Abstract
Magang geese exhibit a unique characteristic of follicular development, with eight largest orderly arranged pre-ovulatory follicles in the abdominal cavity. However, little is known about the mechanisms underlying this follicular development. This study aimed to compare gene expression profiles of granulosa cells (GCs) at different stages of follicular development and provide comprehensive insights into follicle selection and the mechanisms underlying the well-defined follicle hierarchy in Magang geese. GCs of large white follicles (LWFs), small yellow follicles (SYFs), F8, F4, and F1 were used for RNA-seq analysis; 374, 1117, 791, and 593 genes were differentially expressed in stages LWFs to SYFs, SYFs to F8, F8 to F4, and F4 to F1, respectively, suggesting that these genes contribute to follicle selection and development. Reliability of sequencing data was verified through qPCR analysis of 24 genes. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathways revealed a complex mechanism that remodels the extracellular matrix and turnover of extracellular matrix components in follicular development and ovulation and involves multiple pathway, such as focal adhesion, adherens junction, and extracellular matrix–receptor interaction. Some unique characteristics were observed during the different follicular development stages. For instance, some differentially expressed genes were enriched in progesterone-mediated oocyte maturation and steroid biosynthesis from stage SYFs to F8, whereas others were enriched in actin cytoskeleton regulation and vascular smooth muscle contraction from stage F4 to F1. These findings enhance our current understanding of GC function and ovarian follicles during the key stages of follicular development.
Keywords: Follicular development, Follicle selection, Granulosa cells, Magang goose, Transcriptome sequencing
Magang geese are indigenous to Guangdong Province in southern China and exhibit distinct ovarian follicular development characteristics. These geese reportedly exhibit interrupted egg-laying cycles owing to the development of strong broodiness (four to five clutches) during the annual breeding season [1]. Within each egg-laying and incubation cycle, large white follicles (LWFs) develop into approximately 10 small yellow follicles (SYFs) within 20 days before the start of laying. Thereafter, eight SYFs continue to develop into the eight largest orderly arranged pre-ovulatory follicles (F8, F7, F6, F5, F4, F3, F2, F1) and proceed to ovulation, with two or three slowest developing SYFs subsequently being eliminated via atresia [2]. The growth of LYFs is characterized by the accumulation of yolk, which amounts to approximately 60 g in F1 follicles as compared with 2 g in F8 follicles [3]. The selection and development of ovarian follicles are regulated by hormones secreted by the pituitary gland and ovarian follicles [4,5,6] as well as other factors such as tight junctions [7, 8].
Follicle-stimulating hormone (FSH), an important sex hormone, regulates the development of ovarian follicles, particularly of prehierarchical follicles into hierarchical follicles. Anti-Müllerian hormone (AMH) is reportedly an important gene in the selection of prehierarchical follicles owing to its role in regulating FSH sensitivity [9]. SYFs with the highest FSH receptor (FSHR) expression enter the next round of development [10, 11]. Activin and inhibin are important factors regulating follicle selection. Studies have shown that SYFs destined to develop into LYFs exhibit higher mRNA expression levels of activin receptors [12]. Inhibin A (INHA) secreted by large follicles can inhibit the development of prehierarchical follicles, whereas activin and FSH can promote follicular development [13, 14]. Luteinizing hormone receptor (LHR) expression levels increase concomitantly with follicular development and peak during the pre-ovulation period [10, 11]. The surge in LH levels initiates or enhances some critical processes within the steroidogenic pathway via protein kinase A/cAMP signaling, and the mRNA expression levels of steroidogenic acute regulatory protein (StAR), cholesterol side-chain cleavage enzyme (CYP11A1), and 3β-hydroxysteroid (HSD3Β) are sharply increased in granulosa cells (GCs) of pre-ovulatory follicles [15, 16]. Simultaneously, these aromatases regulate the selection and development of follicles.
In recent years, high-throughput RNA sequencing has emerged as a powerful tool for studying the mechanisms underlying the development and selection of follicles in geese. The transcriptome profile of ovarian tissues from laying/brooding geese has revealed that steroid hormone biosynthesis- and dopamine beta-hydroxylase-related genes play important roles in follicular development and the productivity of Zhedong White geese [17]. In Tianfu meat geese, a total of 160 differentially expressed genes (DEGs) contributing to the regulation of nonhierarchical follicular development have been identified using sequencing [18]. Although these studies provide useful information on the development and selection of follicles in geese, the process of follicle selection from a cohort of follicles in Magang geese remains unclear. In this study, we further investigated the transcriptome profile of the granulosa layer of goose ovarian follicles at different stages of development using Illumina sequencing. Furthermore, we compared the gene expression profiles of GCs from different-sized ovarian follicles at different stages of development to obtain comprehensive insights into follicle selection and the mechanisms determining the well-defined follicle hierarchy in Magang geese.
