Abstract
Tandem transcription interference occurs when the act of transcription from an upstream promoter suppresses utilization of a co-oriented downstream promoter. Because eukaryal genomes are liberally interspersed with transcription units specifying long non-coding (lnc) RNAs, there are many opportunities for lncRNA synthesis to negatively affect a neighboring protein-coding gene. Here, I review two eukaryal systems in which lncRNA interference with mRNA expression underlies a regulated biological response to nutrient availability. Budding yeast SER3 is repressed under serine-replete conditions by transcription of an upstream SRG1 lncRNA that traverses the SER3 promoter and elicits occlusive nucleosome rearrangements. SER3 is de-repressed by serine withdrawal, which leads to shut-off of SRG1 synthesis. The fission yeast phosphate homeostasis (PHO) regulon comprises three phosphate acquisition genes – pho1, pho84, and tgp1 – that are repressed under phosphate-replete conditions by 5′ flanking lncRNAs prt, prt2, and nc-tgp1, respectively. lncRNA transcription across the PHO mRNA promoters displaces activating transcription factor Pho7. PHO mRNAs are transcribed during phosphate starvation when lncRNA synthesis abates. The PHO regulon is de-repressed in phosphate-replete cells by genetic manipulations that favor ‘precocious’ lncRNA 3′-processing/termination upstream of the mRNA promoters. PHO lncRNA termination is governed by the Pol2 CTD code and is subject to metabolite control by inositol pyrophosphates.
INTRODUCTION
Transcriptional interference is a widely prevalent biological phenomenon whereby active transcription from one promoter can suppress in cis the transcription from a neighboring promoter. There are many variations on this theme, according to whether the interfering promoter and the target promoter are tandemly oriented or convergently oriented and the degree to which the interfering and target genes overlap one another. Interference in cis can engender dire traffic control problems if boundaries between tandem transcription units are not enforced, especially in the context of compact genomes. At the most basic level, this is achieved by efficient termination of RNA polymerase transcription of an upstream gene before the polymerase traverses the distal gene promoter (1). This safeguard can be breached, and tandem interference imposed, in any of several ways, for example: (i) by mutations in the cis signals or trans factors that elicit 3′-processing/termination (2,3); (ii) loss of intergenic roadblocks that prevent runaway transcription from upstream (4); or (iii) insertion of a mobile promoter-containing DNA element upstream of a native gene promoter (5,6). The unifying feature of interference in cis is that it is predicated on the transcription of the interfering RNA, and not necessarily on the type or abundance of the interfering RNA, or even that the class of RNA polymerase responsible for transcribing the interfering and target RNAs be the same (7). Whereas the target gene is typically protein-coding, the interfering gene need not be protein-coding. Indeed, there is a growing appreciation of biological scenarios in which transcription of an upstream long non-coding RNA negatively influences the expression of a tandemly oriented downstream protein-coding gene. However, influence is not synonymous with regulation. Here, I will review two eukaryal model systems for which there is compelling evidence that lncRNA interference with mRNA expression underlies a regulated physiological response to nutrient availability: SER3 in budding yeast and the phosphate homeostasis (PHO) regulon in fission yeast.
Tandem lncRNA–mRNA transcriptional interference in eukarya: general principles
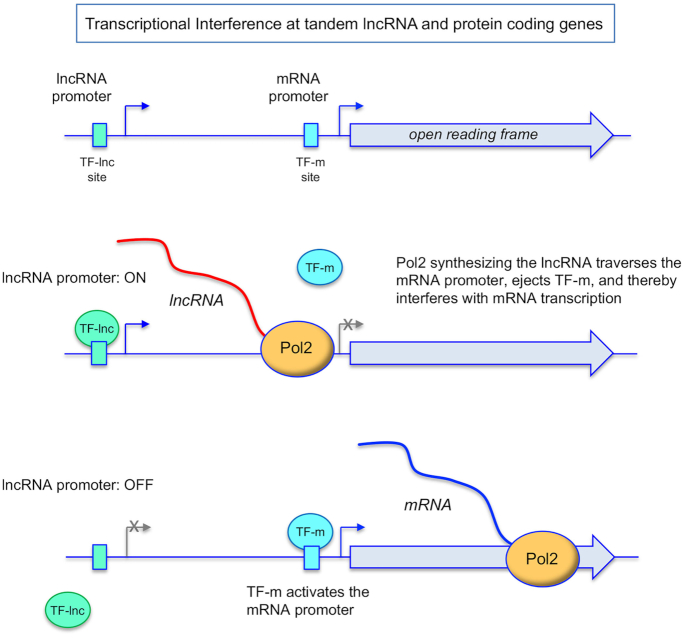
Regulated interference is most simply viewed as a binary switch in promoter utilization by RNA polymerase II (Pol2). As depicted in Figure 1, transcription of the upstream interfering lncRNA gene relies on a particular transcription factor, TF-lnc, that recognizes a specific DNA site (an upstream activating sequence; UAS) in the lncRNA promoter. Transcription of the downstream protein coding gene is governed by a different transcription factor, TF-m, that binds to its cognate site in the mRNA promoter. When the lncRNA promoter is activated, Pol2 initiates lncRNA synthesis. In the event that the Pol2 elongation complex traverses the TF-m site in the downstream mRNA promoter, the interaction of TF-m with its DNA site will be disrupted (at least transiently) and mRNA transcription initiation will be suppressed. Maintenance of the mRNA OFF state requires that the lncRNA promoter remain ON and that lncRNA elongation continually traverses the mRNA promoter. Regulation of interference is achievable by turning lncRNA transcription off, e.g. via interdiction of TF-lnc binding to its site in the lncRNA promoter (per Figure 1) or preventing DNA-bound TF-lnc from recruiting Pol2 and/or other transcription factors to assemble a lncRNA initiation complex. Once lncRNA synthesis abates, the impediment to TF-m binding to the mRNA promoter is lifted and mRNA transcription ensues. A key feature of this scheme of interference is that regulatory potential inheres to the transcription of the upstream lncRNA.
Figure 1.
General scheme of transcriptional interference at tandem lncRNA and protein coding genes. See text for discussion.
One can reasonably invoke regulated interference if the promoter switch occurs in response to specific changes in growth conditions or environmental cues. The directionality of the interference switch during the transition from a physiological ground state to a perturbed state can, in principle, go either way. For example, there might be no interference in the ground state when the product of the coding gene is called for, but the response to a cellular perturbation demands that this protein be depleted. In such a case, the perturbed state would elicit activation of the interfering lncRNA promoter, e.g. by triggering binding of TF-lnc to its DNA site. Alternatively, the need for the coding gene product in the ground state is low and interference by lncRNA synthesis prevails. Upregulation of protein expression in response to a cellular perturbation would then be effected by turning lncRNA synthesis off.
For any given tandem interference system, a full mechanistic framework is contingent on defining the key players and their properties. Ideally, one wants to know the identity of the transcription factors TF-lnc and TF-m, the DNA sites that they recognize, their DNA binding affinities (including their on and off rates), and the relative strengths of the lncRNA and mRNA promoters in vivo. It has been suggested that sensitivity to interference is enhanced when TF-m has a slow spontaneous off rate but is easily dislodged by elongating polymerase (8). Notwithstanding the elegant studies that established the basic properties of several regulated transcriptional interference model systems in yeast, there are still knowledge gaps regarding the transcription factors and their activities.
Whereas lncRNA transcription is necessary to establish regulated interference, a salient question has been how this is achieved and the degree to which it entails events other than, or in addition to, dislodgement of transcription factors from the mRNA promoter. An attractive scenario, for which there is considerable evidence, is that the act of lncRNA transcription alters the chromatin context over the flanking mRNA promoter to attain a repressive state, i.e. one that is relatively non-permissive for binding of TF-m (or re-binding after it is initially ejected). Two main lines of investigation prompt invocation of chromatin-mediated interference: (i) interrogation of chromatin status and the presence of covalent chromatin marks over the regulated gene loci and (ii) testing the effect of mutations in histone modifying factors and chromatin remodelers on the extent of lncRNA interference, e.g. whereby de-repression of the mRNA by such a mutation in the absence of a significant effect on lncRNA synthesis would implicate deposition of specific histone marks (or chromatin re-arrangements) as being key to achieve repression.
