Abstract
DNA nanotechnology is a rapidly advancing field, which increasingly attracts interest in many different disciplines, such as medicine, biotechnology, physics and biocomputing. The increasing complexity of novel applications requires significant computational support for the design, modelling and analysis of DNA nanostructures. However, current in silico design tools have not been developed in view of these new applications and their requirements. Here, we present Adenita, a novel software tool for the modelling of DNA nanostructures in a user-friendly environment. A data model supporting different DNA nanostructure concepts (multilayer DNA origami, wireframe DNA origami, DNA tiles etc.) has been developed allowing the creation of new and the import of existing DNA nanostructures. In addition, the nanostructures can be modified and analysed on-the-fly using an intuitive toolset. The possibility to combine and re-use existing nanostructures as building blocks for the creation of new superstructures, the integration of alternative molecules (e.g. proteins, aptamers) during the design process, and the export option for oxDNA simulations are outstanding features of Adenita, which spearheads a new generation of DNA nanostructure modelling software. We showcase Adenita by re-using a large nanorod to create a new nanostructure through user interactions that employ different editors to modify the original nanorod.
INTRODUCTION
DNA origami is currently one of the most popular techniques for the design of DNA nanostructures (1). It employs a long DNA single-strand, or ‘scaffold’, which is folded into a predefined nanostructure with the help of hundreds of shorter single-strands, or ‘staples’, which bind to the scaffold at specific positions. Although DNA origami was created to build solid 2D faces, it was soon extended to 3D and to wireframe nanostructures (2–4). DNA origami has been successfully applied to create measurement devices (5), enzymatic cascades (6), DNA nanopores (7), biosensing devices (8) and drug delivery vessels (9–11) amongst others.
The construction of DNA origami usually involves the routing of a long scaffold (∼8000 nucleotides), the placement of the staples, and the determination of their sequences. This can be a challenging task for large nanostructures. Computational techniques and software have been developed to tackle this problem. Given the development of increasingly intricate and advanced DNA nanostructures, the need for interactive tools rises that enable the user to effectively visualize the shape and to design structures in 3D space.
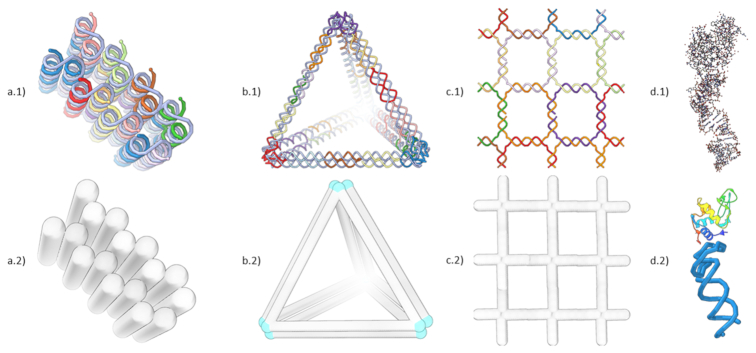
Cadnano is a widely employed software created to assist with the design of lattice-based DNA nanostructures (12). It is highly reliable, as it constrains the cross-section of the design to two lattice types (square and honeycomb) to ensure the proper placement of the crossovers and, therefore, high folding percentages of the DNA nanostructure in vitro. However, it is not possible to create a nanostructure design comprising both types of lattices, it does not provide means to a modular approach, and all DNA double helices must be parallel in a design (Figure 1a). Furthermore, spatial alignment and positioning of several structure in 3D space is not feasible. These limitations reduce significantly the design possibilities. Consequently, alternative DNA nanostructure concepts, such as wireframe DNA origamis, can be hardly realized in Cadnano. In addition, automated design workflows using geometric structures as input and an appropriate visualization of the designs are missing. This represents a significant design challenge with the increasing complexity of nanostructures. In contrast, automated design workflows for wireframed DNA nanostructures are available. To design wireframed DNA origami objects, the target shape is represented as a graph and the problem of tracing the scaffold through is, then, the known NP-complete problem of finding an A-trail along a graph. VHelix is a pipeline of tools that provide a semi-manual interface (3). Its input is a triangular mesh whose edges are partially represented by either one or two double helices (to allow for the routing of the scaffold). It outputs the sequences of the staples for the target wireframe, as well as a model that can be loaded into the commercial 3D-modelling software Maya allowing an inspection after the nanostructure model has been created. Daedalus provides a completely automated tool that can work with nontriangular meshes too (4). Here, the edges are always represented by two double strands (Figure 1b). Nevertheless, it does not provide interactive methods to make a posteriori changes and the final structure can only be inspected using all-atom models with external tools.
