Abstract
Genome-wide passive DNA demethylation in cleavage-stage mouse embryos is related to the cytoplasmic localization of the maintenance methyltransferase DNMT1. However, recent studies provided evidences of the nuclear localization of DNMT1 and its contribution to the maintenance of methylation levels of imprinted regions and other genomic loci in early embryos. Using the DNA adenine methylase identification method, we identified Dnmt1-binding regions in four- and eight-cell embryos. The unbiased distribution of Dnmt1 peaks in the genic regions (promoters and CpG islands) as well as the absence of a correlation between the Dnmt1 peaks and the expression levels of the peak-associated genes refutes the active participation of Dnmt1 in the transcriptional regulation of genes in the early developmental period. Instead, Dnmt1 was found to associate with genomic retroelements in a greatly biased fashion, particularly with the LINE1 (long interspersed nuclear elements) and ERVK (endogenous retrovirus type K) sequences. Transcriptomic analysis revealed that the transcripts of the Dnmt1-enriched retroelements were overrepresented in Dnmt1 knockdown embryos. Finally, methyl-CpG-binding domain sequencing proved that the Dnmt1-enriched retroelements, which were densely methylated in wild-type embryos, became demethylated in the Dnmt1-depleted embryos. Our results indicate that Dnmt1 is involved in the repression of retroelements through DNA methylation in early mouse development.
INTRODUCTION
In mammals, the first wave of global epigenetic reprogramming in the life cycle occurs during early embryogenesis and is important for the establishment of pluripotency (1). Initially, DNA methylation in the paternal chromosomes is drastically reduced shortly after fertilization (active demethylation). During cleavage, a progressive mode of demethylation affects both the maternal and paternal chromosomes, and this passive demethylation is thought to be achieved by the nuclear exclusion of DNA methyltransferase 1 (Dnmt1) in preimplantation-stage embryos (2–7).
Dnmt1 is considered as the main maintenance methyltransferase (8) because of its catalytic preference for hemi-methylated DNA (9) and its localization on the replication fork during the S phase of the cell cycle (10,11). There is accumulating evidence for the role of Dnmt1 in maintaining methylation imprints in somatic cells (12) and preimplantation embryos (5,13). Genomic imprinting patterns play pivotal roles in development and growth; their deregulation greatly impacts the embryonic phenotype and may be the major cause of defects in methylation-deficient mutants (1). Thus, the observed cytoplasmic retention of Dnmt1 indicates that the maintenance of genomic imprints, and possibly other genomic repeats (5,14,15), is required during early embryonic development. In contrast to the early observations of the nuclear exclusion of Dnmt1, a somatic isoform of Dnmt1 (Dnmt1s) has been directly proven to be present in the nucleus of early embryos by means of oocyte-form Dnmt1 (Dnmt1o) knockout mice and a specific antibody discriminating the oocyte isoform from the somatic form (16). This finding has also been supported by other studies (5,13). Moreover, the complete knockout of both the Dnmt1o and Dnmt1s resulted in the complete abolishment of all methylation imprints (13,16,17). Therefore, although it is unclear how local methylation patterns are maintained over genome-wide demethylation, Dnmt1 is thought to contribute to maintaining DNA methylation patterns at certain genomic loci in cleavage-stage preimplantation embryos.
Retrotransposable elements, or retroelements, involving long and short interspersed nuclear elements (SINEs and LINEs, respectively) and long terminal repeats (LTRs) are a significant component of mammalian genomes, occupying ∼40% of mouse genomes (Mouse Genome Sequencing Consortium, 2002). The proven capacity for causing deleterious genomic rearrangements and the potent mechanisms for their silencing of neighboring genes in somatic tissues have led many to view retroelements as harmful parasites (18). In somatic cells, the expression of retroelements is attenuated by DNA methylation (19). However, in germ cells and early embryos where epigenetic reprogramming takes place, the resulting genome-wide demethylation lifts this chief repressive mechanism from transcription- and transposition-competent retroelements. In reality, retroelements are highly expressed in mature oocytes and cleaved embryos in mice, and different retroelements have specific, developmentally regulated expression patterns (20). This coordinated activation and repression of the same type of element with multiple genomic sites may provide robustness to the embryo, thereby allowing for temporal control of the newly formed zygotic genome after fertilization (21). For instance, endogenous retrovirus type L (ERVL) retroelements, which are specifically expressed at the two-cell stage (20), have been proposed to regulate a large number of genes during zygotic genome activation by promoting pervasive transcription into neighboring genes (22,23). In early human embryos, MLT2A1 and THE1A ERVs are expressed in an eight-cell stage-specific fashion and LTR5_Hs is expressed in a morula stage-specific fashion (24). In the case of LINE1 retroelements, they show the highest level of expression at the two-cell stage mouse embryo and function primarily at the chromatin level; in addition, LINE1 retroelements promote global chromatin decondensation and recondensation via transcriptional activity (21). Therefore, it appears persuasive that the transcriptional activation of retrotransposons during the preimplantation development stage may not be a nonspecific side effect of heterochromatic loss in the embryo and that the timely activation and repression of retroelement expression may have great significance for early developmental programs and embryo survival. Therefore, it is relevant to determine the regulatory role of Dnmt1 in this programmed development.
DNA adenine methylase identification (DamID) is one of the most comprehensive and versatile methods available for profiling protein–DNA interactions on a genomic scale (25–27). The Escherichia coli Dam protein methylates adenine (at the N6 position) in the GATC sequence; this protein does not exist in mammals. When fused to a chromatin protein of interest, Dam can be targeted in vivo to the binding sequences of the chromatin protein and can methylate the nearby GATC sequences. The adenine-methylated GATC sequences can be digested by the methylation-sensitive DpnI restriction enzyme, and the resulting DNA fragments can be used in microarrays or can be sequenced for mapping the binding sites. To assess chromatin binding on a genome-wide scale, the leading alternative to DamID is chromatin immunoprecipitation, for which the main limitation is the requirement for a highly specific antibody. In contrast, DamID does not require an antibody for the identification of protein-binding loci.
Because Dnmt1 may act on certain genomic loci to allow the loci to fulfill their roles in cleavage-stage mouse embryos, we attempted to identify genomic Dnmt1-binding loci, particularly focusing on a variety of retroelement families. We first verified the nuclear localization of GFP-Dnmt1 recombinant proteins after microinjection into the cytoplasm of a blastomere. Dnmt1-binding regions were identified using DamID and DpnI fragment enrichment by ligation-mediated PCR sequencing (DELP-seq). We found that Dnmt1 displayed a significant bias toward certain retroelement families that were increasingly transcribed when Dnmt1 was knocked down. Dnmt1 depletion in early mouse embryos via CRISPR/Cas9 technology revealed a considerable demethylation at these retroelement sequences. The identification of the Dnmt1-binding loci and the gross effect of Dnmt1 binding on the repression of retroelements via DNA methylation provide insight into the functions and mechanisms of actions of Dnmt1 during early development.
MATERIALS AND METHODS
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Livestock Research Institute of Korea. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Korea Research Institute of Bioscience and Biotechnology.
Construction of Dnmt1 expression vectors
For cloning mouse Dnmt1s or Dnmt1o sequences, PCR was performed with cDNA obtained from HEK293 cells under the condition of 30 cycles of 94°C/30 s, 57°C/30 s and 72°C/3 m (see Supplementary File S1 for primer information). The resulting PCR products, which were devoid of the catalytic domains (Dnmt1sc− or Dnmt1oc−), were cloned into the EcoRI (NEB) and SalI (NEB) sites of pEGFP-C2 (Clontech) to make pEGFP-Dnmt1sc− and pEGFP-Dnmt1oc− expression vectors. Proper cloning was confirmed by sequencing plasmid clones. For construction of pIND-Dnmt1-V5-Dam, 3.4 kb of Dnmt1 fragment lacking the catalytic domain was amplified from mouse cDNA by PCR (see Supplementary File S1 for primer information) and cloned into pIND-V5-Dam plasmid (Addgene).
Microinjection and in vitro culture of mouse embryos
Mouse zygotes were collected from superovulated BCF1 (C57BL/6 × CBA/CA) females as described previously (28). Briefly, female BCF1 mice at 5 weeks of age were injected with 5 IU of pregnant mare serum gonadotrophin (PMSF), followed by 5 IU of human chorionic gonadotropins (hCG) 48 h apart, and mated with male mice. Successful mating was determined the following morning by detection of a vaginal plug. Mouse zygotes were transferred to M2 medium (Sigma) containing 0.1% (w/v) hyaluronidase to remove cumulus cells and cultured in M16 medium (Sigma) at 37°C, 5% CO2 in air (29).
Microinjection was performed 24 h after human chorionic gonadotropin injection (30). To visualize the pronuclei, cumulus-free zygotes were centrifuged at 13,000 rpm for 5 min. Linearized GFP-Dnmt1sc− or GFP-Dnmt1oc− expression construct (4–20 ng/μl) was microinjected into the male pronuclei of mouse zygotes using an inverted microscope equipped with a micromanipulator (Leica). The injected zygotes were then cultured in M16 media for 48 h. The resulting four- or eight-cell stage embryos were collected, fixed using 4% formaldehyde and mounted with DAPI (Vector). The localization of Dnmt1 isoforms was evaluated by detecting GFP signals under a fluorescence microscope equipped with a microtome (Zeiss).
