Abstract
The COVID-19 pandemic in the United States created a unique situation where multiple molecular SARS-CoV-2 diagnostic assays rapidly received Emergency Use Authorization by the FDA and were validated by laboratories and utilized clinically, all within a period of a few weeks. We compared the performance of four of these assays that were evaluated for use at our institution: Abbott RealTime m2000 SARS-CoV-2 Assay, DiaSorin Simplexa COVID-19 Direct, Cepheid Xpert Xpress SARS-CoV-2, and Abbott ID NOW COVID-19. Nasopharyngeal and nasal specimens were collected from 88 ED and hospital-admitted patients and tested by the four methods in parallel to compare performance. ID NOW performance stood out as significantly worse than the other 3 assays despite demonstrating comparable analytic sensitivity. Further study determined that the use of a nasal swab compared to a nylon flocked nasopharyngeal swab, as well as use in a population chronically vs. acutely positive for SARS-CoV-2, were substantial factors.
Keywords: COVID-19, Molecular diagnostics, SARS-CoV-2, Nasopharyngeal swab, Nasal swab, Ct value
1. Introduction
The rapid onset of COVID-19 in the United States resulted in an accelerated pace of both SARS-CoV-2 nucleic acid amplification test (NAAT) development and FDA Emergency Use Authorization (EUA) approvals. Clinical microbiology laboratories that typically would take weeks to evaluate and verify performance characteristics for a FDA approved diagnostic test had little choice but to perform abbreviated validation and/or verification studies of assays, benchmarked against limited FDA EUA performance data, in a matter of days. The sheer volume of COVID-19 test requests from different patient populations, different specimen types, and with different turnaround time needs demanded that laboratories implement more than one type of NAAT to respond to the crisis.
SARS-CoV-2 testing in our laboratory began with the CDC EUA assay performed on Abbott m2000, but due to that assay’s significant throughput constraints (24 specimens in 8 h), we quickly verified and switched to the Abbott RealTime SARS-CoV-2 EUA Assay (m2000) once released (94 specimens in ~8 h). Although this assay provided capacity for our outpatient testing needs, we also verified the DiaSorin Simplexa COVID-19 Direct (Simplexa) assay, capable of resulting 8 specimens in 90 min, and used this assay as a rapid turn-around time (TAT) option for our inpatient and emergency department (ED) populations. Within a few weeks, additional SARS-CoV-2 NAAT options emerged that were specifically designed for rapid testing of patients in the point of care setting: the Cepheid Xpert Xpress SARS-CoV-2 (Xpert) assay, which could provide results in 45 min, and the Abbott ID NOW COVID-19 (ID NOW) assay, ultimately approved for direct nasal, nasopharyngeal and throat swab testing only, with results in 5–15 min.
In the absence of clinical trials and a gold standard for COVID-19 diagnosis, the clinical performance of SARS-CoV-2 assays is unclear. Anecdotal claims of poor NAAT performance exist in the lay press, and limited studies have shown variable performance of rapid POC tests (Cradic et al., 2020; Harrington et al., 2020; Mitchell and George, 2020; Moore et al., 2020; Rhoads et al., 2020; Smithgall et al., 2020; Visseaux et al., 2020; Wolters et al., 2020; Zhen et al., 2020). As a surrogate for a gold standard, a composite reference standard (CRS) can be used to determine the consensus of comparable assays and identify outlier assays in terms of clinical performance (Schiaffino et al., 2020). Using this approach, our goal was to evaluate the performance—in parallel—of three NAATs from nasopharyngeal (NP) swabs in M4-RT viral transport medium (VTM) (m2000, Xpert, Simplexa) and an NAAT assay performed directly from a nasal swab (ID NOW).