It is noteworthy that different chromatin interventions apply to different model systems of regulated interference in yeast. In the case of repression of the budding yeast IME1 gene by transcription in cis of the upstream IRT1 lncRNA, the act of lncRNA synthesis driven by transcription factor Rme1 results in displacement of a transcriptional activator Pog1 from the IME1 promoter; establishes high nucleosome occupancy over the IME1 promoter; and triggers a repressive chromatin state that depends on the Set2 histone H3K36 methyltransferase and the Set3 histone deacetylase, mutations of which de-repress IME1 (9). By contrast, interference with budding yeast SER3 expression in serine-replete conditions by transcription of an upstream SRG1 lncRNA is associated with high nucleosome occupancy over the SER3 promoter, but does not require histone methyltransferases Set1, Set2, and Dot1 or histone deacetylase subunits Set3 and Rco1 (10). Such variations suggests that a key determinant of tandem lncRNA–mRNA transcriptional repression is a dynamic competition between TF-m binding to, and nucleosome deposition over, the mRNA promoter – whereby differences in TF-m abundance and site affinity can call for less or more stringent chromatin states in order to interdict mRNA promoter firing.
Serine-regulated lncRNA transcriptional interference in budding yeast
The Saccharomyces cerevisiae SER3 gene, which encodes the serine biosynthetic enzyme 3-phosphoglycerate dehydrogenase, is repressed when cells are grown in serine-replete medium and induced in response to serine starvation. A series of insightful studies by the Winston and Martens laboratories identified an upstream flanking lncRNA gene—SRG1—as responsible for SER3 repression in cis via transcriptional interference (Figure 2) (10–12). In serine-replete cells, SRG1 transcription is turned on via a serine-responsive transcription factor Cha4 that recognizes a UAS in the SRG1 promoter (12). Cha4 is a 648-amino acid protein that belongs to the Cys6•Zn2 family of fungal DNA binding transcriptional regulators (13). Cha4 was identified initially as required for the expression of the CHA1 gene specifying the serine-inducible enzyme l-serine deaminase that enables yeast growth on serine as the sole nitrogen source (14). An N-terminal fragment of Cha4 that includes the Cys6•Zn2 module recognizes either of two semi-palindromic UAS elements in the CHA1 promoter, each of which suffices to confer serine-inducible gene expression (14). A related palindromic UAS is present upstream of the TATA-box in the SRG1 promoter (Figure 2). A 5′ truncation of the SRG1 upstream region that deletes this element ablates SRG1 lncRNA production and de-represses SER3 in serine-replete cells (12). The Cha4 DNA-binding domain has not been purified or characterized biochemically/structurally with respect to its mode of DNA recognition or its DNA binding affinity. The inverted palindromic sequences at the putative Cha4 sites would suggest that Cha4 binds as a homodimer, akin to many other Cys6•Zn2 transcription factors (13).
Figure 2.
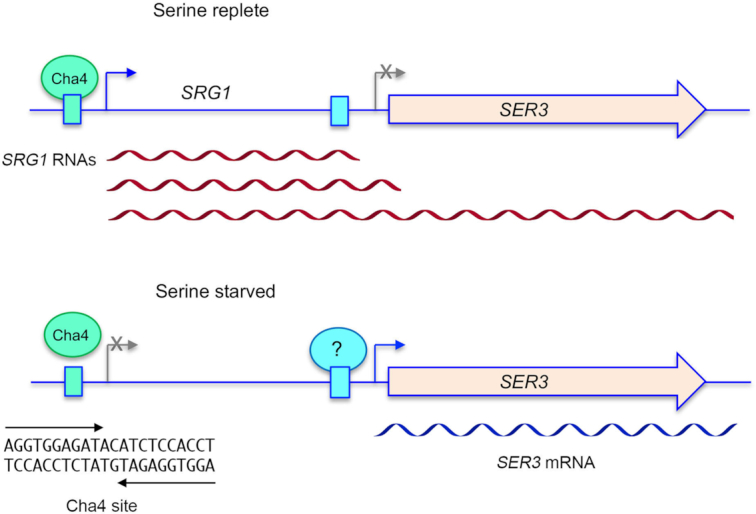
Regulated transcriptional interference at the budding yeast SRG1-SER3 locus in response to serine availability. (Top panel) In serine replete cells, activation of SRG1 transcription by Cha4 results in synthesis of three species of lncRNA (red wavy lines) that are processed at different poly(A) sites. Pol2 synthesizing these lncRNA will traverse the SER3 mRNA promoter. (Bottom panel) SRG1 transcription is turned off quickly in response to serine starvation, which makes the SER3 promoter available for binding to an as yet unknown activating transcription factor and hence synthesis of SER3 mRNA (blue wavy line). It is suggested that Cha4 remains associated with the SRG1 promoter during serine starvation but is unable to activate lncRNA synthesis. The putative Cha4 duplex DNA binding site is shown with the inverted repeat demarcated by arrows.
Transcription from the SRG1 gene in serine-replete cells generates three distinct 5′-co-terminal, 3′ polyadenylated transcripts that differ in their choice of poly(A) sites (15). The 3′ ends of the two SRG1 lncRNAs arise within the SER3 promoter and near the 5′ end of the SER3 mRNA, respectively (Figure 2). Because transcription termination in yeast occurs at an interval downstream of the site of nascent RNA cleavage/polyadenylation, it is certain that Pol2 synthesizing either form of the SRG1 lncRNA will traverse the SER3 promoter. The third species produced from the SRG1 promoter is an SRG1–SER3 read-through transcript that extends to the poly(A) site of the SER3 gene (11,15).
The transcription switch upon serine starvation is rapid, i.e., Northern analysis showed that SRG1 lncRNAs are depleted after 15 min of serine deprivation, by which time the SER3 mRNA attains its peak level (12). A notable finding was that serine starvation elicited only a modest decrement in Cha4 at the SRG1 promoter, as gauged by ChIP, suggesting that regulation by serine availability is achieved at a step subsequent to Cha4 DNA binding, most likely the recruitment of coactivators to the SRG1 promoter (12). Little is known about the mechanism of serine sensing in this system, e.g. whether serine per se, or a metabolite of serine, is the relevant signaling molecule; whether/how serine or its metabolite affect the structure and properties of DNA-bound Cha4; and the transcription activation functions and protein-protein interactions of the Cha4 segments outside of the Cys6•Zn2 module.
As noted in the preceding section, cessation of SRG1 lncRNA synthesis and de-repression of SER3 expression upon serine starvation is associated with a gain of nucleosome occupancy over the SER3 promoter (10). At present, the SER3 UAS has not been finely mapped and there is (to my knowledge) no information as to the identity of the putative TF-m transcription factor that drives SER3 expression. Whereas surrogate SRG1-based transcription interference reporter systems have been devised that replace the SER3 UAS with Gal4 binding sites (11,12), a full understanding of the endogenous regulation will hinge on defining and characterizing TF-m for SER3, insofar as it entails a putative competition between TF-m binding and nucleosome occupancy at the SER3 promoter.
