We read with interest the recent article by Perchetti et al. that describes 4-way pooling to increase throughput for SARS-CoV-2 testing [1]. We agree with these and other authors that pooling strategy in an appropriate population with low pre-test probability is a cost-effective way to increase testing capacity [2], especially during critical shortages in test reagents and availability of trained personnel [3,4].
We instituted a manual pooling strategy to screen asymptomatic healthcare workers.
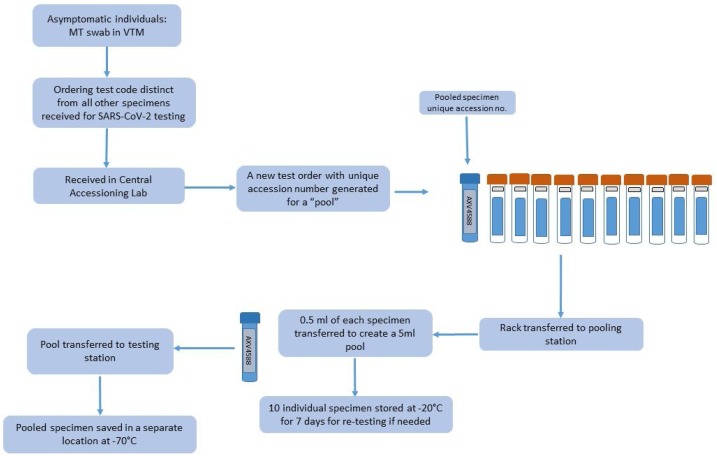
In addition to the validation of optimum pool size to determine any reduction in sensitivity that would potentially result in false negative results at or near the lower limit of detection, we also instituted measures to reduce pre-test errors during accessioning and pooling. (Fig. 1 ).
Fig. 1.
Unidirectional workflow for SARS-CoV-2 testing of 10-specimen pools. Each pool (blue tube) receives a unique accession number (grey label shown as AXV4588). Each individual specimen in the pool (orange cap tubes) receive two identifiers, one unique identifier for the individual specimen (blue label) and one common (grey label) linking each of the ten specimens to the pool. The entire rack of 11 tubes are moved through the multiple stations. Note: blue “pool” tube is empty and added to the rack at the accessioning station while orange cap tubes containing MT swab with VTM from 10 individuals are received and accessioned at the pooling station before generation of a “pool”.
To ascertain the loss in analytical sensitivity, nasal mid-turbinate (MT) and nasopharyngeal (NP) swabs in viral transport medium (VTM) at the lower limit of detection were tested as “contrived pools” using the CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR (CDC assay) and the Panther Fusion® SARS-CoV-2 LDT (Panther) assay.
Three different schemes using ten (n = 5), seven (n = 3) and five specimens (n = 3) per pool were analyzed. Varying number of positive specimens (ranging from one to three) were used to prepare contrived pools. The reduction in Ct values were 2.7-3.6 (in 10-specimen) and 0.2-1.8 (in 7- or 5-specimen) pool, respectively. All pools with a positive specimen with Ct <36 were detected, regardless of pool size.
To further evaluate sensitivity of pooling strategy, 27 ten specimen pools consisting of one positive and nine negative specimens, were evaluated using the CDC (n = 12) and the Panther assay (n = 15). Reduction in Ct was 3.13 ± 0.69 (CDC assay) and 3.47 ± 0.45 (Panther assay).
To streamline the testing algorithm, reduce pre-analytic errors, and minimize cross-contamination during pooling, we instituted physical separation of pooling steps and established unidirectional workflow (Fig. 1).
Each pool was ordered under a unique identifier which linked individual specimens within that pool. This enabled de-convolution of positive pools for subsequent testing of individual specimens. When negative, each specimen in the pool was resulted with a comment that testing was performed as pooled asymptomatic surveillance screen.
Pool size for infectious disease testing is defined based on disease prevalence and test sensitivity [[5], [6], [7]]. In the absence of defined epidemiology for SARS-CoV-2, our decision to pool 10 specimens was based on validation results that detected specimens with Ct <36 when pooled. This reduction in sensitivity was acceptable since the majority of first-time positive patients had a Ct value of 15-33 and with evidence that specimens with Ct>35 are likely non- or significantly less infectious [8,9].