2. Methods
2.1. Accuracy study design
From April 22 to May 5, 2020, specimens were collected from 88 ED and hospital admitted patients and tested for SARS-CoV-2 on the RealTime m2000 SARS-CoV-2 Assay (Abbott Molecular, Des Plaines, IL), Simplexa™ COVID-19 Direct (DiaSorin, Cypress, CA), Xpert® Xpress SARS-CoV-2 (Cepheid, Sunnyvale, CA) and ID NOW COVID-19 (Abbott Molecular, Des Plaines, IL) within 24 h of collection. Each assay was performed according to manufacturer’s EUA instructions.
NP and nasal swabs were collected from 88 patients, of which 75 were patients presenting in the ED and 13 were from a population of recovering COVID-positive inpatients. NP specimen collection, transport to the hospital-based core microbiology laboratory in M4-RT viral transport medium (Thermo Fisher Scientific, San Diego, CA; VTM) and subsequent testing by Simplexa was performed as a part of routine clinical care. Residual NP specimen in VTM was stored at 4°C, transported to our offsite main laboratory, and within 24 h of collection, used for comparative study testing by m2000 and Xpert assays. At the time of NP swab collection, nasal swabs were collected in parallel from each of these patients and transported dry to the offsite main laboratory in a sealed sterile collection bag, stored at 4 °C and tested by ID NOW within 24 h, consistent with the package insert procedure.
In order to determine a percent agreement amongst the methods, we established a composite reference standard as defined by result agreement of SARS-CoV-2 target in at least 2 of 4 NAAT results. Agreement for each individual assay was compared to this standard.
2.2. Analytic limit of detection study design
Dilutions were prepared from ZeptoMetrix inactivated SARS-CoV-2 virus (1.70 × 105 TCID50/mL) that was internally quantified to 109.62 copies/mL relative to a standard curve of AccuPlex™ SARS-CoV-2 Reference Material (SeraCare) constructed on the m2000 assay. From that stock, dilutions were made in VTM (1,042, 521, 260, 130, 65, and 32.5 copies/mL) and tested on the m2000, Simplexa, and Xpert SARS-CoV-2 assays. Five replicates at each concentration were tested for each assay. In contrast to the aforementioned assays which test VTM directly, the test methodology of the ID NOW is designed for a dry nasal swab to be inserted directly into the on board elution buffer (EB) chamber. Had we used a volume of the above standards in VTM to inoculate the EB chamber, the actual concentration tested would be lower due to that dilution of VTM into EB. Instead, a separate set of ZeptoMetrix dilutions in VTM were made so that when added to the 2.5 mL of elution buffer in the ID NOW EB chamber, equivalent concentrations were achieved (1,042, 522, 262, 105, and 53 copies/mL) to that which was tested on the other assays. For example, the first concentration tested was made by diluting 357 μL of VTM at a concentration of 8337 copies/mL into 2.5 mL of EB, resulting in a final concentration tested of 1042 copies/mL. Five replicates at each concentration were tested.
3. Results and discussion
Nasal swabs directly tested on the ID NOW assay had 48% positive agreement compared to the CRS, whereas Simplexa had 88%, m2000 had 96% and Xpert had 100% positive agreement (Table 1a ). While the deficit in positive percent agreement (PPA) seen in ID NOW test results is consistent with other early release studies in the scientific literature (Cradic et al., 2020; Harrington et al., 2020; Mitchell and George, 2020; Moore et al., 2020; Rhoads et al., 2020; Smithgall et al., 2020; Zhen et al., 2020), it is surprising given the ID NOW’s LOD claim of 125 genome equivalents/mL, which is similar to the 100 copies/mL claimed by the m2000 method, 250 copies/mL claimed by Xpress and 242 copies/mL claimed by Simplexa. To clarify this apparent discrepancy, a direct assessment of the analytic sensitivity of all assays in this comparison was performed utilizing dilutions of inactivated SARS-CoV-2 whole virus (ZeptoMetrix). In this limited LOD study, we found each assay had comparable LODs to the reported LOD data in their package insert, including the LOD of the ID NOW assay, as shown in Table 2.