The SRG1–SER3 axis has proven to be a font of insight into gene regulation by testing various mutant yeast strains for de-repression of SER3 under serine-replete conditions. It was thereby shown that repression of SER3 depends on Snf2 (the ATPase subunit of the SWI/SNF chromatin remodeling complex); SAGA complex subunits Spt3, Spt8, Spt7, Spt20, and Ada1; FACT complex subunits Spt16 and Pob3; chromatin reassembly factors Spt6, Spn1, and Spt2; PAF complex subunits Paf1 and Ctr9; and specific amino acids in histones H3 and H4 (10,12,16–20). A pertinent finding was that spt6 and spt16 mutations reduced nucleosome levels over the SER3 promoter without reducing SRG1 transcription (10), signifying that upstream lncRNA synthesis was necessary but not sufficient for downstream mRNA repression.
lncRNA transcriptional interference regulates fission yeast phosphate homeostasis
Inorganic phosphate is an essential nutrient that is assimilated by cells into myriad diverse molecules with distinctive bond chemistries and biological functions. Phosphate availability exerts a profound influence on cellular growth, whereby too much phosphate can promote unwanted cell proliferation and too little phosphate can restrict growth. Cells from all domains of life respond to acute phosphate starvation by inducing the transcription of phosphate acquisition genes encoding secreted or cell-surface associated enzymes that mobilize phosphate from the extracellular environment and transmembrane transporters of inorganic phosphate or simple phosphate-containing compounds. Different taxa rely on diverse strategies to achieve a rapid transcriptional response to acute phosphate starvation (21–23). Phosphate homeostasis in the fission yeast Schizosaccharomyces pombe is achieved by an intricate network of positive and negative influences on the transcription of three genes—comprising a PHO regulon—encoding proteins involved in extracellular phosphate mobilization and uptake: a cell surface acid phosphatase Pho1, an inorganic phosphate transporter Pho84, and a glycerophosphate transporter Tgp1. Pho84 and Pho1 are encoded by adjacent tandemly oriented genes on chromosome II; the gene encoding Tgp1 is located 4-Mb upstream on chromosome II (Figure 3A). Genome-wide microarray analysis showed that expression of the PHO genes is induced over the course of 4 hours following transfer to medium lacking phosphate (24). Colorimetric assay of cell-associated acid phosphatase enzyme activity (by hydrolysis of p-nitrophenylphosphate) provides a quantitative gauge of basal and starvation-induced pho1 expression. Individual yeast colonies can be assayed semi-quantitatively by overlay with a chromogenic substrate α-napthylphosphate. Using the overlay assay for visual screening of a collection of 2813 fission yeast deletion strains, the Wykoff lab identified the pho7 gene as essential for basal Pho1 expression and for Pho1 induction in response to phosphate starvation (25). RT-qPCR and microarray analyses showed that Pho7 is required for upregulation of the pho1, pho84 and tgp1 genes in phosphate-starved cells (24,25). Pho7 is a Cys6•Zn2 family DNA-binding transcription factor and is the TF-m equivalent for the fission yeast PHO regulon.
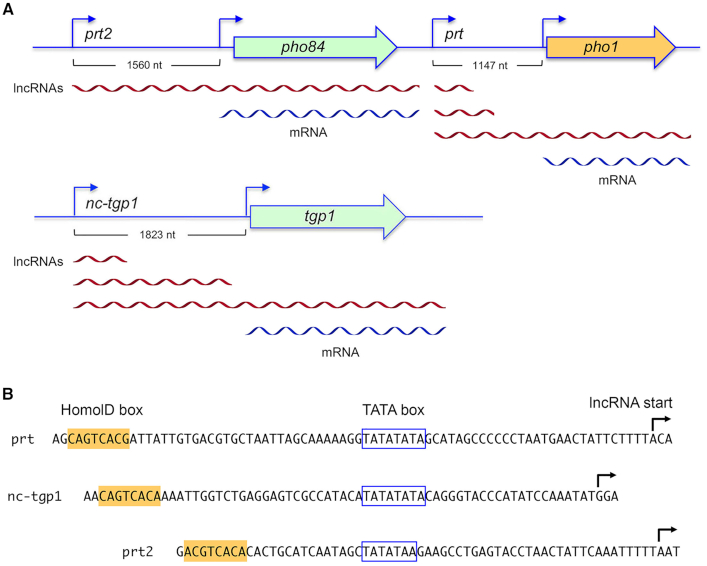
Figure 3.
Regulated transcription interference by upstream lncRNAs underlies phosphate homeostasis in fission yeast. (A) Tandem arrangements of the PHO lncRNA and mRNA genes are shown. Distance between the lncRNA and mRNA transcription start sites are indicated. The prt2, prt, and nc-tgp1 lncRNAs are depicted as red wavy lines. The pho84, pho1 and tgp1 mRNAs are blue wavy lines. (B) The nucleotide sequences and transcription start sites of the PHO lncRNA gene promoters (top strand) are shown. The TATA box and HomolD box elements are highlighted.
The Wykoff lab's deletion library screen also identified several mutants in which Pho1 was expressed constitutively at a high level under phosphate-replete conditions, comparable to the level seen in phosphate-starved wild-type cells (25). Constitutive expression in such mutants required Pho7 in every case. Their findings hinted that pho1 might be actively repressed when phosphate is available.
In the ensuing years, studies from four different groups – alighting on the PHO genes from different angles – have established that the pho1, pho84, and tgp1 genes are indeed actively repressed during growth in phosphate-rich medium by the transcription in cis of a long noncoding RNA from the respective 5′ flanking genes prt, prt2, and nc-tgp1 (26–31) (Figure 3). The transcription start sites of the PHO regulon lncRNAs and mRNAs have been mapped; the spacings between them are indicated in Figure 3A. The PHO lncRNAs have been sized by Northern blotting and their sites of cleavage/polyadenylation have been assigned by 3′-RACE, as depicted in Figure 3A. Analysis of the PHO lncRNAs is complicated by the fact that they are subject to rapid turnover by the nuclear exosome. Defining the population of PHO lncRNAs was facilitated by maneuvers that stabilized them against rapid decay (using mutant yeast strains lacking the Rrp6 subunit of the nuclear exosome or lacking the decay-promoting RNA binding protein Mmi1) or that enhanced the transcriptional signals by increasing the copy number of the tandem PHO lncRNA-mRNA loci. Characterizing the PHO mRNAs in phosphate-starved cells was comparatively straightforward. Note therefore that absolute steady-state abundance of the PHO lncRNAs versus that of the cognate PHO mRNAs is not an accurate gauge of transcription interference. Rather, it is the act of transcribing the lncRNA that establishes PHO gene repression.
The PHO system presents a unique challenge to understand how lncRNA interference coordinately governs three genes in the same physiological pathway of nutrient sensing. As discussed in previous sections, the salient questions are: (i) what cis and trans factors control PHO lncRNA synthesis and how are they responsive to phosphate availability? (ii) how does Pho7 drive PHO mRNA expression and how is Pho7 affected by upstream lncRNA synthesis?; (iii) to what extent does lncRNA-mediated repression of the PHO genes rely on chromatin alterations; (iv) do events other than activity of the interfering lncRNA promoters regulate or impact PHO gene repression in phosphate-replete cells. A corollary question in this multigene system is whether the inputs to control of the three PHO genes are shared or distinctive. Current understanding of these issues is summarized below, and knowledge gaps are highlighted.
Determinants of PHO lncRNA transcription
cis-acting signals for transcription of the PHO lncRNAs were identified by introducing plasmids containing the lncRNA upstream regions and transcription start sites fused to an acid phosphatase reporter gene into cells that had been deleted at the chromosomal prt–pho1 locus (29–31). Serial 5′ truncations demarcated the margins of the prt, prt2, and nc-tgp1 lncRNA promoters to DNA segments extending 110, 62 and 71 nucleotides flanking the lncRNA start sites. The PHO lncRNA promoters share a distinctive bipartite architecture, consisting of a TATA-box element situated 30, 23, or 32 nucleotides upstream of the prt, prt2, and nc-tgp1 start sites and a HomolD-box located 30, 26, or 15 nucleotides upstream of the TATA-box (Figure 3B). A similar bipartite HomolD/TATA promoter structure has been described for the fission yeast U3 snoRNA gene (32). The consensus HomolD element 5′-CAGTCAC(A/G) was identified by the Käufer lab as a Pol2 promoter signal in fission yeast genes encoding ribosomal proteins (33–35). Nucleotide substitutions in either the HomolD box or the TATA-box inactivate the nc-tgp1 and pho84 lncRNA promoters and result in de-repression of the flanking tgp1 and pho84 mRNA promoters. HomolD or TATA mutations additively inactivate the prt promoter and de-repress pho1 mRNA transcription. These effects of lncRNA promoter mutations provide clear evidence that Pho7 and any other components necessary for PHO mRNA transcription are present and available for action under phosphate-replete conditions.