Materials and Methods
Obligatory ethical approval
All experimental procedures were approved by the Research Committee of Jiangsu Academy of Agricultural Sciences and conducted as per the Regulations for the Administration of Affairs Concerning Experimental Animals (Decree No. 63 of the Jiangsu Academy of Agricultural Science on July 8, 2014).
Animals and sample preparation
Magang geese were obtained from Qingxin Jingyu Goose Company Ltd., Guangdong, China. During the experiment, the geese were allowed ad libitum with mixed feed of 12.5% crude protein, supplemented with green grass whenever possible. To obtain as many as 8 follicles, the Magang geese were slaughtered 45 min after they laid their first egg. LYFs of diameter > 9 mm, SYFs of diameter 6–8 mm, and LWFs of diameters < 6 mm were collected [3] (Fig. 1). The granulosa layers of the largest LYFs were separated according to the method of Gilbert et al. [19]. Simply, each follicle was slit with a scalpel blade and inverted with fine forceps over a suitable dish, allowing the follicular content to drop into physiological saline. After removal of the entire theca, the granulosa layer was gently pulled away from the yolk with the forceps. The granulosa layers of the SYFs and LWFs were separated as per the method of Tilly et al. [20]. Briefly, each follicle was slit with a scalpel blade and carefully inverted with fine forceps. The follicular tissue was held in the forceps and was slowly swirled in physiological saline to remove the residual yolk, and the granulosa layer was then gently torn away from the thecal tissue with fine forceps. Once separated, the granulosa layers were snap-frozen in liquid nitrogen and stored at –80°C until RNA extraction.
Fig. 1.
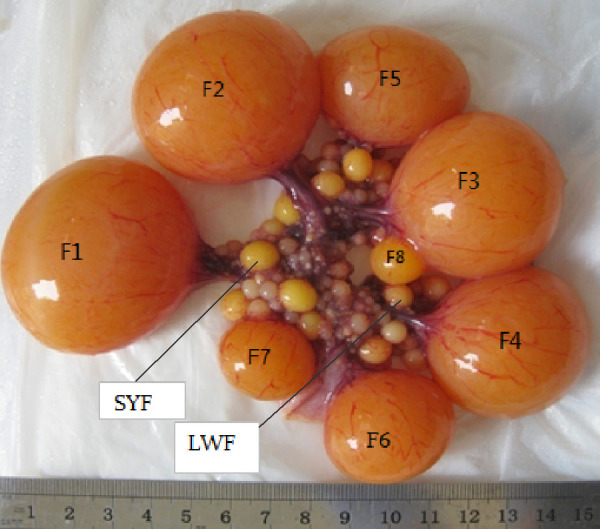
Descriptive summary of avian follicles. F1, F2, F3, F4, F5, F6, F7, and F8 represent the first, second, third, fourth, fifth, sixth, seventh, and eighth large yellow follicles, respectively. Small yellow follicle (SYF) and large white follicle (LWF) represent small yellow follicles with diameter 6–8 mm and large white follicles with diameter < 6 mm, respectively.
Eight large yellow follicles were obtained, and the granulosa layers of LWFs, SYFs, F8, F4, and F1 were separated. Then, the granulosa layers of follicles at the same stage of development from three geese were mixed. Five mixtures of LWFs, SYFs, F8, F4, and F1 were used for transcriptome sequencing.
cDNA library construction and de novo sequencing
Total RNA was extracted using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA), and the concentration and purity of RNA were determined at 260 and 280 nm (A260: 280 ≥ 1.8 and ≤ 2.0) using a NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). RNA integrity (RIN; suitable value: ≥ 7) was assessed using electrophoresis (on a 1% agarose gel) and the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). RNA with good integrity was used to construct a cDNA library, and five cDNA libraries were prepared and sequenced as per the manufacturer’s instructions (HiSeq2500; Illumina, San Diego, CA, USA). Raw data were converted to FASTQ files using bcl2fastq. Clean data were obtained by filtering and removing low-quality sequences and mapped to the reference chicken genome sequences and genes (Gallus_gallus, Galgal4; available at https://www.ncbi. nlm.nih.gov/assembly/GCF_000002315.3) using TopHat 1.3.2. Gene expression levels were assessed using the Reads Per Kb Per Million mapped reads (RPKM) method described by Mortazavi et al. [21].
Differential gene expression profiling and real-time polymerase chain reaction (qPCR) validation
DEGs were detected with the R package DEG seq software (Bioconductor, New York, NY, USA). To screen for DEGs, the following criteria were used: fold change ≥ 1.0 or ≤ 0.67 and P < 0.05 [22]. Using the same RNA samples used for gene expression profiling, qPCR was performed to confirm the results of gene expression profiling.