Production of recombinant GFP-Dnmt1 proteins and microinjection into mouse two-cell embryos
To produce recombinant GFP-Dnmt1 proteins, PCR was performed to obtain the GFP-Dnmt1sc− part using the pEGFP-GFP-Dnmt1sc− plasmid as a template under the condition of 20 cycles of 94°C/30 s, 46°C/45 s and 72°C/2.5 m (MJ gradient cycler). The primers used were 5′-GCGATATCATGGTGAGCAAGG-3′ and 5′-GCGATATCCTAGATCCGGTG-3′. The PCR product was cloned into EcoRV (NEB) site of pET32 vector (Novagen) to make pET-32a(+)-GFP-Dnmt1sc−. GFP-Dnmt1sc− recombinant protein was produced after transformation into E. coli BL21 (DE3) strains and induction using 0.25 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma-Aldrich) for 2 or 4 h. The recombinant protein was purified using Ni2+-nitrilotriacetic acid column (Qiagen) and concentrated by Centricon (Amicon) according to the manufacturer’s instructions. For microinjection, the GFP-Dnmt1sc− recombinant protein at ∼1 μg/μl concentration was injected into the cytoplasm of a single blastomere of two-cell embryo. Three hours later, GFP was observed on the fluorescence microscope as mentioned earlier.
Transfection, western blotting and immunostaining
For western blotting, 293T cells were seeded on a gelatin-coated coverslip a day before transfection. When cells were on a 60% confluency, pIND-Dnmt1-Dam or pIND-Dam vector was co-transfected with pVgRXR into 293T cells using Lipofectamine (Thermo) overnight. Cells were then briefly washed with phosphate-buffered saline (PBS) and cultured before split. Dnmt1-V5-Dam or V5-Dam expression was induced by culturing the cells in the medium supplemented with 5 μM of Ponasterone A (Thermo). Twenty-four hours later, cells were harvested for western blotting. Ponasterone A-treated or untreated 293T cells were lysed with 1× Protease Inhibitor Cocktail (GenDEPOT) and 1 mM PMSF for 30 min on ice and then briefly sonicated. After centrifugation at 4°C for 30 m, the supernatant was transferred to a new 1.5-ml tube. Twenty micrograms of sample protein was run on a 10% SDS-PAGE gel and then transferred to a nitrocellulose membrane (GE). After blocking, anti-V5 antibody (1:3,000) was incubated together with the membrane overnight at 4°C. The membrane was washed with a washing buffer (5% Tween 20 in 1× PBS, PBST), and reacted with a secondary HRP-conjugated anti-mouse antibody (1:6,000, Santa Cruz) at room temperature. After 2 h of wash, the membrane was dropped with ECL solution (GE) and observed on an LAS-3000 imager (Fujifilm).
For immunostaining, transfected 293T cells were fixed with formalin (Sigma) for 20 m at 4°C. After a brief wash using PBST (0.02% Tween 20 in PBS), cells were permeabilized with 2% Triton X-100 (MP Bio) in PBST for 30 m at room temperature. Blocking buffer (2% bovine serum albumin in PBST) was added to each well for 30 m at room temperature. The cells were incubated overnight with mouse anti-V5 antibody (1:300, Invitrogen) at 4°C. Cells were washed and then reacted with Alexa-594-conjugated anti-mouse secondary antibodies (1:300, Santa Cruz). After several washing, coverslips were briefly air-dried. Mounting solution with DAPI (Vector) was dropped on slide glasses, and the cover glasses were mounted on them. The cells were analyzed with an Axiovert 200 fluorescent microscope (Zeiss).
DELP sequencing
A total of 4 ng of either linearized pVgRXR and pIND-Dnmt1-V5-Dam transgenes or pVgRXR and pIND-V5-Dam was injected into the pronucleus of mouse zygote (30). The resulting zygotes were cultured at 37°C in a 5% CO2 incubator. Twenty-four hours later, the embryos were treated with 5 μM Ponasterone A for another 24 h. The resulting four- or eight-cell embryos were harvested and subjected to DELP-seq. Genomic DNAs were isolated from the embryos by incubating them in an embryo lysis buffer (10 mM Tris–HCl, pH 8.0, 0.1 M EDTA, 0.5% SDS, 20 mg/ml RNAse A and 0.4 mg/ml proteinase K) for 3 h at 56°C (31) and serially digested by DpnI (NEB) and FatI (NEB). The digested DNA fragments were ligated with 2.5 μM adapters specific to DpnI- or FatI-digested DNA ends (see Supplementary File S1 for adapter information) and used as a template for suppression PCR. Suppression PCR was performed using a cycling program of 18 cycles of 95°C/15 s, 45°C/2 m and 74°C/1 m (see Supplementary File S1 for primer information). Using 1 μl of suppression PCR products as templates, nested PCR was performed in a PCR program of 15 cycles of 95°C/10 s, 55°C/20 s and 72°C/1 m (see Supplementary File S1 for primer information). Each PCR was triplicated and pooled. For next-generation sequencing, adapters were removed and DNA fragments were end repaired and ligated with Illumina adapters. The NGS libraries were sequenced using HiSeq2000 (Illumina).
Dnmt1 knockdown using siRNAs
For microinjection of siRNAs, 50 μM of Dnmt1 or scrambled control siRNAs (see Supplementary File S1 for siRNA sequences) were injected into the cytoplasm of the zygote, and the embryos were cultured in M16 media until the four/eight-cell stages. The embryos at the desired stages were harvested and kept at −80°C for transcriptomic analysis. Knockdown efficiency was estimated by reverse transcriptase PCR. Pico-profiling products were diluted one hundredth, and 1 μl of the diluted product was used as a template. PCR was performed using AccuPower PCR Premix (Bioneer) along with Dnmt1 or Gapdh primers (see Supplementary File S1 for primer information) in the following PCR setting of 25 cycles of 97°C/30 s, 55°C/1 m and 72°C/1 m.
Generation of Dnmt1-depleted mouse embryos by CRISPR/Cas9 method and genotyping
A pair of single-guide RNAs (sgRNAs) was designed to target exon 32 of Dnmt1 gene and was synthesized (see Supplementary File S1 for sgRNA information; Macrogen, South Korea). Both sgRNAs (40 ng each) and Cas9 recombinant protein (40 ng; Toolgen, South Korea) were mixed and incubated in a microinjection buffer (0.1 mM EDTA, 10 mM Tris–HCl, pH 7.4) for 15 m at 37°C (1). Microinjection was performed 24 h after human chorionic gonadotropin injection. To visualize the pronuclei, cumulus-free zygotes were centrifuged at 13,000 rpm for 5 m, injected with the cocktail of sgRNA and Cas9, and cultured in M16 media for 48 h. Microinjection was performed under an inverted microscope equipped with a micromanipulator and a microinjector (Leica). The resulting four- and eight-cell stage embryos were treated with Tyrode’s solution (Sigma) for 2 min to remove zona pellucida, washed three times with 0.1% PVA–PBS and collected for either genotyping or MBD sequencing.
With a subset of the Dnmt1 CRISPR embryos, gene-editing efficiency was assessed. Individual embryos were placed in separate PCR tubes, and incubated in an embryo lysis buffer (20 mM Tris–HCl, pH 8.0, 0.9% Tween 20, 0.9% NP-40 and 2 mg/ml proteinase K) at 50°C for 30 m. For primary PCR, whole lysates were added to AccuPower PCR Premix (Bioneer, South Korea) with genotyping primers (Supplementary File S1), and subjected to PCR in 25 cycles of 95°C/30 s, 55°C/1 m and 72°C/30 s. Nested PCR was carried out with 0.1 μl of the primary PCR product as a template and internal primers in 35 cycles of 95°C/30 s, 55°C/1 m and 72°C/30 s. The products were visualized on agarose gels and sent for Sanger sequencing to detect any genomic alteration.
Pico-profiling and RNA sequencing library construction
Due to the limited starting amount of materials, we adopted the pico-profiling method as described in previous reports to enrich the transcriptomic materials (32). First, mRNAs were isolated from a pool of siDnmt1 or si-Control injected four/eight-cell embryos using Dynabeads mRNA DIRECT Kit (Invitrogen). Using the extracted mRNAs as templates, pico-profiling-specific adapter-tagged ds-cDNA fragments were generated by reacting mRNA and pico-profiling primers with SuperScript III (Invitrogen) followed by T4 DNA polymerase (NEB). Then, the adapter-tagged ds-cDNAs were amplified by 20 cycles of PCR using the anchor primers for the tagged adapters. The amplicons were digested by MlyI (NEB) to remove the pico-profiling adapters, and then the products were purified using AMPure XP beads (Beckman). Next, RNA-seq libraries were generated using the pico-profiling products according to the protocol suggested by NEBNext Ultra DNA Library Prep Kit for Illumina (NEB) and home-brew method previously reported (32). If not indicated otherwise, all materials were purchased from NEB. Two hundred nanograms of pico-profiling products were treated with T4 DNA polymerase, T4 polynucleotide kinase and Klenow enzymes to repair the ends of amplified DNA fragments. The reaction products were purified using AMPure XP beads (Beckman Coulter) according to the manufacturer’s instructions. To adenylate 3′-ends of the end-repaired products, DNAs were reacted with Klenow fragment and 10 mM dATP for 2 h at 30°C. After purification with AMPure XP beads, NEBNext adapters with unique barcodes were ligated overnight at 16°C in the presence of diluted T4 DNA ligase. The next day, ligation mixtures were purified twice with AMPure XP beads and were enriched by PCR. Paired-end sequencing was performed using NextSeq 550 (Illumina) to generate raw sequencing data.