Whereas the cis signals for PHO lncRNA transcription are well defined, the trans-acting factor(s) corresponding to TF-lnc are not. One presumes there is a positive transcription factor that recognizes the HomolD box in the lncRNA promoters, but its identity is presently unknown. A factor in fission yeast whole-cell extract that binds to a consensus HomolD sequence was purified by serial ion-exchange chromatography steps, followed by HomolD DNA-affinity chromatography. Mass spectrometry identified the RNA polymerase I general transcription factor Rrn7 in the most purified fraction and ChIP analysis indicated association of Rrn7 with the promoter region of an exemplary HomolD-containing ribosomal protein gene (36). A reconstituted fission yeast in vitro system was developed that displayed Rrn7-dependent formation of a Pol2 pre-initiation complex on this ribosomal protein gene promoter (37). Establishing the role of Rrn7 in HomolD-dependent Pol2 transcription in vivo is not straightforward, insofar as Rrn7 is essential for vegetative growth and conditional loss-of-function alleles are lacking. The Maldonado lab reported that Rrn7 is a substrate for phosphorylation on Thr67 by casein kinase 2, that such phosphorylation inhibits Rrn7 binding to a HomolD box in vitro, and that T67A mutation prevents inhibition of DNA binding by casein kinase 2 (38). It was appealing then to think that Rrn7 Thr67 phosphorylation during phosphate starvation might be the basis for shut off of HomolD-driven lncRNA transcription. If so, then prevention of such phosphorylation by T67A mutation might block the starvation induction of phosphate-responsive genes. However, in a strain in which the native rrn7+ gene was deleted and a mutant allele rrn7-T67A was introduced, there was no effect on pho1 induction upon phosphate starvation (30). Thus, Thr67 phosphorylation of Rrn7 is not a decisive event in the phosphate starvation response. Absent convincing evidence in favor of Rrn7, the jury is out concerning the identity of TF-lnc in the PHO regulon.
Defining TF-lnc is the key to understanding how transcription interference is regulated in response to phosphate status, insofar as the most parsimonious model is that simply shutting off the lncRNA promoters upon starvation is principally responsible for turning on PHO mRNA synthesis and re-booting the lncRNA promoters during phosphate repletion is what shuts the PHO genes off. Thus, whatever signals are generated by acute changes in phosphate availability (also an unknown) likely converge on TF-lnc.
Pho7 binding to PHO mRNA promoters
ChIP-seq analysis of tagged Pho7–TAP demonstrated a significant basal level of association of Pho7 with the pho1 and pho84 upstream DNA regions in phosphate-replete cells that was increased several-fold upon phosphate starvation (24). There was virtually no Pho7 ChIP-seq signal upstream of tgp1 in phosphate-replete cells, though Pho7 did accumulate there after 2 h of phosphate starvation. These Pho7 occupancy trends correlate with the relative basal levels of the PHO mRNAs in phosphate-replete cells. Subsequent ChIP studies using Pho7-GFP showed that deletion of the nc-tgp1 promoter and start site elicited a gain of Pho7 occupancy at the tgp1 promoter (28), consistent with lncRNA transcription resulting in ejection of Pho7.
Whereas a Pho7 recognition element could not be discerned at the time from the genome-wide ChIP seq data (24), an updated in silico analysis of the same dataset retrieved a putative 12-nucleotide consensus Pho7 binding site: 5′-TCG(G/C)AxTTTxAA (39). In an independent line of investigation, an autonomous DNA-binding domain (DBD) within Pho7 was delineated (Figure 4A) and its binding to the PHO mRNA promoters was characterized by electrophoretic mobility shift assays (EMSA) and DNase footprinting (31,39). The pho1 promoter contains two Pho7 recognition elements (sites 1 and 2; Figure 4B), separated by a 20-nt spacer. Pho7 binds independently and non-cooperatively to these sites in vitro. The tgp1 promoter contains a single Pho7 recognition site (Figure 4B). Nucleobase mutations in either of the pho1 sites or in the tgp1 site that eliminate Pho7 binding in vitro result in loss of pho1 or tgp1 promoter activity in vivo—to the same extent as does deleting the pho7 gene (39). EMSA data and ChIP seq data indicate that there are multiple Pho7 sites within the region upstream of the pho84 gene, with varying affinity for the Pho7 DBD (24,31). One high-affinity site in the pho84 promoter was mapped by footprinting (31) (Figure 4B). The experimentally assigned Pho7 sites agree with the consensus in silico motif (Figure 4B). The Pho7 DNA recognition site differs from those of other fungal Cys6•Zn2 proteins, which typically recognize pairs of CGG triplets that are arranged as inverted, direct, or everted repeats (13). pho1 promoter site 2, the tgp1 promoter site, and the pho84 promoter site that are recognized by Pho7 contain a single CGG triplet. pho1 promoter site 1 has a variant CGC triplet and binds Pho7 with lower affinity (Kd 130 nM) than pho1 site 2 (Kd 40 nM) in the EMSA assay for binding to 25 nM duplex Pho7-site DNA in the presence of a non-specific DNA poly(dI:dC) (40).
Figure 4.
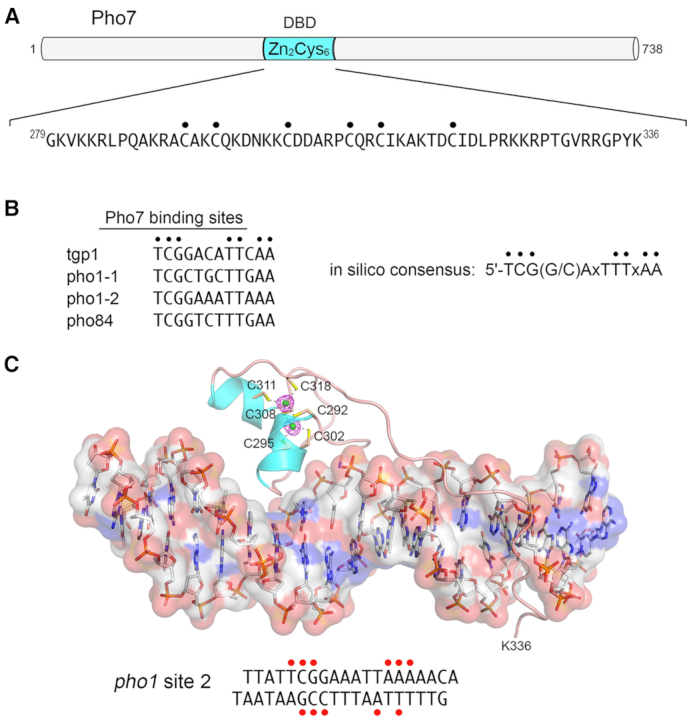
Transcription factor Pho7 recognizes a UAS element in the PHO mRNA promoters. (A) The 738-aa Pho7 polypeptide is depicted as a horizontal bar. The internal DNA-binding domain containing the Cys6•Zn2 module is colored cyan. The primary structure of the minimal DBD is shown below the cartoon, with the Zn-binding cysteines indicated by dots. (B) The nucleobase sequences at the experimentally determined Pho7 binding sites in the indicated PHO mRNA promoters are aligned. Conserved positions are indicated by black dots. The Pho7 binding site consensus sequence identified by in silico analysis of genome-wide Pho7 ChIP-seq data is shown on the right. (C) Structure of Pho7 DBD bound to pho1 site 2 DNA. The Pho7–DNA complex is shown with the DNA depicted as a stick model, with an overlying transparent surface model to highlight the major and minor grooves. The Pho7 protein is rendered as a cartoon trace with cyan α-helices. The two zinc atoms are shown as green spheres and the six zinc-binding cysteines are shown as stick models and labeled. Anomalous difference density for the zinc atoms, contoured at 5σ, is shown in red mesh. The primary structure of the pho1 site 2 DNA ligand is shown at bottom. Red dots indicate DNA nucleobases that are contacted directly by Pho7 amino acids.
