015;59:77–87. doi: 10.1016/j.biomaterials.2015.04.030. [DOI] [PubMed] [Google Scholar]
- 74.Fu W., et al. Targeted delivery of CD44s-siRNA by ScFv overcomes de novo resistance to cetuximab in triple negative breast cancer. Mol. Immunol. 2018;99:124–133. doi: 10.1016/j.molimm.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 75.Esposito C.L., et al. Aptamer chimeras for therapeutic delivery: the challenging perspectives. Genes. 2018;9 doi: 10.3390/genes9110529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou J., et al. Receptor-targeted aptamer-siRNA conjugate-directed transcriptional regulation of HIV-1. Theranostics. 2018;8:1575–1590. doi: 10.7150/thno.23085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhen S., et al. Targeted delivery of CRISPR/Cas9 to prostate cancer by modified gRNA using a flexible aptamer-cationic liposome. Oncotarget. 2017;8:9375–9387. doi: 10.18632/oncotarget.14072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramishetti S., et al. Advances in RNAi therapeutic delivery to leukocytes using lipid nanoparticles. J. Drug Target. 2016;24:780–786. doi: 10.3109/1061186X.2016.1172587. [DOI] [PubMed] [Google Scholar]
- 80.Veiga N., et al. Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat. Commun. 2018;9:4493. doi: 10.1038/s41467-018-06936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Belliveau N.M., et al. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol. Ther. Nucleic Acids. 2012;1:e37. doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Veiga N., et al. Targeted lipid nanoparticles for RNA therapeutics and immunomodulation in leukocytes. Adv. Drug Deliv. Rev. 2020 doi: 10.1016/j.addr.2020.04.002. Published online April 13, 2020. [DOI] [PubMed] [Google Scholar]
- 83.Kumar V., et al. Shielding of lipid nanoparticles for siRNA delivery: impact on physicochemical properties, cytokine induction, and efficacy. Mol. Ther. Nucleic Acids. 2014;3:e210. doi: 10.1038/mtna.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hatakeyama H., et al. Systemic delivery of siRNA to tumors using a lipid nanoparticle containing a tumor-specific cleavable PEG-lipid. Biomaterials. 2011;32:4306–4316. doi: 10.1016/j.biomaterials.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 85.Mui B.L., et al. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol. Ther. Nucleic Acids. 2013;2:e139. doi: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yin H., et al. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 87.Shen W., et al. Screening of efficient polymers for siRNA delivery in a library of hydrophobically modified polyethyleneimines. J. Mater. Chem. B. 2016;4:6468–6474. doi: 10.1039/c6tb01929c. [DOI] [PubMed] [Google Scholar]
- 88.McKinlay C.J., et al. Charge-altering releasable transporters (CARTs) for the delivery and release of mRNA in living animals. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E448–E456. doi: 10.1073/pnas.1614193114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mizrahy S., et al. Current progress in non-viral RNAi-based delivery strategies to lymphocytes. Mol. Ther. 2017;25:1491–1500. doi: 10.1016/j.ymthe.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Biswas S., Torchilin V.P. Dendrimers for siRNA delivery. Pharmaceuticals. 2013;6:161–183. doi: 10.3390/ph6020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dong Y., et al. A dual targeting dendrimer-mediated siRNA delivery system for effective gene silencing in cancer therapy. J. Am. Chem. Soc. 2018;140:16264–16274. doi: 10.1021/jacs.8b10021. [DOI] [PubMed] [Google Scholar]