Figure 1.
Screenshots of different DNA nanostructure design concepts and their visualization in Adenita. (a) A multi-layer DNA brick designed with Cadnano, (b) a tetrahedron designed using Daedalus and (c) a squared lattice DNA Tile that was manually designed. In Cadnano structures, all double helices must be parallel, while DNA wireframe approaches place double strands at the edges of a mesh. DNA tiles require the repetition of one or several small DNA motifs to create a 2D or 3D shape. In this example, we used a four-arm Holliday junction. (d.1) shows the default all-atom model of the PDB structure 4M4O, formed by a protein and an aptamer, in (d.2) a combination of Adenita's visual model and Samson's secondary structure visualization makes it possible to simplify the representation of such molecules.
Another design approach for DNA nanostructures are DNA tiles. This is a modular strategy that employs small motifs with sticky ends that can be used to create higher order 2D and 3D nanostructures (Figure 1c). DNA tiles have been particularly used for the self-assembly of periodic structures, such as 2D lattices (13), 3D crystals (14), and complex shapes (15). The TIAMAT tool enables the design of DNA-based structures, such as DNA tiles (16). In a recent review, the existing in silico tools for the modelling and visualization of DNA nanostructures are discussed in detail (17).
With the aforementioned design concepts, the field of DNA nanotechnology advances rapidly, and the involved DNA nanostructures are ever increasing in size and complexity (18). With the recent developments in hybrid DNA-protein systems, the need for more sophisticated modelling and visualization tools becomes even more apparent (19,20). Thus, we aimed to facilitate the combination of DNA nanostructures with other molecules, such as aptamers, proteins or nanoparticles, in a feasible manner that does not require a large pipeline of tools or the inspection of nanostructures at the atomic scale. A comprehensive review on the development of the DNA nanotechnology domain and the increasing shape space is provided by Nummelin et al. (21).
In this work, we present Adenita, an interactive 3D tool for the design, visualization, and modification of DNA nanostructures, independently of the chosen design paradigm. We provide a semi-manual and highly modular approach that is well-suited not only for multilayer or wireframe DNA origami approaches but also for the use of DNA tiles. We have developed a hierarchical data model that is able to describe arbitrary DNA nanostructures and a sophisticated multiscale visualization method that depicts the nanostructures on multiple levels of detail allowing the user to operate on the desired level of detail for a specific task. Furthermore, real-time feedback of the structural stability is integrated into the visual model.
Through simple 3D interactions and visibility handling, different components or parametrizable predefined structures can be interactively loaded, created or combined into higher-order structures. A straightforward application of this approach allows the user to import Cadnano designs, make free-form designs of DNA tiles, or create wireframe nanostructures using the Daedalus algorithm. Therefore, different approaches can be easily combined in silico. Furthermore, we have developed Adenita as a plugin for SAMSON Connect, a free software for adaptive 3D modelling and simulation of nanosystems, making it possible to edit and work on customized DNA nanostructures while also visualizing and editing other systems, such as aptamers or proteins (Figure 1d).
Adenita has been developed to overcome the design limitations of the existing DNA origami design tools, with a focus on the modelling of nanostructures in more realistic molecular environments. This will significantly facilitate the prediction of intended and unintended interactions. Our contributions can be summarized as: (i) Integration across folding patterns: a unified DNA nanostructure framework that integrates all major folding strategies and allows their smooth combination. (ii) Integration along scales of conceptual organization: a unified modelling concept that seamlessly integrates a wide spectrum of semantic scales on which one can study and manipulate the nanostructure. (iii) Multi-stage DNA-nanotechnology self-assembly: A convincing use case in which elementary pieces can be created in one stage and can be integrated together to form a more complex design in consecutive stages.