Keen insights into Pho7 DNA recognition emerged from high-resolution crystal structures of the Pho7 DBD in complex with its target site in the tgp1 promoter (41) and with pho1 site 2 (40; Figure 4C). The structures underscored distinctive features of the Pho7-DNA interface, whereby: (i) Pho7 DBD binds DNA as a monomer, unlike the DBDs of most other fungal Cys6•Zn2 factors that bind as homodimers; and (ii) Pho7 DBD makes extensive interactions with its asymmetric target sequence over a 14-bp footprint that entails direct and/or water-mediated hydrogen bonding to individual nucleobases and backbone phosphates within, and remote from, the CGG triplet. Figure 4C highlights with red dots the nucleobases in pho1 site 2 that are contacted directly by Pho7. Comparison of the two Pho7–DNA structures revealed shared determinants of target site specificity as well as variations in the protein–DNA interface that accommodate different promoter DNA sequences (40). Structure-guided mutational analysis of the Pho7-DNA interface identified the Pho7 amino acids at which alanine substitutions abolished or attenuated the pho1 phosphate starvation response by virtue of their effect on the binding of Pho7–DBD to recognition site 1 in the pho1 promoter (40,41).
It is not known how Pho7, once engaged to its cognate DNA site, activates transcription of the PHO mRNAs. The large protein regions that flank the Pho7 DBD are predicted (in IUPRED) to be structurally disordered and there has been, as yet, no delineation of transcriptional activation or protein–protein interaction functions within those flanking segments. Nor is it known whether the flanking domains might affect DNA binding. These are issues that warrant attention in order to better understand how transcription interference suppresses the mRNA promoters.
A related issue concerns the relative strengths in vivo of the interfering lncRNA and target mRNA promoters within the tandem gene loci of the PHO regulon. The active accelerated decay of the lncRNAs vitiates RNA levels as a gauge of lncRNA versus mRNA promoter firing. Using reporter assays in which the lncRNA promoter and its transcription start site or the cognate mRNA promoter and its transcription start site drive expression of the same acid phosphatase enzyme in pho1Δ cells, it is possible to compare lncRNA and mRNA promoter strengths in vivo. This approach indicates that: the nc-tgp1 promoter is about 25% more active than the tgp1 promoter; the prt promoter is about half as active as the pho1 promoter; and the prt2 promoter is ∼60% as active as the pho84 promoter (29–31). These relative strengths are compatible with regulation by interference, insofar as it would be challenging to establish interference if the mRNA promoters were many-fold stronger than the upstream lncRNA promoters.
Role of chromatin in PHO mRNA repression
It was proposed initially that prt transcription elicits RNAi-dependent installation of the repressive H3K9me2 facultative heterochromatic mark over the pho1 locus and that this mark was lost during phosphate starvation (27). However, studies from the Allshire lab showed that: (i) levels of H3K9me2 over the prt and pho1 genes were barely above background and did not change upon phosphate starvation; (ii) H3K9me2 was undetectable over the nc-tgp1 and tgp1 genes; (iii) prt, pho1, nc-tgp1, and tgp1 transcript levels were unaffected by loss of RNAi components or the H3K9 methyltransferase Clr4; and (iv) the kinetics of pho1 and tgp1 induction during starvation were unaltered in clr4Δ cells lacking the heterochromatic mark (28). They concluded that RNAi and heterochromatin do not play a significant role in PHO gene regulation. Rather, Ard et al. proposed that lncRNA transcription increases nucleosome density over the pho1 and tgp1 mRNA promoters and that nucleosome density is diminished during phosphate starvation, which allows access of Pho7 to the mRNA promoters.
Their subsequent chromatin-related studies (42) showed that the H3K36me3 mark is present over the tgp1 promoter in phosphate-replete cells and ablation of the Set2 methyltransferase that inscribes this mark leads to partial de-repression of tgp1 (partial in the sense that it is less than the extent of tgp1 de-repression seen when nc-tgp1 lncRNA synthesis is abolished). Partial de-repression of tgp1 was also observed in cells bearing mutations in histone chaperone Spt6 and the FACT complex subunit Spt16. pho1 expression was similarly de-repressed in phosphate-replete cells with Set2, Spt6, and Spt16 mutations (42).
Cascade regulation of the prt2–pho84–prt–pho1 gene cluster
Taking into consideration the locations of the Pho7 binding sites that drive PHO mRNA synthesis, and the sizes and 3′ termini of the lncRNA primary transcripts in phosphate-replete cells (Figure 3A), it is apparent that Pol2 engaged in lncRNA synthesis will traverse the Pho7 binding sites and, as per the general model for interference (Figure 1), transiently dislodge Pho7. In the case of nc-tgp1 repression of tgp1, the cleavage/poly(A) site of the predominant nc-tgp1 lncRNA (5′-UCGGA↓) is located 187 nt upstream of the tgp1 transcription start site, exactly within the Pho7 DNA binding site of the tgp1 promoter (5′-TCGGA↓CATTCAA) (30). The situation is different for the interfering prt2 and prt lncRNAs, which are prt2-pho84 and prt-pho1 read-through transcripts that undergo 3′ end formation at the respective mRNA poly(A) sites (25,26,30) (Figure 3A).
The tightly clustered arrangement of the prt2, pho84, prt, and pho1 transcription units in the fission yeast genome is remarkable, insofar as lncRNA genes alternate with mRNA genes in the same nutrient response pathway (Figure 3A). An intriguing finding regarding the four-gene cluster arrangement was that changes in prt2 lncRNA expression exert a cascade effect on the downstream prt and pho1 mRNA loci, whereby the inactivation of prt2 lncRNA transcription via deletion or mutation of the prt2 promoter elicits upregulation of both pho84 and pho1. The basis for this cascade effect is that increased transcription of the upstream pho84 mRNA gene represses the downstream flanking prt lncRNA gene, by virtue of the fact that the major pho84 mRNA 3′-cleavage/poly(A) site is located just 158 nt upstream of the transcription start site of the flanking prt lncRNA and 83 nt upstream of the HomolD box of the prt promoter (31). Indeed, inactivation of the pho84 promoter by mutating its TATA box suffices to de-repress the flanking prt promoter (31). This illustrates a role reversal for mRNA transcription as the agent of repression by transcriptional interference rather than the passive target of such interference.
PHO lncRNA 3′-processing and termination impact PHO mRNA repression
The simplified general scheme of regulated tandem gene transcription interference in Figure 1 is promoter-centric but has the potential to be embellished by modulating post-initiation events in lncRNA synthesis. In particular, because interference relies on Pol2 traversing the mRNA promoter during lncRNA synthesis, it might be possible to tune interference by increasing or decreasing the frequency with which Pol2 terminates lncRNA transcription prior to encounter with the mRNA promoter. According to this scheme, as it would apply to the PHO regulon, events that enhance ‘precocious’ termination of lncRNA transcription would result in de-repression of PHO mRNA expression in phosphate-replete cells (Figure 5A) and those that reduce the probability of lncRNA termination prior to the mRNA promoter would result in hyper-repression of the flanking PHO mRNAs relative to their basal levels (Figure 5B). A prescient early finding from the Wykoff screen was that deletion of the gene encoding the Ctf1 subunit of the fission yeast cleavage and polyadenylation factor (CPF) complex resulted in reduced pho1 expression in phosphate-replete cells (25). The significance of this result was not evident at the time because the PHO lncRNAs and transcription interference had not yet been discovered as central to phosphate homeostasis.
Figure 5.