MBD sequencing and ChIP sequencing
Embryo sample (∼300 blastomeres per sample group) was transferred to a Bioruptor microtube (Diagenode) with 20 μl lysis buffer (10 mM Tris–HCl, pH 8.0, 100 mM EDTA, pH 8.0, 0.5% SDS and 0.4 mg/ml proteinase K), and incubated sequentially at 56°C for 30 m and at 70°C for 15 m. The lysed chromatins were then sheared by a Bioruptor Pico sonicator (Diagenode). After 30 cycles of 30 s on and 90 s off shearing program, the sheared sample was purified using AMPure XP beads (Beckman) and then subject to MBD-seq library production using MethylMiner Methylated DNA Enrichment Kit (Thermo) according to the manufacturer’s protocol. One microliter of streptavidin beads was precleared and incubated with 1 μl MBD-biotin proteins for 1 h at room temperature. After wash, the MBD–bead complexes were incubated with sheared DNA fragments and methyl/non-methyl spike-in DNAs overnight at 4°C. MBD-seq sensitivity and efficiency for each group were confirmed by estimating the relative amount of spike-in DNAs in elutes and flow-through. Illumina sequencing libraries were constructed using the captured DNA fragments as previously described (33), and sequencing data were generated by NextSeq 550 (Illumina).
For ChIP (chromatin immunoprecipitation) sequencing, 293T cells were plated on a six-well dish and, when they reached at 70–80% confluency, 2 μg of pEGFP-Dnmt1s, pEGFP-Dnmt1sc− or pEGFP-c2 (Clontech) plasmid was transfected into the cells using Lipofectamine LTX (Thermo) according to the manufacturer’s protocol. Cells were harvested 72 h after transfection and 1 × 106 cells were used in ChIP as described previously (34). Briefly, for cross-linking of chromatin and proteins, cells were incubated in 1% formaldehyde solution for 10 m at room temperature, neutralized by glycine treatment and lysed to extract chromatin. The resulting chromatin was sheared to the size ranging from 200 to 500 bp, reacted with 2 μg α-GFP antibodies (Santa Cruz) or IgG control (Cell Signaling), and captured by Protein A magnetic beads (Bio-Rad). After reverse cross-linking and purification, the eluted DNA fragments were proceeded for Illumina sequencing library generation using NEBNext DNA Library Prep modules (NEB) and NEBNext Multiplex Oligos (NEB) according to the manufacturer’s protocol, and paired-end sequencing data were generated by NextSeq 550 (Illumina).
Bioinformatics analyses
Raw Illumina sequencing reads for all experiments were pre-processed to remove low-quality bases and adapter sequences using ‘TrimGalore’ (https://www.bioinformatics.babraham.ac.uk), and aligned on the reference genome using BWA (4) for DELP-seq (mm9), MBD-seq (mm9) and ChIP-seq (hg38), while HISAT2 (mm9) (5) was used for mapping on the transcriptome.
Since we performed the suppression PCR for DELP-seq to obtain the fragments concurrently digested by DpnI/FatI, we only considered the DpnI/FatI-bound genomic loci for peak detection. Peaks were defined as DpnI/FatI loci with sizes of 100–500 bp (3,495,243 loci in mm9), and fold change >30 between DnmtDam and Dam against each other. For peak detection, we first digested the reference genome (mm9) with DpnI and FatI restriction enzymes in silico, and then the fragments with sizes between 100 and 500 bp were extracted and saved as a bed file. Next, we counted the mapped reads of DnmtDam or Dam embryos in the DpnI/FatI-bound loci using ‘bedtools intersect’, and the read enrichments were calculated by dividing the number of reads in DnmtDam by that of Dam in each locus. Finally, a DpnI–FatI locus with >30-fold enrichment in DnmtDam against Dam control was defined, for convenience, as a DnmtDam ‘peak’, and vice versa for a Dam control ‘peak’. Genes (NCBI RefSeq) and retroelements (UCSC RepeatMasker) around peaks were annotated using ‘bedtools closest’ (https://bedtools.readthedocs.io/) to summarize the genomic distributions of peaks. To assess the read coverages over genes, CpG islands (CGIs), and retroelements using ‘deepTools’ (https://deeptools.readthedocs.io/), bed files containing peak intensities were converted to bigWig files using ‘bedGraphToBigWig’ (https://github.com/ENCODE-DCC/kentUtils) in order to input them into ‘computeMatrix’. For retroelements and genes, 3 kb flanking regions from start and end positions were included in the computation, while CGIs were extended ±5 kb. Using the matrix files, heat maps and profile plots were generated using the following tools: ‘plotHeatmap’ and ‘plotProfile’.
For analysis of RNA-seq data from siDnmt1 embryos, gene expression levels were calculated and normalized by HTSeq-DESeq2 pipeline (6,7). For quantification retroelement expression, ‘bedtools intersect’ was used to count only the uniquely mapped reads on specified retroelements (LINE1s >5 kb, ERVs >1 kb). To examine the association of Dnmt1 in coding gene expression, we obtained two publicly available transcriptome data, GSE18290 (35) and GSE66582 (36), and identified the genes possessing DnmtDam or Dam peaks at promoters (TSS ±3 kb). Then, expression levels of the selected genes were profiled based on the number of peaks and presented as box plots.
For MBD-seq data analysis, genome was binned into 500-bp windows, and number of reads in each bin was counted and normalized by total read counts using ‘bedtools intersect’ and custom bash scripts. Additionally, we corrected the data for the MBD captured library sizes to preserve the original DNA methylation states in samples. Using the normalized data, epigenetic alteration on specified retroelements (LINE1s >5 kb, ERVs >1 kb) was analyzed between wild-type (WT) and Dnmt1-depleted embryos by ‘bedtools intersect’. Using ‘deepTools plotProfile’, coverages of MBD-seq reads on coding genes were calculated. The normalized DNA methylation data of each group was converted to a bedGraph file to visualize them on UCSC Genome Browser.
The ChIP-seq data were quantified and normalized using HOMER (Hypergeometric Optimization of Motif EnRichment) (8). Relative coverage depths in 500-bp genomic bins over the GFP control were calculated to identify the full-length or truncated Dnmt1-enriched loci, and the number of overlapping bins between full-length or truncated Dnmt1 was counted to assess their resemblance in chromatin association. A bedGraph file for each sample and control was generated by HOMER.
Generation of all scatter/violin/box plots and statistical tests were executed in the R environment.
RESULTS
Nuclear occupancy of exogenous Dnmt1 in early mouse embryos
Dnmt1s and Dnmt1o are somatic and oocyte-specific forms of Dnmt1, respectively. We removed the C-terminal catalytic domain of Dnmt1s/o to yield truncated forms of GFP-Dnmt1sc− and GFP-Dnmt1oc−. These truncated forms possessed the N-terminal multidomain region that takes part in diverse regulatory functions (37) (Figure 1A). The exclusion of catalytic activity aimed to preclude potential harm due to overmethylation and to prevent other side effects that can interfere with the identification of the Dnmt1-binding regions. To prove a similarity in DNA-binding regions between the full-length GFP-Dnmt1s and the truncated GFP-Dnmt1sc−, we performed ChIP analysis using α-GFP antibody and found that the genome-wide binding profile of the GFP-Dnmt1sc− was very similar to that of the GFP-Dnmt1s in cultured cells (Supplementary Figure S1).
Figure 1.
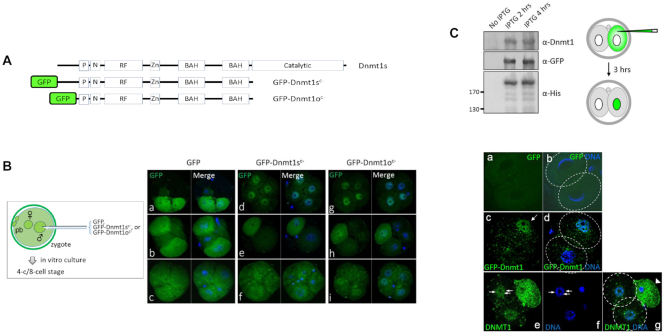
Nuclear localization of Dnmt1 in cleavage-stage mouse embryos. (A) Structure of truncated GFP-Dnmt1sc− and GFP-Dnmt1oc− proteins. The carboxyl-terminal catalytic domain was removed (c−), and GFP was fused to the cDNA amino terminus of the human somatic variant of Dnmt1 (Dnmt1s) and to that of the oocyte-specific variant Dnmt1 (Dnmt1o). P, PCNA binding domain; N, nuclear localization signal; RF, replication foci targeting sequence; Zn, zinc finger-like (CXXC) motif; BAH, bromo-adjacent homology. (B) Variable expression of GFP-Dnmt1oc− and GFP-Dnmt1sc− among the different blastomeres of single embryos and between different embryos. As shown in the box (left), the transgene was microinjected into the male (♂) pronucleus of the mouse zygote; 2 days later, the cleaved embryos were examined for GFP expression (green) with DAPI (blue). GFP transgene alone injected embryos were used as a control. ♀, female pronucleus; pb, polar body. (C) Nuclear occupancy of recombinant Dnmt1 proteins. A 175 kDa His-tagged GFP-Dnmt1sc− recombinant protein produced from E. coli after IPTG induction (left) was introduced into a single blastomere of each two-cell mouse embryo (middle), and 3 h later, the injected embryos were observed for GFP expression (right). The boundaries of the single blastomeres are marked with dotted circles (b, d and g). In (c) and (d), only the Dnmt1-injected blastomere (arrow in c) showed a GFP-positive nucleus, and the noninjected blastomere of the two-cell embryo did not exhibit GFP. Panels (e)–(g) show a four-cell embryo (grown from a Dnmt1-injected two-cell embryo), in which a degenerated blastomere of the two-cell stage (right) and two blastomeres of the four-cell stage carrying nuclear GFP signals exist. In (e) and (f), the GFP-dense dots (arrows) in the nucleus overlap with DAPI-dense heterochromatin regions.