MATERIALS AND METHODS
Dependencies and hardware requirements
We implemented Adenita as a suite of plugins for the computational nanoscience software SAMSON Connect (https://www.samson-connect.net/). Adenita enables the user to create, modify and visualize DNA-based structures. We allow for an optional integration with ntthal from the Primer3 package to compute thermodynamic parameters of the nanostructure (22). Adenita has been developed with the help of Boost (https://www.boost.org/) and Rapidjson (http://rapidjson.org/) libraries. To generate wireframe nanostructures, we employ the Daedalus algorithm (4). A graphics card is highly recommended in order to guarantee interactive framerates and a smooth visualization of the 3D structures.
Experimental methods
The DNA nanostructures were prepared based on protocols already described in the work of Ahmadi et al. (23). These nanorods were initially designed with Cadnano 2.2.0 using the p8064 single stranded scaffold and 236 staple stands (Supplementary Table S1). Subsequently, they were modified with Adenita to form a new nanostructure in a cross shape composed of two nanorods. Each protruding and invading strand necessary to form a cross was assigned to one of the two nanorods composing the design (Supplementary Tables S2 and S3). Individual nanorods were self-assembled separately with a 1:10 scaffold to staple strand ratio in a tris buffer (TB) solution (5 mM Tris, 1 mM EDTA, 5 mM NaCl) containing 18 mM MgCl2. Annealing was performed by exposing the reaction mixture to 65°C for 15 min and then cooling it down from 65 to 25°C by 1°C every 40 min in a one-day thermal ramp. The nanorods were purified using the PEG precipitation method based on a protocol described by Evi Stahl et al. (24). In brief, 100 μl of the nanostructure sample (in TB including 18 mM MgCl2) was mixed with an equivalent volume of 22 mM MgCl2 supplemented TB (100 μl), followed by the addition of 200 μl of purification buffer (15% (w/v) PEG 8000, 5 mM Tris, 1 mM EDTA and 505 mM NaCl). The solution was then mixed gently by tube inversion and centrifuged at 16 000g at r.t. for 25 min. The supernatant was then carefully discarded, and the pellet was dissolved in the TB buffer supplemented with 16 mM MgCl2, followed by incubation for one day at r.t. at 650 rpm. For the super-assembly of the crosses, stoichiometric amounts of purified nanorods were mixed, followed by incubation overnight on a shaker at r.t. and 700 rpm.
TEM images of the crosses and individual rods were obtained by diluting the samples with the folding buffer 1:10 and vortexing them for 10 s. Diluted samples were negatively stained using uranyl acetate on 300-mesh carbon coated grids that had been glow discharged for 40 s and imaged on an FEI Tecnai T12 Spirit electron microscope. Images were collected at a nominal magnification of 1650× using a defocus of 25–40 μm. Fiji was used to analyse the TEM images (25,26).
Software availability
Adenita is open-source and publicly available. It can be downloaded through SAMSON’s Elements store for free (https://www.samson-connect.net/elements.html). Several video tutorials explain the basic features of the tool (Supplementary Table S4). The source code of our design tool can be found at:
RESULTS
Description of the software
Adenita describes arbitrary DNA nanostructures using a data model that comprises two related parent–child hierarchies.
The first hierarchy describes single-stranded DNA. The top element is the single-strand, whose children are the nucleotides, ordered from the 5′ to the 3′ end. Nucleotides are formed by a backbone and a sidechain, which in turn group the atoms.
The second hierarchy describes the geometry of the DNA nanostructure. It is based on a graph model where the double strands are the edges that compose the target geometry. This model is straightforward in the case of wireframe nanostructures, but it can also be applied to any rasterized target shape. The edges or double strands can be considered as the top element whose children are the base pairs that form them. The base pairs can be generalized to also include unpaired regions and motifs, such as the poly-T regions of Daedalus designs illustrated at the vertices in blue in Figure 1b.
The relationship between both hierarchies is established through the nucleotides composing each base pair (Figure 2f). It is determined by the routing of the scaffold and the placement of the staples, which can be done manually by the user or with the help of algorithms, such as Daedalus.