Analysis of Gene Ontology (GO) category and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
Functional analysis of genes was performed with respect to several different aspects based on the sequence similarity search results described above. GO categorization was performed by classifying all expressed genes according to their classification in the GO system [23]. KEGG pathway annotation was performed online (http://www.genome.jp/tools/kaas/) using unigene sequences [24]. GO and KEGG cluster analyses were performed using R package Heatmap, and the expression level was determined using RPKM or Fragments Per Kilobase Million. GO and KEGG enrichment were performed using the online tool DAVID (http://david.abcc.ncifcrf.gov/); the P value was set as 0.05. A corrected P value of 0.05 was set as the threshold to identify significantly different pathways [22].
Statistical analysis
Differences in gene expression levels in GCs were analyzed using Analysis of Variance. The means were compared using the least significant difference method. All values are expressed as the mean ± SEM. All statistical analyses were performed using SAS software version 8.01 (SAS Institute, Cary, NC, USA).
Results
Transcriptome sequencing and de novo assembly
To obtain a transcriptome profile of GCs at different stages of follicular development, five cDNA libraries representing the transcriptome were constructed based on poly (A)-enriched mRNA. Clean data mapping and annotation indicated the high quality of all five libraries. Reads of each sample were separately mapped to the reference genome using the Burrows-wheeler alignment method (BWA). The mapping ratio was found to be similar for the different samples, with an average mapping ratio of 19.8% and a unique mapping ratio of 18.1%. Approximately 8.3% of the mapped reads (approximately 1.7% of the total reads) were observed to align with multiple genomic regions (Table 1). Raw transcript reads containing adaptor sequences, low-quality reads, and duplicated reads were removed, and the remaining 190,396,461 high-stringency reads (78% of total reads) were collected to generate the transcriptome. This resulted in a total of 175,842 contigs containing 94,707 components and 123,582 sub-components. A summary of the transcriptome sequencing is presented in Table 2. The length of contigs ranged from 201 to 17665 bp, with an N50 length of 3075 bp and an average contig length of 1261 bp. Frequency distribution of contig lengths showed that a large number of contigs were distributed in the < 500-bp range; however, contigs with lengths between 1 and 5 kbp contributed to the highest percentage of the total length.
Table 1. Summary of sequencing and mapping information.
| F1g | F4g | F8g | SYFg | LWFg | |
|---|---|---|---|---|---|
| Total reads | 23965070 | 26866716 | 27387506 | 25293626 | 30089652 |
| Mapped reads | 4923192 | 5851526 | 5602924 | 4609840 | 5427645 |
| Unique mapping reads | 4469449 | 5398829 | 5070629 | 4212871 | 4965282 |
| Multiple reads | 453743 | 452697 | 532295 | 396969 | 462363 |
| Total mapping rate | 20.54 | 21.77 | 20.45 | 18.22 | 18.04 |
| Unique mapping rate | 18.64 | 20.09 | 18.51 | 16.65 | 16.50 |
| Multiple mapping rate | 1.89 | 1.68 | 1.94 | 1.56 | 1.54 |
F1g, the granulosa layers of the first largest follicle; F4g, the granulosa layers of the fourth largest follicle; F8g, the granulosa layers of the the eighth largest follicle; SYFg, the granulosa layers of the small yellow follicle; LWFg, the granulosa layers of the large white follicle.
Table 2. Summary of the assembly generated by the program.
| Total reads | 242766608 |
|---|---|
| High-quality reads | 190396461 |
| Contig number | 175842 |
| Length range | 201-17665 bp |
| N50 | 3075 |
| Mean length | 1261 |
Differential gene expression profiles
The DEGs were determined using the R package DEG seq software. In total, 4,427 DEGs were detected at different follicular developmental stages. A significantly larger number of upregulated than downregulated genes was noted at SYFs to F8 stage. Furthermore, 374, 1117, 791, and 593 genes were differentially expressed from LWFs to SYFs, SYFs to F8, F8 to F4, and F4 to F1, respectively (Table 3).
Table 3. Gene number statistics of differentially expressed genes in the different follicular stages.
| Differentially expressed genes | LWF-SYF |
SYF-F8 |
F8-F4 |
F4-F1 |
||||
|---|---|---|---|---|---|---|---|---|
| Up (60) | Down (314) | Up (593) | Down (524) | Up (367) | Down (424) | Up (289) | Down (304) | |
| Known genes | 45 | 266 | 510 | 452 | 332 | 376 | 247 | 275 |
| New genes | 15 | 48 | 83 | 72 | 35 | 48 | 47 | 29 |
| All genes | 374 | 1117 | 791 | 593 | ||||
LWF, large white follicle; SYF, small yellow follicle. F8, the eighth largest follicle; F4, the fourth largest follicle; F1, the first largest follicle.