- 92.Yin W., Rogge M. Targeting RNA: a transformative therapeutic strategy. Clin. Transl. Sci. 2019;12:98–112. doi: 10.1111/cts.12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gómez-Aguado I., et al. Nanomedicines to deliver mRNA: state of the art and future perspectives. Nanomaterials. 2020;10:364. doi: 10.3390/nano10020364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Akinc A., et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eisenstein M. Pharma’s roller-coaster relationship with RNA therapies. Nat. Outlook. 2019 Published online October 16, 2019. www.nature.com/articles/d41586-019-03069-3. [Google Scholar]
- 96.Springer A.D., Dowdy S.F. GalNAc-siRNA conjugates: leading the way for delivery of RNAi therapeutics. Nucleic Acid Ther. 2018;28:109–118. doi: 10.1089/nat.2018.0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Adams D., et al. Trial design and rationale for APOLLO, a phase 3, placebo-controlled study of patisiran in patients with hereditary ATTR amyloidosis with polyneuropathy. BMC Neurol. 2017;17 doi: 10.1186/s12883-017-0948-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ray K.K., et al. Two phase 3 trials of inclisiran in patients with elevated LDL cholesterol. N. Engl. J. Med. 2020;382:1507–1519. doi: 10.1056/NEJMoa1912387. [DOI] [PubMed] [Google Scholar]
- 99.Holdt L.M., et al. Circular RNAs as therapeutic agents and targets. Frontiers in Physiology. 2018;9:1262. doi: 10.3389/fphys.2018.01262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yu A.M., et al. RNA therapy: are we using the right molecules? Pharmacol. Ther. 2019;196:91–104. doi: 10.1016/j.pharmthera.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Setten R.L., et al. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019;18:421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- 102.Kedmi R., et al. A modular platform for targeted RNAi therapeutics. Nat. Nanotechnol. 2018;13:214–219. doi: 10.1038/s41565-017-0043-5. [DOI] [PubMed] [Google Scholar]
- 103.Veiga N., et al. Leukocyte-specific siRNA delivery revealing IRF8 as a potential anti-inflammatory target. J. Control. Release. 2019;313:33–41. doi: 10.1016/j.jconrel.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 104.Ramishetti S., et al. Vol. 9. 2015. Systemic gene silencing in primary T lymphocytes using targeted lipid nanoparticles; pp. 6706–6716. [DOI] [PubMed] [Google Scholar]
- 105.Ramishetti S., et al. A combinatorial library of lipid nanoparticles for RNA delivery to leukocytes. Adv. Mater. 2020;32 doi: 10.1002/adma.201906128. [DOI] [PubMed] [Google Scholar]
- 106.Weinstein S., et al. Harnessing RNAi-based nanomedicines for therapeutic gene silencing in B-cell malignancies. Proc. Natl. Acad. Sci. U. S. A. 2016;113 doi: 10.1073/pnas.1519273113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schneider B., et al. Targeted siRNA delivery and mRNA knockdown mediated by bispecific digoxigenin-binding antibodies. Mol. Ther. Nucleic Acids. 2012;1:e46. doi: 10.1038/mtna.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Okamoto A., et al. Systemic administration of siRNA with anti-HB-EGF antibody-modified lipid nanoparticles for the treatment of triple-negative breast cancer. Mol. Pharm. 2018;15:1495–1504. doi: 10.1021/acs.molpharmaceut.7b01055. [DOI] [PubMed] [Google Scholar]
- 109.Tietze S., et al. A poly(propyleneimine) dendrimer-based polyplex-system for single-chain antibody-mediated targeted delivery and cellular uptake of siRNA. Small. 2017;13 doi: 10.1002/smll.201700072. [DOI] [PubMed] [Google Scholar]
- 110.Min H.S., et al. Tuned density of anti-tissue factor antibody fragment onto siRNA-loaded polyion complex micelles for optimizing targetability into pancreatic cancer cells. Biomacromolecules. 2018;19:2320–2329. doi: 10.1021/acs.biomac.8b00507. [DOI] [PubMed] [Google Scholar]
- 111.Lorenzer C., et al. Going beyond the liver: progress and challenges of targeted delivery of siRNA therapeutics. J. Control. Release. 2015;203:1–15. doi: 10.1016/j.jconrel.2015.02.003. [DOI] [PubMed] [Google Scholar]