Figure 2.
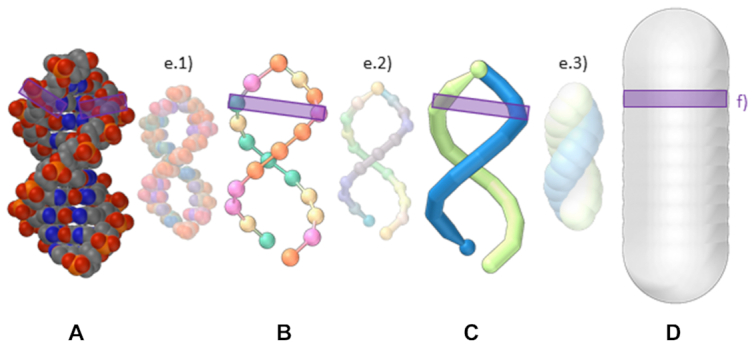
The data model describes every nucleotide using its backbone and side-chain positions fetched from its all-atom representation (A). A single strand is represented as a chain of nucleotides with directionality (B). Double strands can be represented as paired regions of single strands (C) or as the segments that trace the target shape (D). The visual model represents graphically all scales of the data model and allows for a seamlessly transition between them (E). The bottom-up scales (a and b) are related to the top scale (d) through the positioning of the base pairs (F).
Adenita estimates the position of nucleotides using a top-down approach. Once the geometry of the target shape has been specified, the positioning of base pairs and therefore nucleotides is inferred using a model based on B-DNA and idealized base pairs (27). Our model is compatible with other structural data (e.g. the Protein Data Bank) and allows the import of protein and aptamer structures. Thus, we can apply our visualization algorithms also to aptamers and include them during the design process (Figure 1D).
Our visualization concept, depicts the DNA nanostructure in various forms of details, which we call scales (28). Our multiscale approach allows a seamless transition between multiple scales and their related atomic representations as well as the high-level double stranded representation (Figure 2). This provides users with the means to operate at any desired scale and visualize the results at other scales. For better compatibility with 2D designs and Cadnano, the visualization includes a multidimensional approach, which provides a 2D and 1D view for Cadnano designs (29).
Modelling of DNA nanostructures results in an idealized representation of the object that can be experimentally realized. We have implemented a highlight mode that provides immediate feedback to the design process, helping to visually detect interesting patterns in the design, such as single strands with specific lengths, unassigned bases, or crossovers. It is also possible to employ ntthal, from the Primer3 suite to calculate thermodynamic parameters of the binding regions on demand (22). A binding region is defined as consecutive base pairs that are not limited by a strand end or a crossover. All analysis results are colour coded in the visualization. It is also possible to control the visibility of all elements of the data model. By controlling the scale, highlight mode, and visibility, the user can tailor the visualization to be better suited for a specific task.
The combination of the data structure, DNA model, and visualization makes it possible to create, visualize, modify, and analyse DNA nanostructures. The output of these processes can be a re-usable model of the DNA nanostructure, the list of sequences needed for the in vitro self-assembly, or structural files for simulations in oxDNA (30). Basic modification options include the deletion of various elements, concatenation or insertion of DNA strands, and breaking strands by deleting the phosphodiester bond between nucleotides, amongst others.
Users can access all modifications, editors and options through an intuitive user interface. Through parametrized editors, the user can choose predefined shapes, and then vary some parameters to create a customized version of the selected shape. Some shapes provide basic building blocks, like the drawing of simple DNA strands or non-routed nanotubes, multilayer blocks and sheets. Others can provide more complex shapes, such as the wireframed editor, which allows the user to select a target 3D geometry and modify some of its parameters in a controllable manner while visualizing it, after which Daedalus is used to produce the DNA nanostructure.