GO annotation and significant enrichment analysis of DEGs
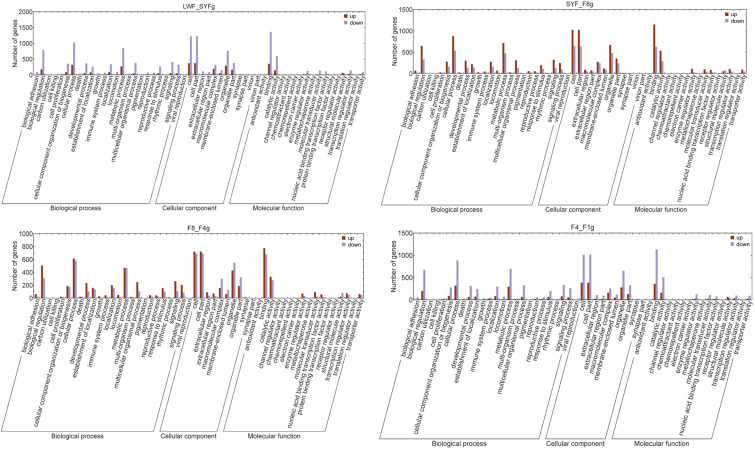
The functional classification GO terms for all DEGs are shown in Fig. 2. In the biological process category, the significantly enriched DEG GO terms were as follows: “biological regulation,” “cellular component organization or biogenesis,” “cellular process,” “developmental process,” “establishment of localization,” “localization,” “metabolic process,” “multicellular organismal process,” “response to stimulus,” “signal,” and “signal process.” In the cellular component category, the enriched GO terms of interest were as follows: “cell,” “cellular part,” “macromolecular complex,” “organelle,” and “organelle part.” In the molecular function category, the significantly enriched GO terms were predominantly associated with “binding” and “catalytic activity”.
Fig. 2.
Functional analysis of the differentially expressed genes in granulosa cells. LWF, large white follicle; SYF, small yellow follicle; F8, the eighth largest follicle; F4, the fourth largest follicle; F1, the first largest follicle.
To gain a better understanding of the biological function of the DEGs identified for each of the different follicular developmental stages, the DEGs were categorized as upregulated and downregulated. The data showed that there was a considerably larger number downregulated than upregulated DEGs in the LWF to SYF stage, whereas there were considerably more upregulated DEGs in the SYF to F8 stage. Moreover, for the F8 to F4 stage, there was little difference in the numbers of up- and downregulated DEGs, whereas for the F4 to F1 stage, a significant decrease in both up- and downregulated DEGs was noted.
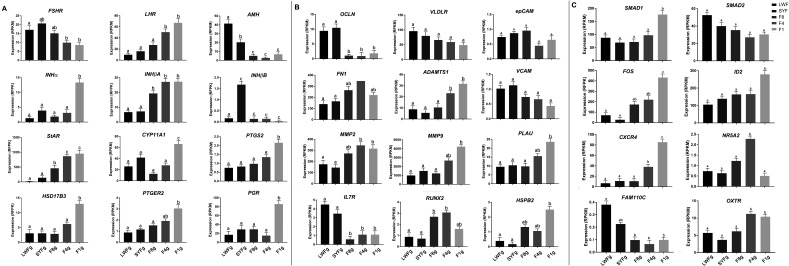
Expression pattern of key genes in follicular development and qPCR validation
To gain further insights into gene expression in follicles, we examined the expression patterns of certain key genes, as summarized in Fig. 3. These genes were divided into three groups, the first of which contained genes associated with sex hormone secretion, namely FSHR, LHR, AMH, INHα, INHβA, INHβB, StAR, CYP11A1, PTGS2, HS17B3, PTGER2, and PGR. The following 12 DEGs were selected for validation by qPCR: FSHR, LHR, AMH, INHα, INHβA, INHβB, StAR, CYP11A1, PTGS2, HSD17B3, PGR, ESRRβ ) (Supplementary Fig. 1: online only). The second group included 12 genes associated with yolk transport, cellular matrix, inflammatory factors, and heat stress (OCLN, VLDLR, epCAM, FN1, ADAMTS1, VCAM, MMP2, MMP9, PLAU, IL7R, RUNX2, and HSPB2). In this group, 6 genes (OCLN, VLDLR, OAZ1, BMPR, BMP1βR, and CCND1) were selected for validation by qPCR (Supplementary Fig. 1). The third group included transcriptionally regulated factors and chemokine factors (SMAD2, SMAD1, FOS, ID2, CXCR4, NR5A2, FAM110C, and OXTR). In this group, 6 genes (SMAD1, SMAD2, SMAD3, ADCY2, ID2, and CXCR4) were selected for validation by qPCR (Supplementary Fig. 1). As determined by qPCR, these 24 genes showed changes in expression patterns similar to those shown by genes in the RNA-seq analysis. The mRNA expression level of FSHR increased from the LWF to SYF stage and then decreased from the SYF to F1 stage and was the highest in SYFs. The expression level of LHR increased from the LWF to F1 stage. Similarly, the expression of StAR was the lowest in LWFs, which thereafter increased and peaked in F1 follicles. Expression of Progesterone receptor (PGR) remained low from the LWF to F4 stage and thereafter increased markedly in F1 follicles. In contrast, occludin (OCLN) expression level was high in LWFs and SYFs and then decreased rapidly at the F8 stage. VLDLR was most highly expressed in LWFs and subsequently showed a gradual decrease. The expression levels of genes related to cellular matrix (FN1, MMP2, MMP9, ADAMTS1, and PLAU) were observed to increase from the LWF to F1 stage; in contrast, there was a decrease in the expression level of cell adhesion-related genes (VCAM and epCAM) from the LWF to F1 stage.