Table 1a.
Agreement of 4 SARS-CoV-2 NAATs relative to the CRS.
| Composite reference standard (CRS) |
Percent agreement with CRS | 95% CI | ||
|---|---|---|---|---|
| Detected | Not detected | |||
| ID NOW | Positive agreement = 48% | 0.30–0.67 | ||
| Detected | 12 | 0 | Negative agreement = 100% | 0.94–1.0 |
| Not detected | 13 | 63 | Overall agreement = 85% | |
| Simplexa | Positive agreement = 88% | 0.70–0.96 | ||
| Detected | 22 | 0 | Negative agreement =100% | 0.94–1.0 |
| Not detected | 3 | 63 | Overall agreement = 97% | |
| m2000 | Positive agreement = 96% | 0.80–1.0 | ||
| Detected | 24 | 0 | Negative agreement =100% | 0.94–1.0 |
| Not detected | 1 | 61 | Overall agreement = 99% | |
| 2 invalids | ||||
| Xpert | Positive agreement = 100% | 0.87–1.0 | ||
| Detected | 25 | 2 | Negative agreement = 97% | 0.87–0.99 |
| Not detected | 0 | 60 | Overall agreement = 98% | |
| 1 invalid | ||||
Table 2.
Comparison of assay analytic limit of detection performance
| Study LOD in M4-RT (copies/mL)⁎ | Package insert LOD | Average Ct at LOD | |
|---|---|---|---|
| m2000 | 32.5 | 100 copies/mL | 26.5⁎⁎ |
| Cepheid | 65 | 250 copies/mL | 36.7 / 39.8 |
| ID NOW | 262 | 125 genome equivalents/mL | N/A |
| Simplexa | 521 | 242 copies/mL | 32.6 / 33.0 |
Defined as lowest dilution in which 5/5 replicates detected.
Reported Ct value for m2000 excludes 10 unread cycles.
Similarities in the LODs among the assays suggest that other factors contribute to the differences in comparative performance when testing clinical specimens. When additional ID NOW testing was performed on the 25 NP VTM specimens that were positive based on the CRS, 6 additional patients were detected that were negative by nasal swab testing. This suggests that testing an NP swab in VTM on ID Now may have better performance than a direct nasal swab, as use of the NP VTM improves the PPA from 48% to 64%. However, testing an NP swab in VTM was recently removed as an approved source from the original ID NOW package insert based on concerns about false negatives, whereas a nasal swab is provided as an approved collection device. Further studies should be conducted directly comparing the performance of direct (not placed in VTM) NP swabs on the ID NOW to NP swabs in VTM tested by other NAATs.
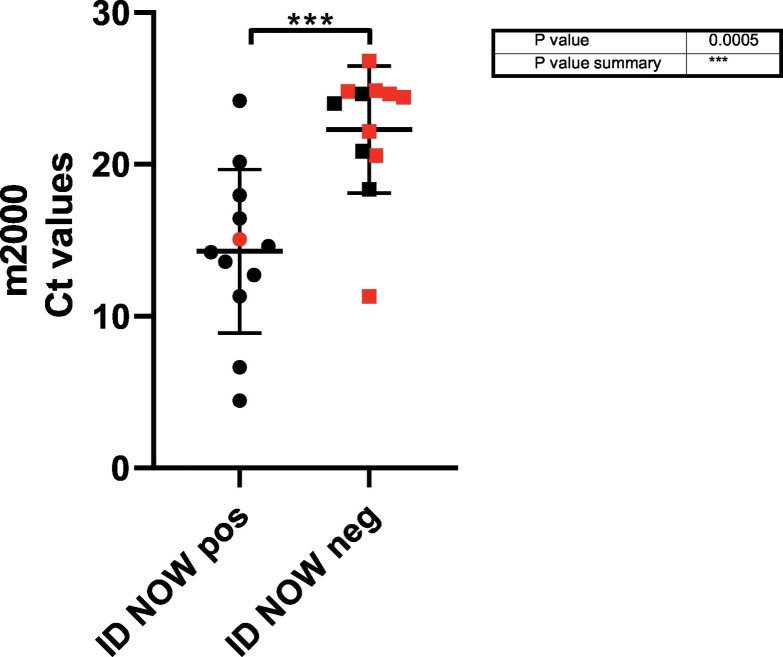
An important variable in our study is that two distinct population groups were analyzed. Thirteen of the 88 patient specimens collected were from an inpatient population of recovering COVID positive patients, with a mean time from initial COVID-19 diagnosis of 25.8 days. A comparison of the m2000 Ct values obtained from m2000 positive samples from inpatients (red) and ED patients (black) and by ID NOW result is shown in Fig. 1 . The mean m2000 Ct value for the ID NOW positive specimens was 14.3 versus a significantly higher mean m2000 Ct value 22.29 (p-value < 0.001) for the ID NOW negative specimens. Inpatients had significantly lower Ct values as measured by Abbott m2000 Ct (n = 9, mean Ct 21.6) than positive specimens collected from patients presenting at the ED (n = 16, mean Ct 16.3, P = 0.04). As the majority of ID NOW negative/m2000 positive specimens (8 of 12) were from this inpatient population of low Ct positives, the overall performance of the ID NOW assay was substantially impacted. To assess test performance in a more typical use case in a POC setting, we reanalyzed percent agreement for all assays using only ED patients. While still notably lower than the other assays, the PPA of ID NOW increased from 48% to 69%, whereas performance of the other assays was nearly identical (Table 1b ).