DNA nanostructure manipulation
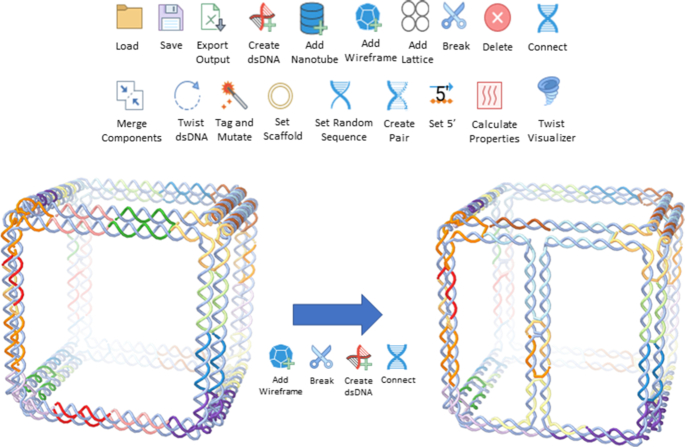
The editors allow the modification of existing DNA nanostructures, as well as the creation of new ones from scratch. It is possible to add single and double strands, straight or circular, cut any strand by either deleting the phosphodiester bond between nucleotides or deleting a nucleotide, and reconnect different strands (Figure 3). To connect strands with each other, the user can either move them in close proximity, thus making a direct connection, or introduce a new strand to link them.
Figure 3.
Depiction of the different editors for the interactive modelling of DNA nanostructures. Here, we demonstrate how a wireframe cube created with the Daedalus algorithm can be further edited to include an extra edge. In this way, it is possible to introduce in silico internal faces to Daedalus designs by creating extra edges on the proper faces, overcoming one of the design limitations of the Daedalus algorithm.
To showcase these editors and evaluate the precision of our data model, we designed cross-shaped nanostructures comprising two individual multilayer DNA origami nanorods. The nanorods consist of around 16 000 nucleotides, have an approximate size of 350 nm × 8 nm × 4 nm (Figure 4A) and were originally designed for other applications (23).
Figure 4.
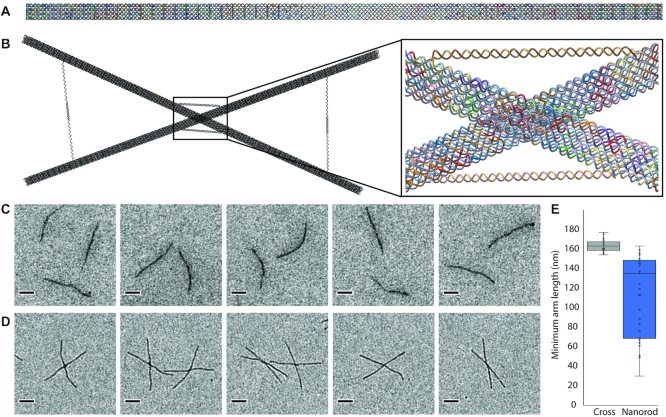
The DNA origami nanorod (A) and the cross (B) as designed in Adenita. (C) Negative stain TEM micrographs taken of the control nanorods. (D) Negative stain TEM images of the cross. (E) Since some images of crosses can also appear when separate nanorods superimpose each other on the grid, a statistical analysis of the cross-arm's length was performed to check that crosses were correctly folded.
We used the nanorods to create a simple cross. Each cross consists of two nanorods that were imported into Adenita as separate components and connected at different points with invading and extruding strands with the help of editors (Figure 4B). Further strands were added to constrain the cross angles and to give further stability. With the help of the visual representation we estimated the connection points and the lengths of the new strands. We selected this example as it demonstrates drawbacks of existing tools. For example, in caDNAno it would not possible to spatially align several structures. The 3D modelling capabilities of Adenita provide straightforward spatial clues, which help the user to select appropriate modifications at specific locations. This opens up the possibility to build up superstructures based on smaller structural components that are connected to each other.
Both, single nanorods and crosses were imaged using negative stain TEM (Figure 4). Due to superimposition of nanorods on the control slide that resulted in structurally deviating crosses, we measured the length of the arms of all observed crosses in the samples and controls. In the case of the samples containing actual crosses, we expected a certain regularity in the length of the arms, as by design each arm should be around half the nanorod length, i.e. 175 nm. In the case of crosses appearing on the TEM images of the controls, we expected to see a greater variation in the length of the arms. These experiments confirmed that our data model allows a realistic in silico manipulation of even long DNA nanostructures (Figure 4E).