Fig. 3.
Expression pattern of key genes in follicular development. LWF, large white follicle; SYF, small yellow follicle; F8, the eighth largest follicle; F4, the fourth largest follicle; F1, the largest follicle. A represents the genes associated with sex hormone secretion; B represents the genes associated with yolk transport, cellular matrix, inflammatory factors, and heat stress; C includes transcriptionally regulated factors and chemokine factors.
KEGG pathway enrichment analysis
All identified genes were assigned to one or more KEGG annotations and mapped to significantly enriched KEGG pathways. Accordingly, 22 significantly enriched KEGG pathways were identified in the LWF to SYF stage, 19 in the SYF to F8 stage, 22 in the F8 to F4 stage, and 24 in the F4 to F1 stage. Among the top 10 KEGG pathways, ribosome, focal adhesion, and extracellular matrix-receptor interaction were associated with all four assessed stages (Table 4). Besides the above pathways, there were some unique KEGG pathways in the different follicular development stages. For the LWF to SYF stage, DEGs were enriched in oxidative phosphorylation, adherens junctions, Notch signaling pathway, Wnt signaling pathway, melanogenesis, and MAPK signaling pathway. For the SYF to F8 stage, DEGs were enriched in the regulation of actin cytoskeleton, ErbB signaling pathway, endocytosis, progesterone-mediated oocyte maturation, and insulin signaling. For the F8 to F4 stage, DEGs were enriched in focal adhesion, regulation of the actin cytoskeleton, cell cycle, and DNA replication. For the F4 to F1 stage, DEGs were enriched in the regulation of actin cytoskeleton, MAPK signaling pathway, Wnt signaling pathway, and vascular smooth muscle contraction.
Table 4. Top ten Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with differentially expressed genes.
| LWF-SYFg | SYF-F8g |
|---|---|
| Ribosome | Ribosome |
| Focal adhesion | Focal adhesion |
| Proteasome | Spliceosome |
| ECM-receptor interaction | Regulation of actin cytoskeleton |
| Oxidative phosphorylation | ECM-receptor interaction |
| Adherens junction | ErbB signaling pathway |
| Notch signaling pathway | Endocytosis |
| Wnt signaling pathway | Progesterone-mediated oocyte maturation |
| MAPK signaling pathway | Insulin signaling pathway |
| Melanogenesis | Lysine degradation |
| F8-F4g | F4-F1g |
| Ribosome | Ribosome |
| Spliceosome | Focal adhesion |
| Proteasome | Regulation of actin cytoskeleton |
| Cell cycle | MAPK signaling pathway |
| Focal adhesion | ECM-receptor interaction |
| Regulation of actin cytoskeleton | Wnt signaling pathway |
| DNA replication | Adherens junction |
| ECM-receptor interaction | Vascular smooth muscle contraction |
| Oocyte meiosis | Spliceosome |
| Oxidative phosphorylation | Endocytosis |
LWF, large white follicle; SYF, small yellow follicle.
Discussion
The profiles and dynamic variation of gene expression were determined in GCs at different stages of follicular development using RNA-seq transcriptome analysis. Accordingly, 4,427 DEGs and some DEGs identified by DGE analysis were further validated by qPCR. Most of the DEGs were mapped to KEGG pathways, including those associated with focal adhesion, extracellular matrix–receptor interaction, GnRH signaling, mitogen activated protein kinase (MAPK) signaling, and TGF-beta signaling. A higher number of genes were differentially expressed in the LWF to SYF stage than in the other stages, and this could be attributed to the fact that the LWF to SYFs stage is particularly important in follicular development and requires the expression of a larger number of genes to facilitate the selection and development of follicles.