Fig. 1.
Comparison of cycle threshold values of CRS positive samples, ID NOW result and patient location. Data points depicted in red indicate inpatient specimens and black are ED specimens.
Table 1b.
Agreement of 4 SARS-CoV-2 NAATs relative to the CRS (ED patients only)
| Composite reference standard (CRS) |
Percent agreement with CRS | 95% CI | ||
|---|---|---|---|---|
| Detected | Not detected | |||
| ID NOW | Positive agreement = 69% | 0.44–0.86 | ||
| Detected | 11 | 0 | Negative agreement = 100% | 0.94–1.0 |
| Not detected | 5 | 59 | Overall agreement = 93% | |
| Simplexa | Positive agreement = 88% | 0.64–0.98 | ||
| Detected | 14 | 0 | Negative agreement = 100% | 0.94–1.0 |
| Not detected | 2 | 59 | Overall agreement = 97% | |
| m2000 | Positive agreement = 94% | 0.72–1.0 | ||
| Detected | 15 | 0 | Negative agreement = 100% | 0.94–1.0 |
| Not detected | 1 | 57 | Overall agreement = 99% | |
| 2 invalids | ||||
| Xpert | Positive agreement = 100% | 0.81–1.0 | ||
| Detected | 16 | 2 | Negative agreement = 97% | 0.89–0.99 |
| Not detected | 0 | 58 | Overall agreement = 98% | |
| 1 invalid | ||||
This comparative analysis of SARS-CoV-2 NAATs utilizing the m2000, Simplexa, Xpert and ID NOW assays demonstrated that significant performance deficits were found in the ID NOW assay when tested in a mixed patient population using both NP and nasal specimens. Based on a CRS, use of the m2000, Xpert, and Simplexa assays for NP specimens in VTM are likely to have similar performance in clinical practice and choice of implementation can be made based on considerations of turnaround time, throughput, work flow and cost. In contrast, despite the ID NOW assay claiming and demonstrating comparable (differences <1 log10) analytic LOD findings to the other assays tested, the lower detection rate of the ID NOW from nasal samples must be considered when deciding on a use case. When limiting our data set to an acute ED patient population and comparing results from the same specimen type (NP in VTM), performance of ID NOW was improved but still demonstrated lower performance compared to the other assays tested, possibly due to the ~14-fold dilution step when specimen is transferred from VTM to ID NOW elution buffer chamber prior to testing. However, the relatively small sample size studied of 88 patients, 25 positive for SARS-CoV-2, is a limitation in our study which prevents attributing statistical significance to the analysis of the results. While this significant of a deficiency in result agreement by the ID NOW is not a novel finding in the literature (Harrington et al., 2020; Mitchell and George, 2020; Moore et al., 2020; Smithgall et al., 2020), we believe that our work alludes to the specimen type and patient population as key components contributing to the deficiency in ID NOW performance. We contend that in situations where the 5–15-min turn-around time of the ID NOW assay may provide distinct advantages to care, there is a critical need for analysis of the most appropriate specimen type, appropriate patient population and need for confirmatory NAAT testing prior to clinical use.