DISCUSSION
Computational support for modelling 3D objects has become standard in many areas in order to facilitate the design and fabrication process. The publication of Cadnano boosted the emerging DNA nanotechnology field providing researchers with a simple tool to create nanoscale multilayer DNA origami objects. Due to limitations associated with the multilayer design concept (e.g. structures cannot fold at physiological ion concentrations), alternative design tools such as Daedalus and vHelix were developed facilitating new design concepts. However, these tools were also limited to a single DNA origami design concept and, in contrast to Cadnano, were lacking an appropriate user-interface and intuitive manual modelling possibilities. Adenita was developed to overcome these limitations. In addition, it addresses the increasing complexity of DNA nanostructures and their envisioned applications by allowing the incorporation of structural data from pdb-files into the modelling. One of the applications that we had in mind during the development of Adenita was the design of a novel class of artificial enzymes that comprise an amino acid based active site in a DNA nanostructure (20). Furthermore, we used Adenita also for the design and modelling of a four-component DNA origami rotor (Supplementary Figures S1–S8), a DNA origami-based enzyme cascade nanoreactor (Supplementary Figures S9–S11), wireframe DNA origami structures (Supplementary Figures S12–S17), a biosensor surface (Supplementary Figure S19), DNA pores (Supplementary Figure S18), DNA bricks (Supplementary Figures S26 and S27), and a DNA robot (Supplementary Figures S20–S25).
The long nanorods were selected to showcase another powerful feature of Adenita. In general, the precise modelling of DNA nanostructures becomes more challenging as the nanostructures increase in size due to the lack of accurate structural prediction. An imprecise model introduces an error at every helix turn, thus, the total error increases as the helix becomes longer. In some cases, this can be overcome by using nanostructures that have been extensively evaluated in the laboratory or with simulations. We took advantage of such a modular approach when designing the DNA origami crosses, as we had previously tested the nanorods. An alternative approach to overcome this problem are simulations that estimate a more realistic in silico model. For this purpose, we have implemented an export function for oxDNA simulations of the nanostructure model. However, in the future our DNA model could be fine-tuned using additional experimental data. More detailed spatial information, e.g. on the helix turns, will further improve the nanostructure designs. Nevertheless, the experiment with the crosses demonstrated that the implemented model is precise enough to modify large structures.
Thanks to its multiscale data structure and 3D CAD-inspired modelling capabilities, DNA nanostructures based on various design paradigms can be generated in Adenita (see supplementary materials). However, the interactive modelling behaviour would need to be adapted in order to create a more optimized workflow for a specific paradigm, with fewer repetitive interactions. Designing DNA bricks can be accelerated by incorporating well-defined snapping interactions between the bricks (31,32). For single-stranded DNA/RNA origami (33), we would need to adapt the colour coding as we currently depict only one colour per single strand. As our software is open-source, we encourage the adaption to specific design paradigms.
Future work will also involve optimizing the computational performance, so Adenita will be capable of working more smoothly with larger designs or with the new Gigadalton structures. This can be achieved by incorporating a representation of the nanostructure at the Gigadalton scale or by modifying the visualization to handle global and local representations.
Adenita is not only a framework capable of handling different design paradigms, but also introduces novel concepts to the modelling of DNA nanostructures, such as a modular approach, a novel visualization, and an environment capable of handling also other types of molecules, e.g. proteins or aptamers. We have shown that Adenita is capable of handling large structures and that the combination of its data model and the novel visualization gives the user the ability to edit and visualize nanostructures effectively. It combines in one tool several steps of current DNA nanostructure design pipelines. The use of several scales in the data model as well as in the visualization allows the user to work with the DNA nanostructure at different resolutions in parallel. Furthermore, this can be combined with editors and analysis options, extending the design possibilities much further than any other existing tool. At the same time, we recognize the strengths of the current methods, and we have found a way to incorporate them into Adenita's workflow.
We foresee that the combination of a user-friendly environment with a modular approach will foster a sharing-economy in the DNA nanotechnology community.