KEGG analysis showed that the DEGs were significantly enriched in the extracellular matrix, including focal adhesion and extracellular matrix-receptor interaction at the four assessed developmental stages. The gene expression data highlighted several genes encoding ECM proteins such as collagen (COL1A2, COL11A1, and COL6A1), FN1, and integrin receptors (ITGA8 and ITGAV) [25]. By binding transmembrane receptors such as integrins, matrix molecules such as FN1 influence cell signal transduction, ultimately regulating cellular signaling and remodeling extracellular matrix components of ovarian follicles [26]. In the present study, we detected high levels of the matrix metalloproteases (MMP-2 and MMP-9) and ADAMT-1 mRNAs in the GCs of F1 follicles. Previous studies have revealed significant differences in the mRNA levels of MMP-2 and -9 expressed in chicken post-ovulatory follicles, whereas analysis of ovarian function has revealed that ADAMTS-1 levels increased prior to follicular rupture [27,28,29,30]. The plasminogen activator system (PLAU) level is reportedly the highest during pre-ovulation and the upregulation of plasminogen [31], and this is consistent with our findings that the levels of PLAU increase concomitantly with follicular development and then peak in F1 follicles. Collectively, these observations indicate that elevated expression levels of MMP-2 and -9, PLAU, and ADAMTS-1 promotes the remodeling of the extracellular matrix during follicular development and ovulation. Therefore, the remodeling of the extracellular matrix and turnover of extracellular matrix components were important in follicular development and ovulation.
KEGG analysis also showed that there were some unique characteristics in the different follicular development stages. From the LWF to SYF stage, DEGs were enriched in oxidative phosphorylation, Notch signaling pathway, Wnt signaling pathway, melanogenesis, and MAPK signaling pathway. These results showed that energy is required to supply ATP for DNA and protein synthesis in the LWF to SYF stage, consistent with the findings of a previous report that lipid metabolism is involved in nonhierarchical follicular development in Tianfu meat geese [18]. DEGs were enriched in progesterone-mediated oocyte maturation and steroid biosynthesis in the SYF to F8 stage. The findings of the present study are consistent with those reported by Du et al. who stated that hormone biosynthesis, positive regulation of hormone secretion, and steroid biosynthesis are involved in FSH-mediated proliferation of goose GCs of SYFs [32]. These results further highlight the important roles played by sex and ovarian hormones in promoting the successful selection of SYFs for subsequent development to F8. Similarly, energy is required to supply ATP for cell cycle and DNA replication due to the rapid growth of the follicles from the F8 to F4 stage. These results are consistent with those reported by Seol [33]. From the F4 to F1 stage, DEGs were enriched in the regulation of actin cytoskeleton, MAPK signaling pathway, Wnt signaling pathway, and vascular smooth muscle contraction. The blood vessels on the follicles increased significantly to provide nutrients for the rapid development of follicles at this stage, and the accumulation of yolk was finally completed.
The accumulation of yolk requires the participation of multiple genes and proteins. As a major component of tight junctions, OCLN mediates the transfer of yolk material to the follicle surface by sealing intercellular spaces [7, 8]. In avian species, activin A and FSH have been shown to control tight junction activity by regulating OCLN expression via the MAP kinase (MEK)/ERK signaling pathway [7]. VLDLR, an endocytotic membrane receptor belonging to the LDLR superfamily, mediates the endocytosis of yolk protein into growing follicles and is thereby involved in the regulation of oocyte growth [34]. In the present study, we found that the mRNA expression of OCLN, VLDLR, and vascular cell adhesion protein 1 (VCAM1) was higher in LWFs and SYFs and subsequently decreased concomitant with follicular growth. Tight junctions have been demonstrated to prevent the access of vitellogenins to VLDLR on the surface of oocytes in small follicles [35] via the downregulation of OCLN expression. The decreased expression of VCAM1 suggests that the adhesion between cells decreases with follicular development, thereby facilitating the access of yolk precursors to oocyte VLDLR and in turn promoting rapid yolk incorporation and follicle growth [36].
In conclusion, 4,427 DEGs were identified in the present study, and these genes may contribute to the selection and development of follicles in Magang geese. The reliability of the sequencing data was verified through qPCR analysis of the 24 genes. These data depict a complex mechanism that remodels the extracellular matrix and turnover of extracellular matrix components in follicular development and ovulation and involves multiple processes, such as focal adhesion, adherens junction, and extracellular matrix-receptor interaction. Importantly, progesterone-mediated oocyte maturation and steroid biosynthesis were involved in follicular development from the SYF to F8 stage, and the regulation of actin cytoskeleton and vascular smooth muscle contraction has been associated with follicular development from the F4 to F1 stage. Thus, these results provide useful information for future in-depth studies concerning nonhierarchical follicular development in goose reproduction. Nevertheless, despite these valuable insights, as a complex process, follicular development remains inadequately characterized and thus warrants further studies.
Conflict of Interests
The authors declare that they have no competing interests.
Supplementary
Acknowledgments
This study was supported by National Science Foundation of China (Grant no. 31972551 and 31501946), China Agriculture Research System (grant number CARS-42-20), Natural Science Foundation of Jiangsu Province (BK20160592), and Henan Science and Technology Projects (182102410032).