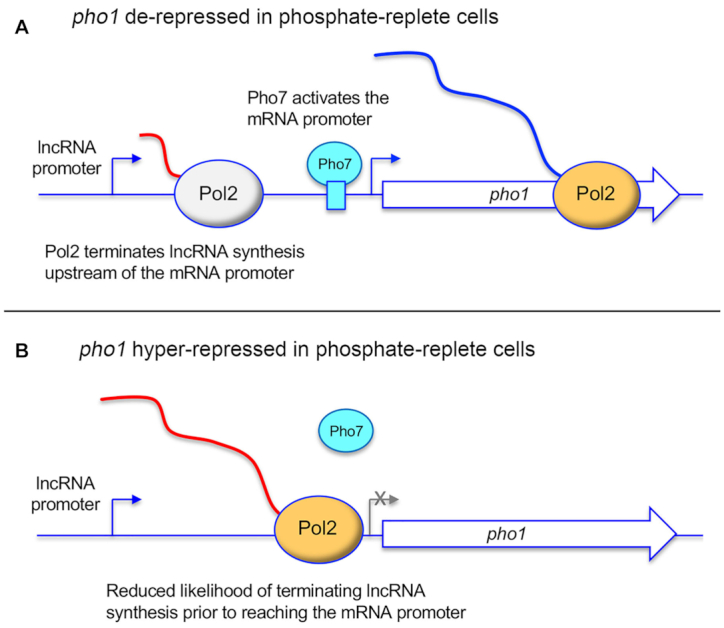
Modulation of PHO lncRNA termination can impact PHO mRNA repression. (A) Precocious termination of lncRNA synthesis upstream of the mRNA promoter can result in de-repression of mRNA transcription. (B) Decreased termination of lncRNA synthesis can increase interference with the mRNA promoter and hyper-repress mRNA expression. See text for discussion.
Appreciation of lncRNA termination as a governor of PHO gene transcription interference emerged in a roundabout way from studies of the effects of mutations in the Pol2 CTD (carboxyl terminal domain) on fission yeast gene expression. The CTD of the Pol2 Rpb1 subunit comprises tandemly repeated heptapeptides of consensus sequence Y1S2P3T4S5P6S7 that serve as a scaffold to recruit proteins that regulate transcription, adjust chromatin structure, and catalyze or regulate mRNA processing. The primary structure of the CTD, which is dynamically sculpted by serine, threonine, and tyrosine phosphorylation, conveys information about the status of the transcription machinery—a CTD code—that is read by CTD-interacting proteins and RNA processing assemblies. Initial genome-wide RNA-seq and ensuing gene-specific analyses showed that pan-substitution of fission yeast CTD residues Ser7 or Ser5 with alanine elicited de-repression of pho1, pho84 and tgp1 expression in phosphate-replete cells, whereas pan-replacement of Thr4 with alanine resulted in pho1 and pho84 hyper-repression (29–31,43). An important finding was that these CTD mutations did not affect the activities of the PHO lncRNA or mRNA promoters per se. Two parallel lines of investigation supported the termination-centric mechanisms depicted in Figure 5.
The first pertains to genetic perturbations of the fission yeast machinery for nascent RNA 3′ processing and Pol2 termination. Cleavage and Polyadenylation Factor (CPF) is a 13-subunit protein assembly responsible for the cotranscriptional 3′ processing of RNA polymerase II (Pol2) transcripts that precedes Pol2 transcription termination (44). Holo-CPF consists of two component complexes: a 10-subunit CPF core composed of proteins Ysh1 (the cleavage endonuclease), Pla1 (the poly(A) polymerase), Pta1, Yth1, Pfs2, Iss1, Cft1, Cft2, Ctf1, and Ssu72 (a phosphoprotein phosphatase that acts on the Pol2 CTD); and a 3-subunit DPS complex comprising Dis2 (a phosphoprotein phosphatase), Ppn1, and Swd22. Eight of the CPF core subunits, including the cleavage endonuclease and poly(A) polymerase, are essential for fission yeast viability. By contrast, two of the core subunits (Ctf1 and Ssu72) and all three DPS complex subunits (Dis2, Ppn1, and Swd22) are dispensable for growth. Rhn1 is a transcription termination factor that binds the phospho-CTD. It was found that de-repression of pho1 by CTD-S7A mutation is erased by deletion or loss-of-function mutations of CPF subunits Ctf1, Ssu72, Ppn1, Swd22 and Dis2 and termination factor Rhn1 (45), consistent with the idea that CTD-S7A elicits precocious lncRNA termination as modeled in Figure 5A. Conversely, it was noted that the CPF/Rhn1 mutations per se resulted in hyper-repression of pho1 in phosphate replete cells, as predicted by the scheme in Figure 5B and akin to the effects of CTD-T4A. The findings that CTD-T4A is synthetically lethal with ppn1Δ and swd22Δ indicate that Thr4 and the Ppn1•Swd22 module of CPF play important, functionally redundant roles in promoting Pol2 termination (45).
The genetic evidence is fortified by analyses of the prt and nc-tgp1 transcripts—entailing 3′-RACE, Northerns, and probe protection—that document the existence of short forms of these lncRNAs that are cleaved and polyadenylated at sites well upstream of the Pho7 binding sites in the pho1 and tgp1 mRNA promoters (Figure 3). The short nc-tgp1 lncRNA is cleaved/polyadenylated at position +504 of the transcription unit, which is 17-nt downstream of a consensus fission yeast polyadenylation signal. A key finding was that mutation of the poly(A) signal for the short nc-tgp1 lncRNA abolished de-repression of the downstream tgp1 mRNA promoter in a CTD-S5A strain background (30). There are two classes of short prt lncRNAs (Figure 3) that are cleaved and polyadenylated at sites +351 and +589 in response to two different upstream poly(A) signals (45). Simultaneous mutations of the short prt poly(A) sites resulted in hyper-repression of the flanking pho1 promoter and reversal of the de-repression of pho1 promoter activity by the CTD-S7A allele (45).
Collectively, these studies establish the prt–pho1 tandem interference system as a sensitive in vivo readout of genetic influences on 3′ processing and termination, one that has shed light on the CTD code and a network of processing factors that interact functionally with the CTD. That said, a key question is the degree to which lncRNA termination contributes to phosphate nutrient sensing, e.g. whether phosphate starvation increases the frequency of precocious PHO lncRNA termination and thereby abets up-regulation of PHO mRNA production. One suggestion that this might be the case is the finding that mutating the poly(A) signal of the short nc-tgp1 lncRNA delays the induction of the tgp1 promoter during phosphate starvation (30).
A very recent study from the Bachand lab approaches this issue from a different angle, by surveying the effects of a N494D mutation in the Rpb1 subunit of fission yeast Pol2 that slows its rate of elongation (46). Genome-wide RNA-seq revealed that the slow Pol2 mutation resulted in up-regulation of the tgp1 mRNA in phosphate-replete cells, which was associated with a shift in nc-tgp1 poly(A) site utilization in favor of the short isoform and a concomitant increase in Pho7 ChIP signal over the tgp1 promoter. The rationale is that slowing Pol2 elongation during lncRNA synthesis expands the kinetic window for 3′ processing at the short lncRNA poly(A) site and precocious termination, which alleviates transcriptional interference at the mRNA promoter. The slow Pol2 mutation also leads to de-repression of pho1 and pho84, albeit to a lesser extent than tgp1. Relevance of poly(A) site choice to phosphate nutrient sensing was suggested by the finding that ChIP signals for 3′ processing factor Rna14 and the termination factor Seb1 increased over the short nc-tgp1 poly(A) site 4 hours after acute phosphate starvation (46).
Metabolite control of phosphate homeostasis by inositol pyrophosphates
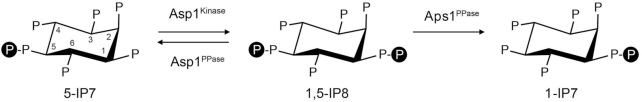
Inositol pyrophosphates (IPPs) IP7 and IP8 are signaling molecules implicated in a broad range of physiological processes in eukaryal cells (47). There are two cellular forms of IP7 that differ according to whether the pyrophosphate moiety is at the 1 or 5 position of the inositol ring; IP8 is pyrophosphorylated at both ring positions (Figure 6). IPPs participate in phosphate homeostasis in Saccharomyces cerevisiae and Arabidopsis thaliana (48). The first clues that IPPs might be involved in fission yeast phosphate homeostasis emerged (once again!) from the Wykoff lab screen and follow-up studies in which a deletion of asp1 (which encodes a kinase that synthesizes IPPs) was found to hyper-repress pho1 under phosphate-replete conditions and a deletion of aps1 (which encodes a Nudix-family IPP pyrophosphatase) de-repressed pho1 under phosphate-replete conditions (25,49). Subsequent studies showed that Asp1 is actually a bifunctional enzyme composed of an N-terminal IPP kinase domain that converts 5-IP7 to 1,5-IP8 and a C-terminal IPP pyrophosphatase domain that converts 1,5-IP8 back to 5-IP7 (50,51) (Figure 6). Asp1 can also phosphorylate IP6 to yield 1-IP7 and de-phosphorylate 1-IP7 back to IP6. The in vivo effect of an asp1Δ null allele or a kinase-dead asp1-D333A allele is to eliminate intracellular IP8 and 1-IP7 and to increase the level of 5-IP7; the in vivo effect of a pyrophosphatase-defective asp1-H397A allele is to increase the level of IP8 (49,50). Aps1 is an IPP pyrophosphatase that converts 1,5-IP8 to 1-IP7 (52) (Figure 6).