Supplementary Material
ACKNOWLEDGEMENTS
We want to thank Doris Rakoczy for proofreading the manuscript and all Adenita beta users for their feedback.
Contributor Information
Elisa de Llano, Center for Health and Bioresources, AIT Austrian Institute of Technology, Austria; Computational Physics Group, University of Vienna, Austria.
Haichao Miao, Center for Health and Bioresources, AIT Austrian Institute of Technology, Austria; Institute for Computer Graphics, TU Wien, Austria.
Yasaman Ahmadi, Center for Health and Bioresources, AIT Austrian Institute of Technology, Austria.
Amanda J Wilson, Department of Life Sciences, Imperial College London, UK.
Morgan Beeby, Department of Life Sciences, Imperial College London, UK.
Ivan Viola, Visual Computing Center, King Abdullah University of Science and Technology, Saudi Arabia.
Ivan Barisic, Center for Health and Bioresources, AIT Austrian Institute of Technology, Austria.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union's Horizon 2020 Research and Innovation programme [686647]; WWTF under the ILLVISATION [VRG11-010]; King Abdullah University of Science and Technology (KAUST) Office of Sponsored Research (OSR) [OSR-2019-CPF-410]. Funding for open access charge: H2020 Project MARA [686647].
Conflict of interest statement. None declared.
REFERENCES
- 1. Rothemund P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature. 2006; 440:297–302. [DOI] [PubMed] [Google Scholar]
- 2. Douglas S.M., Dietz H., Liedl T., Högberg B., Graf F., Shih W.M.. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009; 459:414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benson E., Mohammed A., Gardell J., Masich S., Czeizler E., Orponen P., Högberg B.. DNA rendering of polyhedral meshes at the nanoscale. Nature. 2015; 523:441–444. [DOI] [PubMed] [Google Scholar]
- 4. Veneziano R., Ratanalert S., Zhang K., Zhang F., Yan H., Chiu W., Bathe M.. Designer nanoscale DNA assemblies programmed from the top down. Science (New York, N.Y.). 2016; 352:1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nickels P.C., Wünsch B., Holzmeister P., Bae W., Kneer L.M., Grohmann D., Tinnefeld P., Liedl T.. Molecular force spectroscopy with a DNA origami-based nanoscopic force clamp. Science (New York, N.Y.). 2016; 354:305–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Linko V., Eerikäinen M., Kostiainen M.A.. A modular DNA origami-based enzyme cascade nanoreactor. Chem. Commun. (Camb.). 2015; 51:5351–5354. [DOI] [PubMed] [Google Scholar]
- 7. Bell N.A.W., Engst C.R., Ablay M., Divitini G., Ducati C., Liedl T., Keyser U.F.. DNA origami nanopores. Nano Lett. 2012; 12:512–517. [DOI] [PubMed] [Google Scholar]
- 8. Selnihhin D., Sparvath S.M., Preus S., Birkedal V., Andersen E.S.. Multifluorophore DNA origami beacon as a biosensing platform. ACS Nano. 2018; 12:5699–5708. [DOI] [PubMed] [Google Scholar]
- 9. Jiang Q., Song C., Nangreave J., Liu X., Lin L., Qiu D., Wang Z.-G., Zou G., Liang X., Yan H. et al.. DNA origami as a carrier for circumvention of drug resistance. J. Am. Chem. Soc. 2012; 134:13396–13403. [DOI] [PubMed] [Google Scholar]
- 10. Zhang Q., Jiang Q., Li N., Dai L., Liu Q., Song L., Wang J., Li Y., Tian J., Ding B. et al.. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS nano. 2014; 8:6633–6643. [DOI] [PubMed] [Google Scholar]
- 11. Zhao Y.-X., Shaw A., Zeng X., Benson E., Nyström A.M., Högberg B.. DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano. 2012; 6:8684–8691. [DOI] [PubMed] [Google Scholar]
- 12. Douglas S.M., Marblestone A.H., Teerapittayanon S., Vazquez A., Church G.M., Shih W.M.. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009; 37:5001–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Winfree E., Liu F., Wenzler L.A., Seeman N.C.. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998; 394:539–544. [DOI] [PubMed] [Google Scholar]