References
- 1.Shi ZD, Huang YM, Wu W. Review on the egg laying cycle and its physiological regulation mechanism. China Poultry 2008; 30: 1–5 (In Chinese). [Google Scholar]
- 2.Huang YM, Shi ZD, Liu Z, Liu Y, Li XW. Endocrine regulations of reproductive seasonality, follicular development and incubation in Magang geese. Anim Reprod Sci 2008; 104: 344–358. [DOI] [PubMed] [Google Scholar]
- 3.Qin Q, Sun A, Guo R, Lei M, Ying S, Shi Z. The characteristics of oviposition and hormonal and gene regulation of ovarian follicle development in Magang geese. Reprod Biol Endocrinol 2013; 11: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lovell TM, Gladwell RT, Groome NP, Knight PG. Ovarian follicle development in the laying hen is accompanied by divergent changes in inhibin A, inhibin B, activin A and follistatin production in granulosa and theca layers. J Endocrinol 2003; 177: 45–55. [DOI] [PubMed] [Google Scholar]
- 5.Johnson AL, Woods DC. Dynamics of avian ovarian follicle development: cellular mechanisms of granulosa cell differentiation. Gen Comp Endocrinol 2009; 163: 12–17. [DOI] [PubMed] [Google Scholar]
- 6.Onagbesan O, Bruggeman V, Decuypere E. Intra-ovarian growth factors regulating ovarian function in avian species: a review. Anim Reprod Sci 2009; 111: 121–140. [DOI] [PubMed] [Google Scholar]
- 7.Schuster MK, Schmierer B, Shkumatava A, Kuchler K. Activin A and follicle-stimulating hormone control tight junctions in avian granulosa cells by regulating occludin expression. Biol Reprod 2004; 70: 1493–1499. [DOI] [PubMed] [Google Scholar]
- 8.Stephens CS, Johnson PA. Occludin expression and regulation in small follicles of the layer and broiler breeder hen. Gen Comp Endocrinol 2017; 248: 106–113. [DOI] [PubMed] [Google Scholar]
- 9.Durlinger AL, Gruijters MJ, Kramer P, Karels B, Ingraham HA, Nachtigal MW, Uilenbroek JT, Grootegoed JA, Themmen AP. Anti-Müllerian hormone inhibits initiation of primordial follicle growth in the mouse ovary. Endocrinology 2002; 143: 1076–1084. [DOI] [PubMed] [Google Scholar]
- 10.Woods DC, Johnson AL. Regulation of follicle-stimulating hormone-receptor messenger RNA in hen granulosa cells relative to follicle selection. Biol Reprod 2005; 72: 643–650. [DOI] [PubMed] [Google Scholar]
- 11.Dong X, Liu HH, Wang JW, Xiao QH, Yuan X, Li L, Xia L, Li L. Histological and developmental study of prehierarchical follicles in geese. Folia Biol (Krakow) 2014; 62: 171–177. [DOI] [PubMed] [Google Scholar]
- 12.Lovell TM, Knight PG, Gladwell RT. Differential expression of mRNAs encoding the putative inhibin co-receptor (betaglycan) and activin type-I and type-II receptors in preovulatory and prehierarchical follicles of the laying hen ovary. J Endocrinol 2006; 188: 241–249. [DOI] [PubMed] [Google Scholar]
- 13.Johnson AL, Haugen MJ, Woods DC. Role for inhibitor of differentiation/deoxyribonucleic acid-binding (Id) proteins in granulosa cell differentiation. Endocrinology 2008; 149: 3187–3195. [DOI] [PubMed] [Google Scholar]
- 14.Lovell TM, Al-Musawi SL, Gladwell RT, Knight PG. Gonadotrophins modulate hormone secretion and steady-state mRNA levels for activin receptors (type I, IIA, IIB) and inhibin co-receptor (betaglycan) in granulosa and theca cells from chicken prehierarchical and preovulatory follicles. Reproduction 2007; 133: 1159–1168. [DOI] [PubMed] [Google Scholar]
- 15.Johnson AL, Bridgham JT. Regulation of steroidogenic acute regulatory protein and luteinizing hormone receptor messenger ribonucleic acid in hen granulosa cells. Endocrinology 2001; 142: 3116–3124. [DOI] [PubMed] [Google Scholar]
- 16.King SR, LaVoie HA. Gonadal transactivation of STARD1, CYP11A1 and HSD3B. Front Biosci (Landmark Ed) 2012; 17: 824–846. [DOI] [PubMed] [Google Scholar]
- 17.Xu Q, Zhao W, Chen Y, Tong Y, Rong G, Huang Z, Zhang Y, Chang G, Wu X, Chen G. Transcriptome profiling of the goose (Anser cygnoides) ovaries identify laying and broodiness phenotypes. PLoS One 2013; 8: e55496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Zhang W, Li Q, Liu J, Zhang T, Zhou T, Li L, Wang J, Xu H, He H. The comprehensive mechanisms underlying nonhierarchical follicular development in geese (Anser cygnoides). Anim Reprod Sci 2015; 159: 131–140. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert AB, Evans AJ, Perry MM, Davidson MH. A method for separating the granulosa cells, the basal lamina and the theca of the preovulatory ovarian follicle of the domestic fowl (Gallus domesticus). J Reprod Fertil 1977; 50: 179–181. [DOI] [PubMed] [Google Scholar]