Figure 6.
Enzymes of inositol pyrophosphate IP7 and IP8 metabolism in fission yeast. The chemical structures of 5-IP7 (at left, with the positions of the myo-inositol ring numbered), 1,5-IP8 (middle), and 1-IP7 (at right) are shown. Fission yeast Asp1 is a bifunctional enzyme composed of an N-terminal IPP kinase domain that converts 5-IP7 to 1,5-IP8 and a C-terminal IPP pyrophosphatase domain that converts 1,5-IP8 back to 5-IP7. Fission yeast Aps1 is an IPP pyrophosphatase that converts 1,5-IP8 to 1-IP7.
Recent work has revealed that metabolite control of phosphate homeostasis by IPPs is exerted through the 3′-processing/termination machinery and the Pol2 CTD code (53). Increasing 1-IPPs (IP8, and possibly 1-IP7) via an Asp1 IPP pyrophosphatase active site mutation de-repressed the PHO regulon (and led to precocious termination of prt lncRNA synthesis) in a manner dependent on CPF subunits, termination factor Rhn1, and CTD Thr4. Also, pho1 de-repression by CTD-S7A depended on 1-IPP synthesis by the Asp1 kinase. Simultaneous inactivation of the Asp1 and Aps1 IPP pyrophosphatases was lethal, signifying that too much IP8 (or 1-IP7) is toxic, but this lethality was suppressed by mutations of CPF subunits Ppn1, Swd22, Ssu72, and Ctf1 and CTD mutation T4A. Failure to synthesize 1-IPPs via Asp1 kinase mutation resulted in pho1 hyper-repression. Thus, IPP status is an integral part of the termination-centric governance of transcriptional interference depicted in Figure 5. Findings of synthetic lethality of asp1Δ with Ppn1, Swd22 and Ssu72 mutations argues that IP8 (or 1-IP7) plays an important role in essential 3′-processing/termination events, albeit in a manner genetically redundant to CPF. A key issue for future study is the degree to which IPP dynamics and IPP levels in fission yeast are responsive to phosphate nutrient status.
Transcriptional profiling delineated an IPP-responsive regulon composed of genes overexpressed when 1-IPP levels are increased (53). This gene set includes the phosphate acquisition regulon, which is repressed by upstream lncRNA synthesis, as well as other genes that are plausible candidates for similar lncRNA control. The component(s) of the fission yeast Pol2 transcription complex and/or 3′ processing and termination machinery that are targeted by IPPs, and how IPPs exert their effects, are wide open questions at this point. An intriguing possibility is that increased IP8 exerts its effects by slowing the rate of Pol2 elongation.
Concluding remarks
Budding yeast SRG1-SER3 and the fission yeast PHO regulon are bona fide examples of how cells can respond to nutrient status by tuning transcriptional interference at tandem lncRNA and mRNA genes. Interrogation of these systems has enhanced appreciation of gene regulation at the levels of transcription-coupled nucleosome occupancy, transcription factor ejection, and 3′ processing/termination. Many mechanistic details remain to be elucidated, especially how serine and phosphate status are sensed and transmitted to achieve the requisite changes in lncRNA transcription. Other examples of regulated lncRNA–mRNA transcriptional interference are known (9) and it is safe to say that many more await discovery.
FUNDING
National Institute of General Medical Sciences [R35-GM126945]. Funding for open access charge: National Institute of General Medical Sciences [R35-GM126945].
Conflict of interest statement. None declared.
REFERENCES
- 1. Proudfoot N.J. Transcriptional interference and termination between duplicated α-globin gene constructs suggests a novel mechanism for gene regulation. Nature. 1986; 322:562–565. [DOI] [PubMed] [Google Scholar]
- 2. Greger I.H., Proudfoot N.J.. Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 1998; 17:4771–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Greger I.H., Aranda A., Proudfoot N.J.. Balancing transcriptional interference and initiation on the GAL7 promoter of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:8415–8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Colin J., Candelli T., Porrua O., Boulay J., Zhu C., Lacroute F., Steinmetz L.M., Libri D.. Roadblock termination by Reb1p restricts cryptic and readthrough transcription. Mol. Cell. 2014; 56:667–680. [DOI] [PubMed] [Google Scholar]
- 5. Silverman S.J., Fink G.R.. Effects of Ty insertions on HIS4 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 1984; 4:1246–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hirschman J.E., Durbin K.J., Winston F.. Genetic evidence for promoter competition in Saccharomyces cerevisiae. Mol. Cell. Biol. 1988; 8:4608–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gerber A., Ito K., Chu C.S., Roeder R.G.. Gene-specific control of tRNA expression by RNA polymerase II. Mol. Cell. 2020; 78:765–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hao N., Palmer A.C., Dodd I.B., Shearwin K.E.. Directing traffic of DNA… how transcription factors relieve of induce transcriptional interference. Transcription. 2017; 8:120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Werven F.J., Neuert G., Hendrick N., Lardenois A., Buratowski S., van Oudenaarden A., Primig M., Amon A.. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012; 150:1170–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hainer S.J., Pruneski J.A., Mitchell R.D., Monteverde R.M., Martens J.A.. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 2011; 25:29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martens J.A., Laprade L., Winston F.. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004; 429:571–574. [DOI] [PubMed] [Google Scholar]
- 12. Martens J.A., Wu P.J., Winston F.. Regulation of an intergenic transcript controls adjacent gene activation in Saccharomyces cerevisiae. Genes Dev. 2005; 19:2695–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. MacPherson S., Larochelle M., Tyrcotte B.. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 2006; 70:583–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holmberg S., Schjerling P.. Cha4p of Saccharomyces cerevisiae activates transcription via serine/threonine response elements. Genetics. 1996; 144:467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thompson D.M., Parker R.. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007; 27:92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pruneski J.A., Hainer S.J., Petrov K.O., Martens J.A.. The Paf1 complex represses SER3 transcription in Saccharomyces cerevisiae by facilitating intergenic transcription-dependent nucleosome occupancy of the SER3 promoter. Eukaryot. Cell. 2011; 10:1283–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hainer S.J., Charsar B.A., Cohen S.B., Martens J.A.. Identification of mutant versions of the Spt16 histone chaperone that are defective for transcription-coupled nucleosome occupancy in Saccharomyces cerevisiae. G3. 2012; 2:555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thebault P., Boutin G., Bhat W., Rufiange A., Martens J., Nourani A.. Transcription regulation by the noncoding RNA SRG1 requires Spt2-dependent chromatin deposition in the wake of RNA polymerase II. Mol. Cell. Biol. 2011; 31:1288–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hainer S.J., Martens J.A.. Identification of histone mutants that are defective for transcription-coupled nucleosome occupancy. Mol. Cell. Biol. 2011; 31:3557–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hainer S.J., Martens J.A.. Regulation of chaperone binding and nucleosome dynamics by key residues within the globular domain of histone H3. Epigenet. Chromatin. 2016; 9:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jain A., Nagarajan V.K., Raghothama K.G.. Transcriptional regulation of phosphate acquisition by higher plants. Cell. Mol. Life Sci. 2012; 69:3207–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tomar P., Sinha H.. Conservation of PHO pathway in ascomycetes and the role of Pho84. J. Biosci. 2014; 39:525–536. [DOI] [PubMed] [Google Scholar]