Both GFP-Dnmt1sc− and GFP-Dnmt1oc− showed a high level of similarity with the WT Dnmt1 in the cell cycle-dependent pattern of localization in the somatic cell nucleus (Supplementary Figure S2), agreeing with previous reports (10,38,39). To compare the locations of GFP-Dnmt1sc− and GFP-Dnmt1oc− proteins in mouse early embryos, we microinjected the expression plasmid into the zygote. The inspection of the resultant four- and eight-cell embryos revealed various GFP patterns that were not different between the GFP-Dnmt1sc− and GFP-Dnmt1oc− embryos (Figure 1B and Supplementary Figure S3). In most embryos, GFP signals were dispersed throughout the whole blastomere, including in the nucleus, while 25–30% of the embryos were marked by the nucleus-dense GFP signal (Figure 1B, d and g; also see the table in Supplementary Figure S3). The random incongruous localization of the GFP signal between the cytoplasm and the nucleus among embryos supports the temporary, cyclic localization of Dnmt1 to the nucleus in early embryos. The nuclear occupancy of exogenous Dnmt1 partially relied on the concentration of the injected vectors in that the fraction of nuclear GFP-positive embryos tended to increase with the concentration of microinjected DNA (data not shown), which agrees with the result of a previous study (3). To exclude the possibility that the presence of nuclear GFP-Dnmt1 results from passive entrapment within the nucleus during nuclear envelope reassembly, we produced a 175-kDa His-tagged GFP-Dnmt1sc− recombinant protein and injected it into one of the blastomeres in a two-cell embryo, as illustrated in Figure 1C. When observed 3 h later, GFP-Dnmt1sc− proteins, injected into the cytoplasm, were detected in the nuclei in 50% of the microinjected embryos (Supplementary Figure S3). It demonstrated the active movement of GFP-Dnmt1sc− from the cytoplasm into the nucleus without cell cycle progression (Figure 1C, c and d). The noninjected blastomere of the two-cell embryos did not exhibit a GFP signal in the nuclei. Some GFP-Dnmt1-intense spots overlapped with DAPI-dense regions (Figure 1C, e and f), indicating that Dnmt1 is associated with specific chromosomal domains in the nucleus. In summary, these results suggest that the absence of a catalytic domain does not affect Dnmt1 localization in either the embryonic cells or the somatic cells and that the GFP-Dnmt1sc− and GFP-Dnmt1oc− proteins are comparable for the behaviors in the nucleus. Based on these results and the recent observations that this Dnmt1s somatic variant was present in the nucleus of early embryos (16,17), we decided to use the Dnmt1sc− form in subsequent experiments.
Identification of Dnmt1-binding regions using DamID and DELP sequencing
The observation of ectopic Dnmt1 in the nucleus of early embryos prompted us to map the Dnmt1-binding regions. For this, we carried out a DamID experiment, which was used to identify the genomic targets of DNA-binding proteins (27). We constructed a vector expressing V5-Dam fused Dnmt1sc− (DnmtDam; Figure 2A). The proper expression of the DnmtDam protein was observed in 293T cells (Supplementary Figure S4A). The expression mechanism of the inducible DamID system involving retinoic acid X receptor and VP16 transactivator is shown in Supplementary Figure S4B. The proper induction of DnmtDam by Ponasterone A treatment was confirmed by immunostaining (Supplementary Figure S4C) and western blotting (Supplementary Figure S4D). The Dnmt1 moiety guides the fusion proteins to the binding regions, and the Dam moiety, which does not exist in eukaryotes, subsequently methylates the surrounding adenines in the 5′-GATC-3′ sequence. The DpnI restriction enzyme specifically recognizes and cuts the Dam-methylated sequence (Supplementary Figure S4E).
Figure 2.
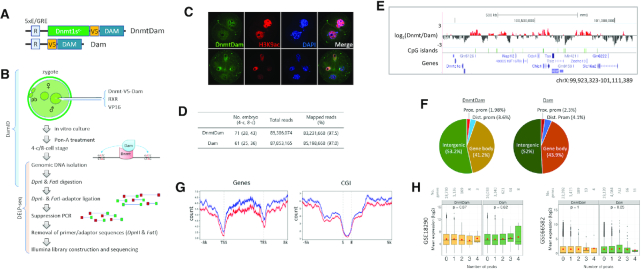
Identification and characterization of the Dnmt1-binding regions in early mouse embryos by DamID and DELP sequencing. (A) Structure of Dnmt1–Dam (DnmtDam) fusion protein. V5 is an epitope for immunostaining. Refer to Supplementary Figure S4 for the regulatory region (R). (B) DamID and DELP-seq (DpnI fragment enrichment by ligation-mediated PCR sequencing) procedures. Mouse zygotes were injected with the DamID toolkit and were cultured until the four- and eight-cell stages. The embryos were treated with Ponasterone A for 24 h to induce the expression of DnmtDam for the methylation of nearby DpnI sites (5′-GATC-3′). Genomic DNA from the four/eight-cell embryos was subjected to DELP-seq. (C) Nuclear localization of DnmtDam in mouse embryos. Two- and four-cell nuclei were stained with the anti-V5 antibody (DnmtDam, green). The nuclei were stained for histone H3-lysine 9 acetylation (H3K9ac, red) and were counterstained with DAPI (blue). (D) Collection of DamID embryo samples and DELP-seq statistics. The table shows the number of cleavage embryos used for DamID/DELP-seq and the number of sequencing reads and mapped reads. The mapping efficiencies are shown in parentheses. (E) A genome browser snapshot for DELP-seq results (Xist locus). The relative enrichment of DnmtDam over Dam is depicted as red bars, while Dam-enriched loci are colored in gray. CGIs (green) and genes (blue) in this region are also displayed. The genomic coordinates are indicated below. (F) Genomic distributions of DnmtDam or Dam peaks. The proportion of DnmtDam or Dam peaks over genomic features, including proximal (from TSS/−3 kb to TSS) and distal promoters (from TSS/−10 kb to TSS/3 kb), gene bodies and intergenic regions. TSS and TES refer to transcription start and end sites, respectively. (G) Read coverages of DnmtDam (red) and Dam (blue) over genes and CGIs. The genes and CGIs of various lengths were scaled to fit into a fixed width surrounded by TSS/TES or S/E (start/end), respectively. For comprehensive analysis, the analytic regions were extended 5 kb from both the TSS and TES for genes and the S and E for CGIs. (H) Transcriptional profiles of genes having DnmtDam or Dam peaks at promoters. Consulting two publicly available transcriptomic datasets (GSE18920 and GSE66582), the expression of genes with an increasing occurrence of DnmtDam or Dam peaks at the promoter regions (TSS/±3 kb) was profiled. The x-axes indicate the number of corresponding peaks at the promoters and the y-axes represent the expression levels of the associated genes. The number of genes with a specified number of peaks is denoted on each box plot. For statistical analysis, one-way ANOVA tests were performed.
We microinjected the DamID expression toolkit into mouse zygotes and cultured them in vitro until the four/eight-cell stage (Figure 2B). The immunostaining of DamID-injected embryos using an anti-V5 antibody revealed a weak DnmtDam signal in the nuclei (Figure 2C). We used 71 DnmtDam and 61 Dam control embryos in library construction and sequencing (Figure 2D). Genomic DNA was isolated from the embryos and was subjected to DELP-seq, which began with the double digestion of genomic DNA with DpnI and FatI (5′-C/ATG-3′). There are ∼2-fold more FatI sites (∼2.5 per kb) than the number of DpnI sites (∼1.1) in the mouse genome, and a simulation with the reference genome (mm9) in the UCSC Genome Browser identified a 10-fold or greater enrichment of optimally sized DNA fragments (<500 bp in length) in the double digestion than those identified in the DpnI digestion alone (Supplementary Figure S5). The DNA fragments with DpnI-generated blunt ends and FatI-generated sticky ends were ligated with DpnI- and FatI-specific adapters, respectively, and were amplified via suppression PCR (40,41) to enrich those carrying different adapters at both ends before constructing the sequencing libraries (Figure 2B). Sequencing produced ∼86 million reads each from Dam and DnmtDam samples, and the reads were mapped at a 97% efficiency on mm9 (Figure 2D and also see Supplementary File S1). For instance, Figure 2E shows the read coverages around the Xist X chromosome locus on the UCSC Genome Browser. However, we confess that there is a possibility that this Dnmt1-binding profile may not fully recapitulate that of full-length Dnmt1 as it was obtained from the truncated Dnmt1sc−, even though the high similarity in the relative genomic coverage was shown between the full-length and the truncated Dnmt1 (Supplementary Figure S1).
No correlation between DnmtDam peaks at promoters and the regulation of gene expression
For ‘peak’ identification, we considered only those reads mapped to the DpnI- and FatI-bound genomic regions of length ranging from 100 to 500 bp in length and equivalent to the range of the sequencing library sizes in this study. In DELP-seq, differing from typical ChIP-seq, no conventional peak calling algorithms are applicable since genomic regions to be analyzed are predetermined by DpnI–FatI restriction enzyme sites. For convenience, we defined a DpnI–FatI-bound locus where DnmtDam or Dam reads were predominantly enriched against each other (fold change >30) as a DnmtDam ‘peak’ or a Dam ‘peak’. We detected the Dam peaks for a nonspecific control. Of these 3.5 million DpnI–FatI loci in total, 41,975 and 80,806 were identified as the DnmtDam- and Dam-specific peaks, respectively, when the peak criterion of >30-fold enrichment of reads against each other was arbitrarily applied. Both the DnmtDam and Dam peaks were mainly found in intergenic regions (53.2% and 52%, respectively) and in gene bodies (41.2% and 43.9%, respectively), and they were only marginally found in proximal gene promoters (1.98% and 2.3%, respectively; Figure 2F). We evaluated the coverages of the DnmtDam and Dam reads over the genic regions and CGIs and failed to see any preferential occupancy of DnmtDam reads over Dam reads in these regions (Figure 2G).