- 14. Zheng J., Birktoft J.J., Chen Y., Wang T., Sha R., Constantinou P.E., Ginell S.L., Mao C., Seeman N.C.. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 2009; 461:74–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wei B., Dai M., Yin P.. Complex shapes self-assembled from single-stranded DNA tiles. Nature. 2012; 485:623–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Williams S., Lund K., Lin C., Wonka P., Lindsay S., Yan H.. Goel A., Simmel F.C., Sosík P.. Tiamat: A three-dimensional editing tool for complex DNA structures. DNA Computing. 2009; Berlin, Heidelberg: Springer Berlin Heidelberg; 90–101. [Google Scholar]
- 17. Kekic T., Barisic I.. In silico modelling of DNA nanostructures. Comput. Struct. Biotechnol. J. 2020; 18:1191–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wagenbauer K.F., Sigl C., Dietz H.. Gigadalton-scale shape-programmable DNA assemblies. Nature. 2017; 552:78–83. [DOI] [PubMed] [Google Scholar]
- 19. Kosuri P., Altheimer B.D., Dai M., Yin P., Zhuang X.. Rotation tracking of genome-processing enzymes using DNA origami rotors. Nature. 2019; 572:136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kekic T., Ahmadi Y., Barisic I.. An enzymatic active site embedded in a DNA nanostructure. 2019; bioRxiv doi:13 January 2020, preprint: not peer reviewed 10.1101/804518. [DOI]
- 21. Nummelin S., Kommeri J., Kostiainen M.A., Linko V.. Evolution of structural DNA nanotechnology. Adv. Mater. (Deerfield Beach, Fla.). 2018; 30:e1703721. [DOI] [PubMed] [Google Scholar]
- 22. Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G.. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012; 40:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ahmadi Y., Llano E. de, Barišić I.. (Poly)cation-induced protection of conventional and wireframe DNA origami nanostructures. Nanoscale. 2018; 10:7494–7504. [DOI] [PubMed] [Google Scholar]
- 24. Stahl E., Martin T.G., Praetorius F., Dietz H.. Facile and scalable preparation of pure and dense DNA origami solutions. Angew. Chem. Int. Ed. Engl. 2014; 53:12735–12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rueden C.T., Schindelin J., Hiner M.C., DeZonia B.E., Walter A.E., Arena E.T., Eliceiri K.W.. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017; 18:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu X.-J., Olson W.K.. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003; 31:5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miao H., Llano E. de, Sorger J., Ahmadi Y., Kekic T., Isenberg T., Groller M.E., Barisic I., Viola I.. Multiscale visualization and scale-adaptive modification of DNA nanostructures. IEEE Trans. Vis. Comput. Graph. 2018; 24:1014–1024. [DOI] [PubMed] [Google Scholar]
- 29. Miao H., Llano E. de, Isenberg T., Gröller M.E., Barišić I., Viola I.. DimSUM: dimension and scale unifying map for visual abstraction of DNA origami structures. Comput. Graph. Forum. 2018; 37:403–413. [Google Scholar]
- 30. Šulc P., Romano F., Ouldridge T.E., Rovigatti L., Doye J.P.K., Louis A.A.. Sequence-dependent thermodynamics of a coarse-grained DNA model. J. Chem. Phys. 2012; 137:135101. [DOI] [PubMed] [Google Scholar]
- 31. Ke Y., Ong L.L., Shih W.M., Yin P.. Three-dimensional structures self-assembled from DNA bricks. Science (New York, N.Y.). 2012; 338:1177–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ong L.L., Hanikel N., Yaghi O.K., Grun C., Strauss M.T., Bron P., Lai-Kee-Him J., Schueder F., Wang B., Wang P. et al.. Programmable self-assembly of three-dimensional nanostructures from 10,000 unique components. Nature. 2017; 552:72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Han D., Qi X., Myhrvold C., Wang B., Dai M., Jiang S., Bates M., Liu Y., An B., Zhang F. et al.. Single-stranded DNA and RNA origami. Science (New York, N.Y.). 2017; 358:eaao2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.