- 23. Gardner S.G., McCleary W.R.. Control of the phoBR regulon in Escherichia coli. EcoSal Plus. 2019; 8:10.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carter-O’Connell I., Peel M.T., Wykoff D.D., O’Shea E.K.. Genome-wide characterization of the phosphate starvation response in Schizosaccharomyces pombe. BMC Genomics. 2012; 13:697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Henry T.C., Power J.E., Kerwin C.L., Mohammed A., Weissman J.S., Cameron D.M., Wykoff D.D.. Systematic screen of Schizosaccharomyces pombe deletion collection uncovers parallel evolution of the phosphate signal transduction pathway in yeasts. Eukaryot. Cell. 2011; 10:198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee N.N., Chalamcharia V.R., Reyes-Turce F., Mehta S., Zofall M., Balachandran V., Dhakshnamoorthy J., Taneja N., Yamanaka S., Zhou M., Grewal S.. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell. 2013; 155:1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shah S., Wittmann S., Kilchert C., Vasiljeva L.. lncRNA recruits RNAi and the exosome to dynamically regulate pho1 expression in response to phosphate levels in fission yeast. Genes Dev. 2014; 28:231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ard R., Tong P., Allshire R.C.. Long non-coding RNA-mediate transcriptional interference of a permease gene confers drug tolerance in fission yeast. Nature Comm. 2014; 5:5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chatterjee D., Sanchez A.M., Goldgur Y., Shuman S., Schwer B.. Transcription of lncRNA prt, clustered prt RNA sites for Mmi1 binding, and RNA polymerase II CTD phospho-sites govern the repression of pho1 gene expression under phosphate-replete conditions in fission yeast. RNA. 2016; 22:1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sanchez A.M., Shuman S., Schwer B.. Poly(A) site choice and Pol2 CTD Serine-5 status govern lncRNA control of phosphate-responsive tgp1 gene expression in fission yeast. RNA. 2018; 24:237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garg A., Sanchez A.M., Shuman S., Schwer B.. A long noncoding (lnc) RNA governs expression of the phosphate transporter Pho84 in fission yeast and has cascading effects on the flanking prt lncRNA and pho1 genes. J. Biol. Chem. 2018; 293:4456–4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nabavi S., Nazar R.N.. U3 snoRNA promoter reflects the RNA’s function in ribosome biogenesis. Curr. Genet. 2008; 54:175–184. [DOI] [PubMed] [Google Scholar]
- 33. Witt I., Straub N., Käufer N.F., Gross T.. The CAGTCACA box in the fission yeast Schizosaccharomyces pombe functions like a TATA element and binds a novel factor. EMBO J. 1993; 12:1201–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Witt I., Kwart M., Groβ T., Käufer N.F.. The tandem repeat AGGGTAGGGT is, in the fission yeast, a proximal activation sequence and activates basal transcription mediated by the sequence TGTGACTG. Nucleic Acids Res. 1995; 23:4296–4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gross T., Käufer N.F.. Cytoplasmic ribosomal protein genes of the fission yeast Schizosaccharomyces pombe display a unique promoter type: a suggestion for nomenclature of cytoplasmic ribosomal proteins in databases. Nucleic Acids Res. 1998; 26:33019–33322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rojas D.A., Moreira-Ramos S., Zock-Emmenthal S., Urbina F., Contreras-Levicoy J., Käufer N.F., Maldonado E.. Rrn7 protein, an RNA polymerase I transcription factor, is required for RNA polymerase II-dependent transcription directed by core promoters with a HomolD box sequence. J. Biol. Chem. 2011; 286:26480–26486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Montes M., Moreira-Ramos S., Rojas D.A., Urbina F., Käufer N.F., Maldonado E.. RNA polymerase II components and Rrn7 form a preinitiation complex on the HomolD box to promote ribosomal protein expression in Schizosaccharomyces pombe. FEBS J. 2017; 284:615–633. [DOI] [PubMed] [Google Scholar]
- 38. Moreira-Ramos S., Rojas D.A., Montes M., Urbina F., Miralles V.J., Maldonado E.. Casein kinase 2 inhibits HomolD-directed transcription by Rrn7 in Schizosaccharomyces pombe. FEBS J. 2015; 282:491–503. [DOI] [PubMed] [Google Scholar]
- 39. Schwer B., Sanchez A.M., Garg A., Chatterjee D., Shuman S.. Defining the DNA binding site recognized by the fission yeast Zn2Cys6 transcription factor Pho7 and its role in phosphate homeostasis. mBio. 2017; 8:e01218-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garg A., Goldgur Y., Sanchez A.M., Schwer B., Shuman S.. Structure of fission yeast transcription factor Pho7 bound to pho1 promoter DNA and effect of Pho7 mutations on DNA binding and phosphate homeostasis. Mol. Cell. Biol. 2019; 39:e00132-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Garg A., Goldgur Y., Schwer B., Shuman S.. Distinctive structural basis for DNA recognition by the fission yeast Zn2Cys6 transcription factor Pho7 and its role in phosphate homeostasis. Nucleic Acids Res. 2018; 46:11262–11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ard R., Allshire R.C.. Transcription-coupled changes to chromatin underpin gene silencing by transcriptional interference. Nucleic Acids Res. 2016; 44:10619–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schwer B., Bitton D.A., Sanchez A.M., Bähler J., Shuman S.. Individual letters of the RNA polymerase II CTD code govern distinct gene expression programs in fission yeast. Proc. Natl Acad. Sci. USA. 2014; 111:4185–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vanoosthuyse V., Legros P., van der Sar S.J., Yvert G., Toda K., Le Bihan T., Watanabe Y., Hardwick K., Bernard P.. CPF-associated phosphatase activity opposes condensin-mediated chromosome condensation. PLos Genet. 2014; 10:e1004415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sanchez A.M., Shuman S., Schwer B.. RNA polymerase II CTD interactome with 3′ processing and termination factors in fission yeast and its impact on phosphate homeostasis. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E10652–E10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yague-Sanz C., Vanrobaeys Y., Fernandez R., Duval M., Larochelle M., Beaudoin J., Berro J., Labbe S., Jaques P.E., Bachand F.. Nutrient-dependent control of RNA polymerase II elongation rate regulates specific gene expression programs by alternative polyadenylation. Genes Dev. 2020; 34:883–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee S., Kim M.G., Ahn H., Kim S.. Inositol pyrophosphates: signaling molecules with pleiotropic actions in mammals. Molecules. 2020; 25:2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Azevedo C., Saiardi A.. Eukaryotic phosphate homeostasis: the inositol pyrophosphate perspective. Trends Biochem. Sci. 2017; 42:219–231. [DOI] [PubMed] [Google Scholar]
- 49. Estill M., Kerwin-Iosue C.L., Wykoff D.D.. Dissection of the PHO pathway in Schizosaccharomyces pombe using epistasis and the alternative repressor adenine. Curr. Genet. 2015; 61:175–183. [DOI] [PubMed] [Google Scholar]
- 50. Pascual-Ortiz M., Saiardi A., Walla E., Jakopec V., Künzel N.A., Span I., Vangala A., Fleig U.. Asp1 bifunctional activity modulates spindle function via controlling cellular inositol pyrophosphate levels in Schizosaccharomyces pombe. Mol. Cell. Biol. 2018; 38:e00047-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dollins D.E., Bai W., Fridy P.C., Otto J.C., Neubauer J.L., Gattis S.G., Mehta K.P., York J.D.. Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling. Proc. Natl Acad. Sci. U.S.A. 2020; 117:9356–9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Safrany S.T., Ingram S.W., Cartwright J.L., Falck J.R., McLennan A.G., Barnes L.D., Shears S.B.. The diadenosine hexaphosphate hydrolase from Schizosaccharomyces pombe and Saccharomyces cerevisiae are homologues of the human disphoshoinositol polyphosphate phosphohydrolase: overlapping substrate specificities in a MutT-type protein. J. Biol. Chem. 1999; 274:21735–21740. [DOI] [PubMed] [Google Scholar]
- 53. Sanchez A.M., Garg A., Shuman S., Schwer B.. Inositol pyrophosphates impact phosphate homeostasis via modulation of RNA 3′ processing and transcription termination. Nucleic Acids Res. 2019; 47:8452–8469. [DOI] [PMC free article] [PubMed] [Google Scholar]