Footnotes
Declarations of interest: none
References
- Cradic K., Lockhart M., Ozbolt P., Fatica L., Landon L., Lieber M. Clinical evaluation and utilization of multiple molecular in vitro diagnostic assays for the detection of SARS-CoV-2. Am J Clin Pathol. 2020 doi: 10.1093/ajcp/aqaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington A., Cox B., Snowdon J., Bakst J., Ley E., Grajales P. Comparison of Abbott ID Now and Abbott m2000 methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from symptomatic patients. J Clin Microbiol. 2020 doi: 10.1128/jcm.00798-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S.L., George K.S. Evaluation of the COVID19 ID NOW EUA assay. J Clin Virol. 2020;128:104429. doi: 10.1016/j.jcv.2020.104429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore N.M., Li H., Schejbal D., Lindsley J., Hayden M.K. Comparison of two commercial molecular tests and a laboratory-developed modification of the CDC 2019-nCoV RT-PCR assay for the detection of SARS-CoV-2. J Clin Microbiol. 2020 doi: 10.1128/jcm.00938-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D.D., Cherian S.S., Roman K., Stempak L.M., Schmotzer C.L., Sadri N. Comparison of Abbott ID Now, Diasorin Simplexa, and CDC FDA EUA methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from individuals diagnosed with COVID-19. J Clin Microbiol. 2020 doi: 10.1128/jcm.00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S., Tritella S., Cozzi A., Carriero S., Blandi L., Ferraris L. Diagnostic performance of chest x-ray for COVID-19 pneumonia during the SARS-CoV-2 pandemic in Lombardy Italy. J Thorac Imaging. 2020 doi: 10.1097/rti.0000000000000533. [DOI] [PubMed] [Google Scholar]
- Smithgall M.C., Scherberkova I., Whittier S., Green D.A. Comparison of Cepheid Xpert Xpress and Abbott ID Now to Roche Cobas for the rapid detection of SARS-CoV-2. J Clin Virol. 2020;128:104428. doi: 10.1016/j.jcv.2020.104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visseaux B., Le Hingrat Q., Collin G., Bouzid D., Lebourgeois S., Le Pluart D. Evaluation of the QIAstat-Dx Respiratory SARS-CoV-2 Panel, the first rapid multiplex PCR commercial assay for SARS-CoV-2 detection. J Clin Microbiol. 2020 doi: 10.1128/jcm.00630-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters F., van de Bovenkamp J., van den Bosch B., van den Brink S., Broeders M., Chung N.H. Multi-center evaluation of Cepheid Xpert® Xpress SARS-CoV-2 point-of-care test during the SARS-CoV-2 pandemic. J Clin Virol. 2020;128:104426. doi: 10.1016/j.jcv.2020.104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen W., Smith E., Manji R., Schron D., Berry G.J. Clinical evaluation of three sample-to-answer platforms for the detection of SARS-CoV-2. J Clin Microbiol. 2020 doi: 10.1128/jcm.00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]