A total of 700 pools (7000 individual specimens) were tested between May 21st to July 31st 2020, using the Panther assay (n = 332) and the CDC assay (n = 368). Eight positive pools were detected, equivalent to 0.11% positivity rate in asymptomatic healthcare workers. Every positive pool yielded only one positive specimen upon deconvolution (Table 1 ).
Table 1.
Results of Positive Asymptomatic Prospective Screening in Healthcare Workers: Comparison of Ct of Pooled specimens versus Individual Specimens Tested.
| Pool No. | Ct of pool (0.5 ml per sample to make a 5 ml pool) |
Ct of individual positive in that pool (each specimen tested individually) |
Difference in Ct‡ | ||
|---|---|---|---|---|---|
| Ct ORF1a/b (Panther Assay) | Ct N1/N2 (CDC assay) | Ct ORF1a/b (Panther Assay) | Ct N1/N2 (CDC assay) | ||
| POOL1 | 34.2/40 | 31.4/35.7 | 2.8 | ||
| POOL2 | 19.2/21.3 | 16.7/18.2 | 2.5 | ||
| POOL3 | 19.7/22.6 | 17.7/17.6 | 2 | ||
| POOL4 | 28.4 | 25.4/27.8 | 3 | ||
| POOL5 | 21.3/23.4 | 18.6/20.1 | 2.7 | ||
| POOL6* | 38.4 | 35.2 | 3.2 | ||
| POOL7 | 23 | 22.1/25.1 | 19.6 | 3.4/2.5 | |
| POOL8 | 28.7 | 25.4 | 3.3 | ||
Values of Ct ORF1a/b and Ct N1 targets were used for calculation of mean of difference in Ct; for Pool 7 difference in Ct between individual and pool shown for both platforms.
The positive individual of this pool returned with symptoms 3 days later. Diagnostic testing of their NP swab was positive (Ct 19.2), indicating that our pooling algorithm effectively detected this pre-symptomatic individual.
We conclude that 10-specimen manual pooling algorithm maintaining a unidirectional workflow is effective for surveillance testing of SARS-CoV-2 in asymptomatic healthcare workers.
Funding
This work was supported by the Department of Laboratory Medicine, Clinical Center, National Institutes of Health.
References
- 1.Perchetti G.A., Sullivan K.W., Pepper G., et al. Pooling of SARS-CoV-2 samples to increase molecular testing throughput. J Clin Virol. 2020;131(104570) doi: 10.1016/j.jcv.2020.104570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McMahan C.S., Tebbs J.M., Bilder C.R. Informative Dorfman screening. Biometrics. 2012;68:287–296. doi: 10.1111/j.1541-0420.2011.01644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdalhamid B., Bilder C.R., McCutchen E.L., et al. medRxiv; 2020. Assessment of Specimen Pooling to Conserve SARS CoV-2 Testing Resources. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hogan C.A., Sahoo M.K., Pinsky B.A. Sample Pooling as a Strategy to Detect Community Transmission of SARS-CoV-2. JAMA. 2020 doi: 10.1001/jama.2020.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dize L., West S.K., Mkocha H., et al. Evaluation of pooled ocular and vaginal swabs by the Cepheid GeneXpert CT/NG assay for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae compared to the GenProbe Aptima Combo 2 Assay. Diagn Microbiol Infect Dis. 2015;81:102–104. doi: 10.1016/j.diagmicrobio.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hitt B.D., Bilder C.R., Tebbs J.M., et al. The objective function controversy for group testing: Much ado about nothing? Stat Med. 2019;38:4912–4923. doi: 10.1002/sim.8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van TT Miller J, Warshauer D.M., et al. Pooling nasopharyngeal/throat swab specimens to increase testing capacity for influenza viruses by PCR. J Clin Microbiol. 2012;50:891–896. doi: 10.1128/JCM.05631-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullard J., Dust K., Funk D., et al. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.La Scola B., Le Bideau M., Andreani J., et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]