We next tested whether there was any correlation between the number of DnmtDam peaks at promoters and the expression levels of the corresponding genes harboring the peaks. We listed the genes with DnmtDam peaks at promoters and grouped them by the peak numbers (0–4) before comparing the gene expression levels among the groups using early embryo RNA-seq datasets, GSE18290 (35) and GSE66582 (36). As shown in Figure 2H, there were no noticeable differences in the mean expression levels among the peak groups in both datasets between the DnmtDam and Dam control groups, showing no relation of the promoter occupancy of Dnmt1 to the regulation of its associated gene. Therefore, this result refutes that Dnmt1 generally localizes at gene promoters and CGIs to fulfill transcriptional regulation in mouse cleavage-stage embryos.
Preferential binding of DnmtDam to LINE1 and ERVK retroelements
The occurrence of the genome-wide events of epigenetic reprogramming during early development in mammals leads to the derepression of a variety of genomic retroelements and leads to genomic instability; in addition, the way that early embryos survive this instability is to quickly re-establish a set of defensive epigenetic mechanisms, including DNA methylation (21,42–44). We investigated whether Dnmt1 was involved in the regulation of retroelements in early mouse embryos. When the read coverages over various types of retroelements were plotted, the DnmtDam signal was denser in the LINE1 subfamilies (>5.0 kb in length; Figure 3A) and in IAPEz copies (>1.0 kb), the largest subfamily group in the ERVK (endogenous retrovirus type K) family (Figure 3B; for genomic copy numbers in individual subfamilies, see Supplementary Figure S6). In contrast, there was no DnmtDam signal enrichment in the ERV1 and ERVL families (>1.0 kb; Figure 3C). Based on this binding propensity of DnmtDam to certain retroelements, we attempted to make a list of the types of retroelements that DnmtDam binds by computing the read coverages of the whole genomic copies of individual retroelement subfamilies. Overall, the mean counts of DnmtDam reads over the LINE1 subfamilies were generally high and were significantly different (P = 0.016; Wilcoxon’s test) compared with those of the Dam reads (Figure 3D). However, there was no such difference (P > 0.38) among the other retroelement groups. We selected 10 subfamilies from each of the retroelement families based on their copy abundance in the mouse genome. The enrichment of DnmtDam reads was shown in all the selected LINE1 subfamilies at the P < 0.0001 level of significance (except for Lx, P < 0.01). Certain ERVK subfamilies, such as IAPEz, MMETn and RLTR10, were also shown to be predisposed by DnmtDam (P < 0.01). This result indicates that Dnmt1 binds to specific kinds of retroelements, such as LINE1 and ERVK sequences, in early mouse embryos.
Figure 3.
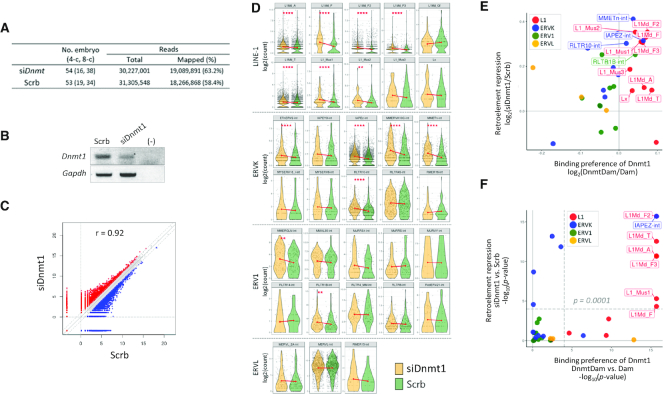
Preferential binding of DnmtDam to LINE1 and ERVK retroelements in mouse embryos. (A) Enrichment of DnmtDam signals over LINE1 subfamilies. DnmtDam and Dam coverages over L1Md_T and L1_Mus1 (upper) and flanking regions (±3 kb); k-means (k = 2) clustered the DnmtDam reads over the indicated LINE1 subfamilies (lower). The elements of various lengths are scaled to fit into the fixed width surrounded by S (start) and E (end). (B) DamID signals over ERVK elements. The read coverages over IAPEz-int (upper) and the rest of the ERVK subfamilies (lower). (C) DamID signals over the ERV1 and ERVL elements. (D) Comparison of DnmtDam and Dam enrichment over the subfamilies of retroelements. Box and violin plots showing the distributions of the DnmtDam (pink) or Dam (blue) enrichment levels of individual retroelement subfamilies (black dots). Red diamonds indicate the trimmed means, and the corresponding subfamilies are linked by gray lines across DnmtDam and Dam charts for comparison. For statistical analysis, Wilcoxon signed-rank tests were performed. (E) DamID enrichment signals in each subfamily. In each of the LINE1, ERVK, ERV1 and ERVL retroelement families, the subfamilies of the top 10 highest copy numbers were selected to compare the DnmtDam (pink) and Dam (blue) enrichment over the individual genomic copies (dots). In each facet, the red dots in the middle of the violin plots indicate the trimmed means of enrichment levels, and they are linked by red lines to clarify the differences between the samples. In panels (A)–(E), only large-sized retroelement copies in the genome (>5 kb for LINE1s and >1 kb for ERVs) are included. Statistical significances were computed using Wilcoxon signed-rank tests. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Inverse relation between DnmtDam binding and the expression of LINE1 and IAPEz retroelements in mouse early embryos
We next addressed whether DnmtDam could transcriptionally repress retroelements in mouse early embryos. We microinjected a set of Dnmt1 (siDnmt1) or scrambled (Scrb) siRNAs into the zygote and grew the zygotes in vitro. We obtained mRNAs from the resulting four/eight-cell embryos (n = 54 for siDnmt1 and n = 53 for Scrb; Figure 4A), verified the reduction of Dnmt1 transcript in the knockdown embryo (Figure 4B) and further processed mRNA for pico-profiling (45,46), Illumina library construction and RNA sequencing. A similar number of reads were mapped at ∼60% alignment efficiency. A comparison of transcriptomes using a scatter plot showed a similarity (R = 0.92) between the siDnmt1 and Scrb embryo samples (Figure 4C). For reference, a total of 323 genes were upregulated (FC >5, read count >10), whereas 468 genes were downregulated in the siDnmt1 embryos. As for imprinted genes, 37 out of 134 genes were expressed (read count >0 in either siDnmt1 or Scrb), and the number of genes with increased expression levels was twice that of the decreased (FC >2; 15 versus 8 genes) in the siDnmt1 embryos (Supplementary File S1).
Figure 4.
Inverse correlation between DnmtDam binding and the expression of retroelements in mouse embryos. (A) Dnmt1 knockdown embryos were produced after the microinjection of short interference RNA (siRNA) for Dnmt1 (siDnmt1) or scrambled sequences (Scrb). Following in vitro culture, the embryos at the four- and eight-cell stages (the numbers in the parentheses) were used in the RNA-seq experiment. The numbers of the total and mapped reads are denoted along with the mapping efficiencies in the parentheses. (B) Semiquantitative PCR analysis of Dnmt1 expression in siDnmt1- and Scrb-injected embryos. Gapdh was amplified along with Dnmt1 and was used as an internal control. (–) indicates a no-template control. (C) Differential expression between siDnmt1 and Scrb embryos. A scatter plot depicting the transcriptomic differences of coding genes between RNA-seq samples in log2-transformed read count values. Dotted diagonal lines indicate the 2-fold upregulation against each other. The Pearson correlation coefficient (r) is denoted on the plot. (D) Expression profiles of retroelements by subfamily in the siDnmt1 and Scrb samples. Within each LINE1, ERVK, ERV1 and ERVL retroelement family, the subfamilies of the top 10 highest copy numbers were selected to compare the expression of retroelements in siDnmt1 (yellow) or Scrb (green) embryos. Black dots represent the individual copies of elements. In each facet, red dots in the middle of violin plots indicate the trimmed means of enrichment levels, and they are linked by red lines to clarify the differences between samples. Statistical significances were computed using Wilcoxon signed-rank tests. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (E) Relationship between retroelement repression (siDnmt1/Scrb) and DnmtDam enrichment (DnmtDam/Dam). The mean DnmtDam enrichment levels in retroelement subfamilies over Dam (x-axis) are plotted against the mean expression levels in siDnmt1 over Scrb embryos (y-axis). (F) Comparison of statistical significance between retroelement expression (siDnmt1 versus Scrb) and DnmtDam enrichment (DnmtDam versus Dam). The P-values (Wilcoxon signed-rank tests) from DELP-seq and RNA-seq were compared by a scatter plot. In (E) and (F), solid gray lines represent x/y-axes on the quadrant plane. Dotted gray lines in (E) are the significance threshold (P-value = 0.0001). The names of the elements on quadrant 1 are labeled and colored by their family type.