- 20.Tilly JL, Kowalski KI, Johnson AL. Stage of ovarian follicular development associated with the initiation of steroidogenic competence in avian granulosa cells. Biol Reprod 1991; 44: 305–314. [DOI] [PubMed] [Google Scholar]
- 21.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008; 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 22.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res 2012; 22: 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris MA, Parkinson H. Standards and ontologies for functional genomics 2. Comp Funct Genomics 2004; 5: 618–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005; 21: 3787–3793. [DOI] [PubMed] [Google Scholar]
- 25.van der Flier A, Sonnenberg A. Function and interactions of integrins. Cell Tissue Res 2001; 305: 285–298. [DOI] [PubMed] [Google Scholar]
- 26.Patterson ML, Atkinson SJ, Knäuper V, Murphy G. Specific collagenolysis by gelatinase A, MMP-2, is determined by the hemopexin domain and not the fibronectin-like domain. FEBS Lett 2001; 503: 158–162. [DOI] [PubMed] [Google Scholar]
- 27.Hrabia A, Socha JK, Sechman A. Involvement of matrix metalloproteinases (MMP-2, -7, -9) and their tissue inhibitors (TIMP-2, -3) in the regression of chicken postovulatory follicles. Gen Comp Endocrinol 2018; 260: 32–40. [DOI] [PubMed] [Google Scholar]
- 28.Lind T, Birch MA, McKie N. Purification of an insect derived recombinant human ADAMTS-1 reveals novel gelatin (type I collagen) degrading activities. Mol Cell Biochem 2006; 281: 95–102. [DOI] [PubMed] [Google Scholar]
- 29.Peluffo MC, Murphy MJ, Baughman ST, Stouffer RL, Hennebold JD. Systematic analysis of protease gene expression in the rhesus macaque ovulatory follicle: metalloproteinase involvement in follicle rupture. Endocrinology 2011; 152: 3963–3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandy JD, Westling J, Kenagy RD, Iruela-Arispe ML, Verscharen C, Rodriguez-Mazaneque JC, Zimmermann DR, Lemire JM, Fischer JW, Wight TN, Clowes AW. Versican V1 proteolysis in human aorta in vivo occurs at the Glu441-Ala442 bond, a site that is cleaved by recombinant ADAMTS-1 and ADAMTS-4. J Biol Chem 2001; 276: 13372–13378. [DOI] [PubMed] [Google Scholar]
- 31.Leonardsson G, Peng XR, Liu K, Nordström L, Carmeliet P, Mulligan R, Collen D, Ny T. Ovulation efficiency is reduced in mice that lack plasminogen activator gene function: functional redundancy among physiological plasminogen activators. Proc Natl Acad Sci USA 1995; 92: 12446–12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du L, Gu T, Zhang Y, Huang Z, Wu N, Zhao W, Chang G, Xu Q, Chen G. Transcriptome profiling to identify key mediators of granulosa cell proliferation upon FSH stimulation in the goose (Anser cygnoides). Br Poult Sci 2018; 59: 416–421. [DOI] [PubMed] [Google Scholar]
- 33.Seol HS, Sato K, Murakami H, Toyomizu M, Akiba Y. Changes in gene expression involved in energy utilization during chicken follicle development. Anim Reprod Sci 2006; 95: 283–294. [DOI] [PubMed] [Google Scholar]
- 34.Gotthardt M, Trommsdorff M, Nevitt MF, Shelton J, Richardson JA, Stockinger W, Nimpf J, Herz J. Interactions of the low density lipoprotein receptor gene family with cytosolic adaptor and scaffold proteins suggest diverse biological functions in cellular communication and signal transduction. J Biol Chem 2000; 275: 25616–25624. [DOI] [PubMed] [Google Scholar]
- 35.Hu S, Liu H, Pan Z, Xia L, Dong X, Li L, Xu F, He H, Wang J. Molecular cloning, expression profile and transcriptional modulation of two splice variants of very low density lipoprotein receptor during ovarian follicle development in geese (Anser cygnoide). Anim Reprod Sci 2014; 149: 281–296. [DOI] [PubMed] [Google Scholar]
- 36.Solano ME, Sander VA, Ho H, Motta AB, Arck PC. Systemic inflammation, cellular influx and up-regulation of ovarian VCAM-1 expression in a mouse model of polycystic ovary syndrome (PCOS). J Reprod Immunol 2011; 92: 33–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.