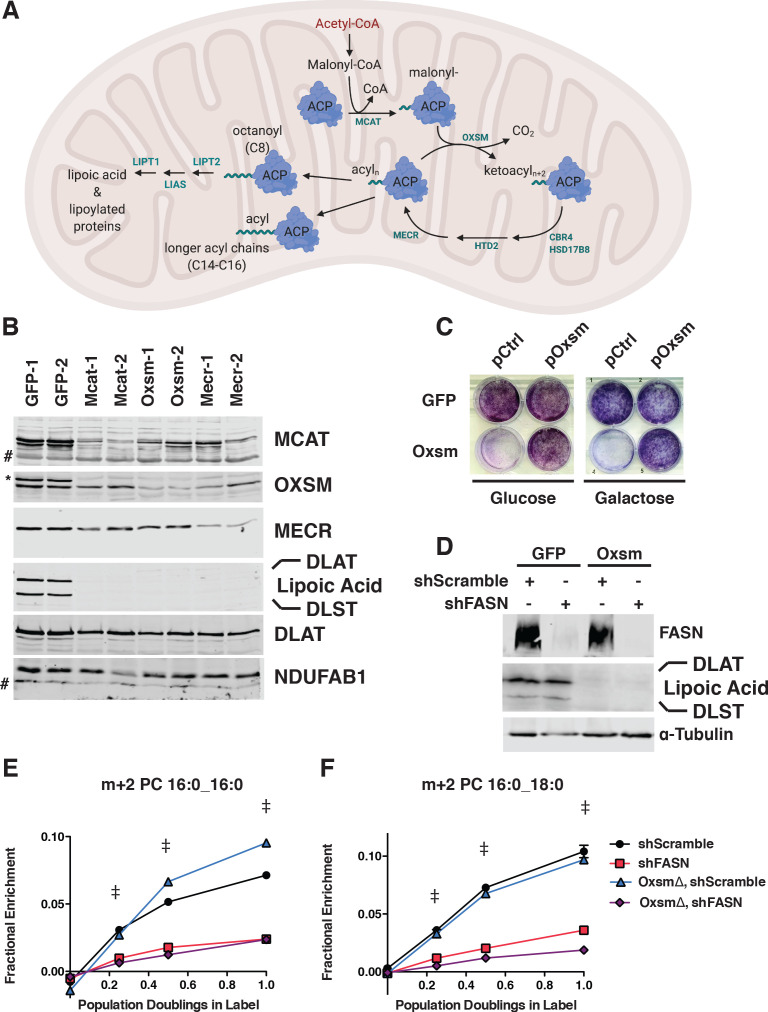
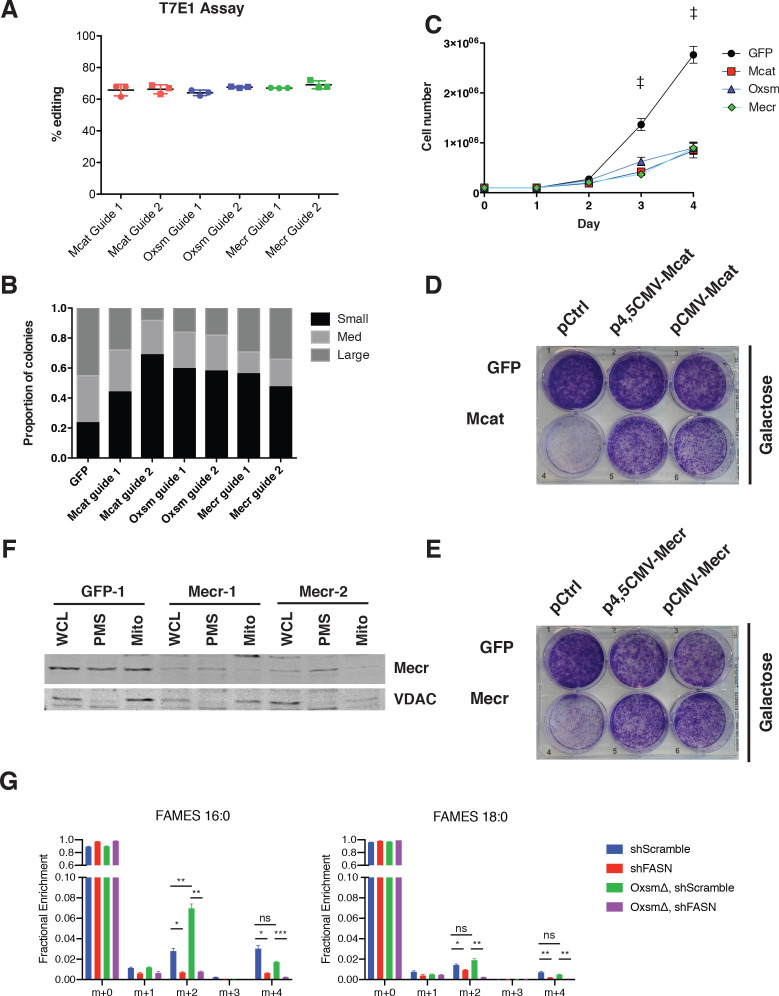
Figure 1. MtFAS is an essential pathway in mammalian skeletal myoblasts but does not contribute to synthesis of major cellular lipids.
(A) Schematic of the mitochondrial fatty acid synthesis pathway and downstream lipoic acid synthesis. (B) Crude isolated mitochondrial fractions from duplicate single cell clones of Mcat, Oxsm, and Mecr mutants, compared with GFP control clonal cell lines, were separated via SDS-PAGE and immunoblotted for the indicated targets. *=Lipoic acid band (reprobe of earlier blot). #=non specific bands C. GFP control and Oxsm mutant cells were infected with retroviral control plasmid (pCtrl) or a plasmid expressing Oxsm off the CMV promoter (pOxsm), plated at equal densities in normal growth medium with either 4.5 g/L glucose or 10 mM galactose and grown for 3 or 4 days, respectively, then stained with crystal violet. (D) Whole cell lysates from stable cell lines generated by infecting Oxsm mutant cells (OxsmΔ) or GFP controls with shRNA constructs targeting FASN (shFASN) or scramble control (shScramble) were separated by SDS-PAGE and immunoblotted for FASN, lipoylated proteins, or tubulin. (E-F) Stable cell lines created in (D) were incubated with U13C-glucose for the indicated number of doublings, harvested, lipids extracted, and analyzed via LC-MS. Shown are quantitation of m+2 isotopologues for two representative phospholipid species, PC 16:0_16:0 and PC 16:0_18:0. †=p < 0.001, ‡=p < 0.0001, error bars are SEM.