A variety of retroelements showed a significant increase in the expression level in siDnmt1 embryos that belong to either the LINE1 family (L1Md_A, L1Md_F, L1Md_F2, L1Md_T, L1Md_F3, L1_Mus1 and L1_Mus2) or the ERVK family (ETnERV2, IAPEz, MMERVK10C, MMETn and RLRT10; Figure 4D). As expected, these groups of retroelements with increased expression in the siDnmt1 embryos mostly corresponded to those preferred by Dnmt1 (see Figure 3E); this result exhibits the strong correlation between the two (Figure 4E). As shown from the direct comparison of DamID and the RNA-seq results using scatter plots for P-values, the L1Md_F2, L1Md_T, L1Md_A, L1Md_F3, L1_Mus1, L1Md_F and IAPEz sequences were shown to have the highest statistical significance (Figure 4F). Meanwhile, the retroelements, including L1Md_Gf, L1_Mus3, Lx and RMER16, appeared to display some disjunction between Dnmt1 binding and transcriptional repression, which may be due to their intrinsically low copy numbers and low expression levels in early embryos (Figure 4D); this renders the statistical evaluation of them quite tricky. Our result suggests that Dnmt1 specifically binds and transcriptionally represses a set of retroelements belonging to the LINE1 or ERVK family in early mouse embryos.
A profound loss of DNA methylation at retroelement sequences in Dnmt1-depleted mouse embryos
We next investigated whether Dnmt1 binding was associated with DNA methylation at the retroelement sequences. Using the CRISPR/Cas9 genome editing technology, we disrupted the exon 32 of Dnmt1 gene encoding DNA methylase domain to remove its enzymatic activity (Supplementary Figure S7A). With a subset of four/eight-cell embryos obtained after microinjection of CRISPR/Cas9 toolkit, we first examined mutation frequency by PCR genotyping (47), and identified that all the CRISPR (CR) embryos had random deletions at the target regions (Supplementary Figure S7B). For MBD-seq, we used ∼2 ng genomic DNA (∼300 blastomeres per group) pooled from WT and CR embryos (n = 44 for WT embryos and n = 41 and 45 for CR1 and CR2 groups, respectively, as biological replicates; Supplementary Figure S7C), and performed MBD-based capture to enrich methylated DNA fragments before library construction for Illumina sequencing (Figure 5A). About 9.3 × 107 reads per group were mapped with ∼91% alignment efficiency. The mean read counts per 500-bp genomic bin were very low in the CR groups compared to the control (Figure 5B). The histograms for the distribution of bins by FC values between the CR groups versus the control revealed a global loss of DNA methylation in the CR embryos (Figure 5C).
Figure 5.
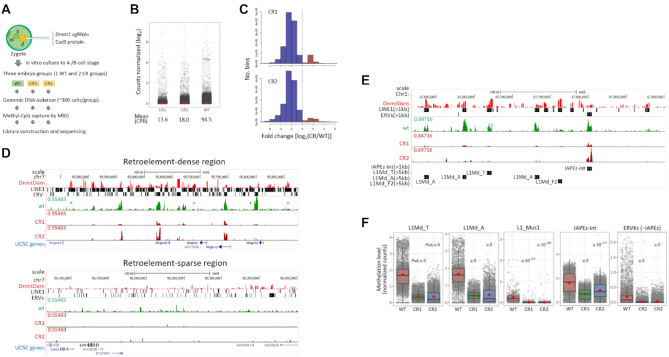
Dnmt1 is involved in DNA methylation of genomic retroelements in mouse early embryos. (A) Generation of Dnmt1-depleted embryos using CRISPR/Cas9 method and methyl-CpG capture for sequencing. Mouse zygotes were injected with Cas9 recombinant protein and a pair of sgRNAs targeting exon 33 of Dnmt1 gene and were cultured to the four- and eight-cell stages. Single WT embryo group and two Dnmt1 CRISPR embryo groups (CR1 and CR2) were separately subjected to methyl-CpG capture and library construction. MBD, methyl-CpG-binding protein. (B) Distribution of log2-transformed normalized count in MBD-seq samples. Each gray dot represents a 500-bp genomic bin containing a number of reads normalized in counts per million. The mean count in each embryo group is shown in red dot with the actual mean values in counts per billion below. (C) Global loss of DNA methylation in CR embryos compared to the WT embryos. Histograms show the relative methylation levels in 500-bp genomic bins (x-axes) versus the frequency of bins (y-axes) in CR1 and CR2 embryos against the control (WT). Blue and red bars represent fractions of bins with methylation decrease and increase in CR embryos, respectively. (D) UCSC Genome Browser snapshots showing both DELP-seq (red) and MBD-seq signals (green for WT and deep red for CR groups) on a retroelement-dense or -sparse region chosen arbitrarily. Notably, retroelement loci with DnmtDam enrichment do not always show a dense methylation (see the loci marked with asterisk). (E) Dependence of DNA methylation levels on the type and the size of retroelement copy. UCSC Genome Browser snapshot shows only the large LINE1 (>1.0 or >5.0 kb in length as indicated) and ERV copies (>1.0 kb) extracted from the RepeatMasker (mm9) track. Arrows indicate the orientation of the retroelements. (F) Demethylation of retroelement copies in the CR1 and CR2 embryo groups. Box plots show difference in the DNA methylation levels between WT and CR embryos. Red dot represents the mean count (DNA methylation level) in each embryo group (see the explanation in B). Adjusted P-values (Padj) are indicated.
Visual inspection on the UCSC Genome Browser indicated that, in the WT embryos, methylated DNA fragments were largely enriched in retroelement-dense genomic stretches and usually spotted alongside DnmtDam peak clusters (Figure 5D). Those retroelements harboring DnmtDam signals were not evenly methylated; certain retroelement sequences were much less methylated or not methylated at all, depending on the size and the family they belonged to or possibly on the genomic context (see marked regions in Figure 5D). For instance, the full-length L1Md_A and L1Md_T sequences (>5.0 kb in length), LINE1 subfamilies that are considered as evolutionarily young and capable of independent mobilization (48), were relatively strongly methylated, whereas a large fraction of full-length L1Md_F2 copies, depending on the genomic loci, were weakly methylated even though they were also closely occupied by DnmtDam signals, reflecting the notion that the recruitment of Dnmts did not necessarily lead to local methylation (49,50) (Figure 5E). However, in the CR1 and CR2 embryos, heavy DNA methylation signals at these retroelements were, in general, markedly reduced (Figure 5D and E). IAPEz sequences, which also carried dense DnmtDam signals, were demethylated in the CR embryos but only partially (Figure 5C) as observed in Dnmt1-null mouse embryonic stem cells (51,52), suggesting a joint participation of other Dnmt(s) in the methylation of IAPEz sequences. Figure 5F summarizes the degree of demethylation among the DnmtDam-rich retroelement families (Figure 3A and B) determined by calculating the mean read counts over the corresponding retroelement families. The results indicated that Dnmt1 was involved in the maintenance of DNA methylation at these retroelement sequences. Meanwhile, the profile of the mean DNA methylation levels of genic regions spanning between the transcription start and end sites also showed a demethylation of single-copy sequences in the CR embryos (Supplementary Figure S8A). With a special interest in imprinted genes, we looked into differentially methylated regions (DMRs) of imprinted loci. We found that both DnmtDam and methylation signals were co-localized at H19, Zsrs1, Zim3 and Kcqn1 gene DMRs (Supplementary Figure S8B) although the general weakness of both signals at unique genomic sequence leaves assignment of a detailed mechanistic study on Dnmt1 involvement at DMRs during the cleavage stage. In conclusion, we showed that Dnmt1 can bind and transcriptionally repress various retroelement sequences through DNA methylation mechanism during preimplantation period of mouse development.
DISCUSSION
Successful ChIP requires a good quality antibody and sufficient genomic material. If the number of cells is very limited, ChIP demands even higher quality antibody with extra specificity and sensitivity. ChIP experiments with early embryo samples are a challenge due to the above-mentioned reasons; at least tens of thousands of cells are demanded in recent low-cell-number ChIP-seq protocols (53) for typical transcription factors. We only had 500 diploid cells (blastomeres), an incomparably small scale for conventional ChIP with usual antibody. In these circumstances, DamID was a good alternative because of its enzyme-vouched specificity and fidelity in selecting global target loci free from uncertainty due to the use of different antibodies with variable performances. For your information, several studies have directly compared DamID with ChIP profiles, all of which found a strong similarity between the two methods (54–57). In this study, we identified and characterized the Dnmt1-binding genomic regions in mouse early embryos and used the DamID method as a tool for profiling the Dnmt1-binding sites. The distance between two DpnI sites on the mouse chromosomes is ∼1 kb on average and is greatly variable depending on the chromosomal locations. If the length between two DpnI sites is too long, the resulting DNA fragment is inadequate for sequencing. To overcome the size problem, we double digested the genome with DpnI and FatI enzymes; the latter cuts the genome 2-fold more frequently than the former. In addition, we performed suppression PCR to prevent the ligated sequences with the same DpnI or FatI adapter at both ends from being amplified and enriched those ligated sequences only with different adapters. Thus, we were able to reduce the average length of the amplicon library to 300–500 bp, which is appropriate for Illumina sequencing. This modified protocol is particularly useful for the analysis of samples, such as early mammalian embryos, because sequencing libraries prepared from such samples typically suffer from shallow read coverage and insufficient statistics, and this method produces an exhaustive collection of embryonic DNA libraries with high efficiency and minimal loss.
In typical somatic cells, Dnmt1 occupies replication forks to form replication machinery and continues to methylate newly synthesized DNA strands. Therefore, the Dnmt1-DamID experiment may have a high chance of producing nonspecific peaks over the whole chromosomes, which logically yields unintelligible results. In cleavage-stage mouse embryos, both global and passive modes of DNA demethylation occur, and the maintenance of DNA methylation is uncoupled from DNA replication, which greatly reduces the risk of generating nonspecific data from the Dnmt1-DamID experiment. However, the long-lasting scenario of the nuclear exclusion of early embryonic Dnmt1 is now being challenged. A fraction of Dnmt1 proteins are present in the nucleus and play a role in sustaining methylation imprints (5,13,16,17). Although Dnmt1 is nuclear, its full-time association with the replication fork during the S phase of cell cycle is quite unlikely if considering the global progressive demethylation in this period. Additionally, the nuclear Dnmt1 activity is limited and temporary based on our results (Figure 1B) and those of others (2–7). Therefore, the early embryo is a relevant and reasonable choice for Dnmt1-DamID experiments.
The transcriptomes of repetitive elements in preimplantation-stage embryos in humans (24) and mice (58) are characterized by dynamic changes over time that impart a stage-specific signature to the early embryo. The transcriptional activities of both the LINE1 and IAP retroelements of the ERVK family peaked at the two-cell stage and then continuously decreased as embryos advanced to the blastocyst stage in mice (21,58). This observation is consistent with the early study reporting that mouse oocytes and early embryos provide an environment unsuitable for the transcription of IAP and LINE1 retroelement families and provide an environment that is particularly suitable for the transcription of the MaLR and ERVL families (20). The unsuitable nuclear environment for LINE1 transcription notwithstanding, the requirement of LINE1 transcription for progression to the cleavage stage is interesting (21,59). The sharp decrease in the levels of LINE1 and IAP transcripts from the four-cell stage indicates the progressive repression of these retroelements during early development (58).
These early observations are consistent with our results of the enrichment of Dnmt1 at LINE1 and IAP retroelements in four/eight-cell embryos (Figure 3). Given the role of DNA methylation in maintaining genome stability by suppressing the transposition of retroelements (1), DNA methylation can serve as a critical mechanism of repression of retroelements in early embryos. Indeed, several Dnmts are involved in the suppression of retroelements in germ cells. In Dnmt3l-deficient germ cells, every type of retroelement is derepressed (60). IAP sequences are reactivated in Dnmt1-deficient fetal cells (61) and in Dnmt3a-deficient germ cells (62). LINE1s are demethylated in the absence of Dnmt3c in germ cells (63). In this study, we found that the Dnmt1-enriched LINE1 and IAPEz families that were relatively highly methylated in WT embryos were markedly demethylated in the Dnmt1-depleted embryos (Figure 5). Since these retroelements were the target loci of Dnmt1, and became demethylated and derepressed in the Dnmt1-depleted embryos, it indicated the involvement of Dnmt1 in the suppression of the LINE1 and IAP sequences via DNA methylation in early embryos. Meanwhile, several studies have reported a steady-state level of CpG methylation at the retroelement loci during the early developmental period (14,64,65), indicating that DNA methylation at these sequences is stably maintained against the genome-wide event of passive demethylation occurring in preimplantation-stage embryos (66,67). Thus, for now, there is little evidence for methylation change at these retroelements during this period, which leaves a question whether Dnmt1 has a role in the epigenetic regulation of retroelements that display a progressive repression from the four-cell stage on (58). To explain the potential role of Dnmt1 upon binding to retroelements, we noted that DNA methylation change occurred quite limitedly at a group of retroelement copies. For example, full-length LINE1 sequences, unlike most other 5′-truncated versions, have intact 5′-UTRs through which epigenetic regulators and transcription factors such as NANOG (24) control LINE1 expression. Consistently, DNA methylation was found highly concentrated on the large-sized LINE1 copies (Figure 5D and E). Despite the transcriptional competency of the full-length LINE1 copies, their extremely small fraction among the whole LINE1 population in the genome seldom allows methylation changes to be perceived with a global methylation assay even though DNA methylation changes at these LINE1 5′-UTRs. We indeed observed, from a separate study with mouse skeletal muscle tissues, an age-associated, size-dependent change of DNA methylation among the retroelement copies, which was otherwise not unearthed from the whole copy-based analysis (33).
Notably, the four-cell stage in mice is the time when retroelement expression begins to decline and the paternal genome of the zygote becomes densely modified with H3K9 methylation; the paternal genome, differing from the maternal genome carrying dense H3K9 methylation, stays seemingly unmethylated for H3K9 until the two-cell stage (29,68). It hints at a correlation between the establishment of global H3K9 methylation and the suppression of retroelements. In reality, in somatic and embryonic stem cells, SETDB1/Trim28/KRAB-ZNF-mediated H3K9 methylation, separately from or in combination with DNA methylation, represses certain retroelement families (69,70). Additionally, LINE1 sequences are repressed in a target-specific fashion in the experiment using the TALE-L1-KRAB construct in mouse embryos (21). Therefore, it is likely that DNA methylation cooperates with SETDB1/Trim28/KRAB-ZNF-mediated H3K9 methylation to transcriptionally regulate certain retroelement families in early mouse embryos. This may explain the uncomfortable gap between the extensive DNA demethylation at L1Md_A and L1Md_T families and their relatively small increase of transcription in the siDnmt1 embryos (Figures 4D and 5F).
DATA AVAILABILITY
Raw sequencing data for DELP-seq and RNA-seq were deposited under GSE111266:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE111266
UCSC Genome Browser session link for DELP-seq and MBD-seq:
http://genome.ucsc.edu/s/mbk0asis/DELPseq_MBDseq_Min_et_al_NAR
Supplementary Material
Contributor Information
Byungkuk Min, Development and Differentiation Research Center, Korea Research Institute of Bioscience Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, South Korea.
Jung Sun Park, Development and Differentiation Research Center, Korea Research Institute of Bioscience Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, South Korea.
Young Sun Jeong, Development and Differentiation Research Center, Korea Research Institute of Bioscience Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, South Korea.
Kyuheum Jeon, Development and Differentiation Research Center, Korea Research Institute of Bioscience Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, South Korea; Department of Functional Genomics, Korea University of Science and Technology, Daejeon 34113, South Korea.
Yong-Kook Kang, Development and Differentiation Research Center, Korea Research Institute of Bioscience Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 34141, South Korea; Department of Functional Genomics, Korea University of Science and Technology, Daejeon 34113, South Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Korea Research Institute of Bioscience and Biotechnology Initiative Program; National Research Foundation of Korea [NRF-2019R1A2C1083916]. Funding for open access charge: Korea Research Institute of Bioscience and Biotechnology.
Conflict of interest statement. None declared.
REFERENCES
- 1. Messerschmidt D.M., Knowles B.B., Solter D.. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014; 28:812–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Branco M.R., Oda M., Reik W.. Safeguarding parental identity: Dnmt1 maintains imprints during epigenetic reprogramming in early embryogenesis. Genes Dev. 2008; 22:1567–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cardoso M.C., Leonhardt H.. DNA methyltransferase is actively retained in the cytoplasm during early development. J. Cell Biol. 1999; 147:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carlson L.L., Page A.W., Bestor T.H.. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: implications for genomic imprinting. Genes Dev. 1992; 6:2536–2541. [DOI] [PubMed] [Google Scholar]
- 5. Howell C.Y., Bestor T.H., Ding F., Latham K.E., Mertineit C., Trasler J.M., Chaillet J.R.. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001; 104:829–838. [DOI] [PubMed] [Google Scholar]
- 6. Jeong Y.S., Oh K.B., Park J.S., Kim J.S., Kang Y.K.. Cytoplasmic localization of oocyte-specific variant of porcine DNA methyltransferase-1 during early development. Dev. Dyn. 2009; 238:1666–1673. [DOI] [PubMed] [Google Scholar]
- 7. Ratnam S., Mertineit C., Ding F., Howell C.Y., Clarke H.J., Bestor T.H., Chaillet J.R., Trasler J.M.. Dynamics of Dnmt1 methyltransferase expression and intracellular localization during oogenesis and preimplantation development. Dev. Biol. 2002; 245:304–314. [DOI] [PubMed] [Google Scholar]
- 8. Jones P.A., Liang G.. Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 2009; 10:805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hermann A., Goyal R., Jeltsch A.. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 2004; 279:48350–48359. [DOI] [PubMed] [Google Scholar]
- 10. Leonhardt H., Page A.W., Weier H.U., Bestor T.H.. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992; 71:865–873. [DOI] [PubMed] [Google Scholar]
- 11. Vertino P.M., Sekowski J.A., Coll J.M., Applegren N., Han S., Hickey R.J., Malkas L.H.. DNMT1 is a component of a multiprotein DNA replication complex. Cell Cycle. 2002; 1:416–423. [DOI] [PubMed] [Google Scholar]
- 12. Li E., Beard C., Jaenisch R.. Role for DNA methylation in genomic imprinting [see comments]. Nature. 1993; 366:362–365. [DOI] [PubMed] [Google Scholar]
- 13. Hirasawa R., Chiba H., Kaneda M., Tajima S., Li E., Jaenisch R., Sasaki H.. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008; 22:1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim S.H., Kang Y.K., Koo D.B., Kang M.J., Moon S.J., Lee K.K., Han Y.M.. Differential DNA methylation reprogramming of various repetitive sequences in mouse preimplantation embryos. Biochem. Biophys. Res. Commun. 2004; 324:58–63. [DOI] [PubMed] [Google Scholar]
- 15. McGraw S., Oakes C.C., Martel J., Cirio M.C., de Zeeuw P., Mak W., Plass C., Bartolomei M.S., Chaillet J.R., Trasler J.M.. Loss of DNMT1o disrupts imprinted X chromosome inactivation and accentuates placental defects in females. PLoS Genet. 2013; 9:e1003873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cirio M.C., Ratnam S., Ding F., Reinhart B., Navara C., Chaillet J.R.. Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC Dev. Biol. 2008; 8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kurihara Y., Kawamura Y., Uchijima Y., Amamo T., Kobayashi H., Asano T., Kurihara H.. Maintenance of genomic methylation patterns during preimplantation development requires the somatic form of DNA methyltransferase 1. Dev. Biol. 2008; 313:335–346. [DOI] [PubMed] [Google Scholar]
- 18. Walsh C.P., Bestor T.H.. Cytosine methylation and mammalian development. Genes Dev. 1999; 13:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goodier J.L., Kazazian H.H. Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008; 135:23–35. [DOI] [PubMed] [Google Scholar]
- 20. Peaston A.E., Evsikov A.V., Graber J.H., de Vries W.N., Holbrook A.E., Solter D., Knowles B.B.. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell. 2004; 7:597–606. [DOI] [PubMed] [Google Scholar]
- 21. Jachowicz J.W., Bing X., Pontabry J., Boskovic A., Rando O.J., Torres-Padilla M.E.. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet. 2017; 49:1502–1510. [DOI] [PubMed] [Google Scholar]
- 22. Ishiuchi T., Enriquez-Gasca R., Mizutani E., Boskovic A., Ziegler-Birling C., Rodriguez-Terrones D., Wakayama T., Vaquerizas J.M., Torres-Padilla M.E.. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol. 2015; 22:662–671. [DOI] [PubMed] [Google Scholar]
- 23. Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S.L.. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012; 487:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goke J., Lu X., Chan Y.S., Ng H.H., Ly L.H., Sachs F., Szczerbinska I.. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell. 2015; 16:135–141. [DOI] [PubMed] [Google Scholar]
- 25. Aughey G.N., Southall T.D.. Dam it’s good! DamID profiling of protein–DNA interactions. Wiley Interdiscip. Rev. Dev. Biol. 2016; 5:25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Steensel B., Delrow J., Henikoff S.. Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet. 2001; 27:304–308. [DOI] [PubMed] [Google Scholar]
- 27. van Steensel B., Henikoff S.. Identification of in vivo DNA targets of chromatin proteins using tethered Dam methyltransferase. Nat. Biotechnol. 2000; 18:424–428. [DOI] [PubMed] [Google Scholar]
- 28. Hogan B. Manipulating the Mouse Embryo: A Laboratory Manual. 1994; 2nd edn.NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 29. Yeo S., Lee K.K., Han Y.M., Kang Y.K.. Methylation changes of lysine 9 of histone H3 during preimplantation mouse development. Mol. Cells. 2005; 20:423–428. [PubMed] [Google Scholar]
- 30. Kang Y.K., Park J.S., Lee C.S., Yeom Y.I., Chung A.S., Lee K.K.. Efficient integration of short interspersed element-flanked foreign DNA via homologous recombination. J. Biol. Chem. 1999; 274:36585–36591. [DOI] [PubMed] [Google Scholar]
- 31. Kang Y.K., Koo D.B., Park J.S., Choi Y.H., Kim H.N., Chang W.K., Lee K.K., Han Y.M.. Typical demethylation events in cloned pig embryos. Clues on species-specific differences in epigenetic reprogramming of cloned donor genome. J. Biol. Chem. 2001; 276:39980–39984. [DOI] [PubMed] [Google Scholar]
- 32. Min B., Cho S., Park J.S., Lee Y.G., Kim N., Kang Y.K.. Transcriptomic features of bovine blastocysts derived by somatic cell nuclear transfer. G3 (Bethesda). 2015; 5:2527–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Min B., Jeon K., Park J.S., Kang Y.K.. Demethylation and derepression of genomic retroelements in the skeletal muscles of aged mice. Aging Cell. 2019; 18:e13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cho S., Park J.S., Kang Y.K.. AGO2 and SETDB1 cooperate in promoter-targeted transcriptional silencing of the androgen receptor gene. Nucleic Acids Res. 2014; 42:13545–13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xie D., Chen C.C., Ptaszek L.M., Xiao S., Cao X., Fang F., Ng H.H., Lewin H.A., Cowan C., Zhong S.. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res. 2010; 20:804–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu J., Huang B., Chen H., Yin Q., Liu Y., Xiang Y., Zhang B., Liu B., Wang Q., Xia W et al.. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016; 534:652–657. [DOI] [PubMed] [Google Scholar]
- 37. Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018; 19:81–92. [DOI] [PubMed] [Google Scholar]
- 38. Easwaran H.P., Schermelleh L., Leonhardt H., Cardoso M.C.. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 2004; 5:1181–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Margot J.B., Cardoso M.C., Leonhardt H.. Mammalian DNA methyltransferases show different subnuclear distributions. J. Cell. Biochem. 2001; 83:373–379. [DOI] [PubMed] [Google Scholar]
- 40. Siebert P.D., Chenchik A., Kellogg D.E., Lukyanov K.A., Lukyanov S.A.. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995; 23:1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Diatchenko L., Lau Y.F., Campbell A.P., Chenchik A., Moqadam F., Huang B., Lukyanov S., Lukyanov K., Gurskaya N., Sverdlov E.D et al.. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl Acad. Sci. U.S.A. 1996; 93:6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gerdes P., Richardson S.R., Mager D.L., Faulkner G.J.. Transposable elements in the mammalian embryo: pioneers surviving through stealth and service. Genome Biol. 2016; 17:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garcia-Perez J.L., Widmann T.J., Adams I.R.. The impact of transposable elements on mammalian development. Development. 2016; 143:4101–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gifford W.D., Pfaff S.L., Macfarlan T.S.. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol. 2013; 23:218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Min B., Park J.S., Jeon K., Kang Y.K.. Characterization of X-chromosome gene expression in bovine blastocysts derived by in vitro fertilization and somatic cell nuclear transfer. Front. Genet. 2017; 8:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Min B., Park J.S., Kang Y.K.. Determination of oocyte-manipulation, zygote-manipulation, and genome-reprogramming effects on the transcriptomes of bovine blastocysts. Front. Genet. 2018; 9:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jeon J., Park J.S., Min B., Chung S.K., Kim M.K., Kang Y.K.. Retroelement insertion in a CRISPR/Cas9 editing site in the early embryo intensifies genetic mosaicism. Front. Cell Dev. Biol. 2019; 7:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sookdeo A., Hepp C.M., McClure M.A., Boissinot S.. Revisiting the evolution of mouse LINE-1 in the genomic era. Mob. DNA. 2013; 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jeltsch A., Jurkowska R.Z.. Allosteric control of mammalian DNA methyltransferases: a new regulatory paradigm. Nucleic Acids Res. 2016; 44:8556–8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Laisne M., Gupta N., Kirsh O., Pradhan S., Defossez P.A.. Mechanisms of DNA methyltransferase recruitment in mammals. Genes (Basel). 2018; 9:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Damelin M., Bestor T.H.. Biological functions of DNA methyltransferase 1 require its methyltransferase activity. Mol. Cell. Biol. 2007; 27:3891–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tsumura A., Hayakawa T., Kumaki Y., Takebayashi S., Sakaue M., Matsuoka C., Shimotohno K., Ishikawa F., Li E., Ueda H.R et al.. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006; 11:805–814. [DOI] [PubMed] [Google Scholar]
- 53. Nakato R., Shirahige K.. Recent advances in ChIP-seq analysis: from quality management to whole-genome annotation. Brief. Bioinform. 2017; 18:279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cheetham S.W., Gruhn W.H., van den Ameele J., Krautz R., Southall T.D., Kobayashi T., Surani M.A., Brand A.H.. Targeted DamID reveals differential binding of mammalian pluripotency factors. Development. 2018; 145:dev170209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Moorman C., Sun L.V., Wang J., de Wit E., Talhout W., Ward L.D., Greil F., Lu X.J., White K.P., Bussemaker H.J et al.. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc. Natl Acad. Sci. U.S.A. 2006; 103:12027–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Negre N., Hennetin J., Sun L.V., Lavrov S., Bellis M., White K.P., Cavalli G.. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006; 4:e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Southall T.D., Gold K.S., Egger B., Davidson C.M., Caygill E.E., Marshall O.J., Brand A.H.. Cell-type-specific profiling of gene expression and chromatin binding without cell isolation: assaying RNA Pol II occupancy in neural stem cells. Dev. Cell. 2013; 26:101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fadloun A., Le Gras S., Jost B., Ziegler-Birling C., Takahashi H., Gorab E., Carninci P., Torres-Padilla M.E.. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 2013; 20:332–338. [DOI] [PubMed] [Google Scholar]
- 59. Beraldi R., Pittoggi C., Sciamanna I., Mattei E., Spadafora C.. Expression of LINE-1 retroposons is essential for murine preimplantation development. Mol. Reprod. Dev. 2006; 73:279–287. [DOI] [PubMed] [Google Scholar]
- 60. Kato Y., Kaneda M., Hata K., Kumaki K., Hisano M., Kohara Y., Okano M., Li E., Nozaki M., Sasaki H.. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum. Mol. Genet. 2007; 16:2272–2280. [DOI] [PubMed] [Google Scholar]
- 61. Walsh C.P., Chaillet J.R., Bestor T.H.. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998; 20:116–117. [DOI] [PubMed] [Google Scholar]
- 62. Kaneda M., Okano M., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H.. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004; 429:900–903. [DOI] [PubMed] [Google Scholar]
- 63. Barau J., Teissandier A., Zamudio N., Roy S., Nalesso V., Herault Y., Guillou F., Bourc’his D.. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science. 2016; 354:909–912. [DOI] [PubMed] [Google Scholar]
- 64. Arand J., Wossidlo M., Lepikhov K., Peat J.R., Reik W., Walter J.. Selective impairment of methylation maintenance is the major cause of DNA methylation reprogramming in the early embryo. Epigenetics Chromatin. 2015; 8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Smith Z.D., Chan M.M., Mikkelsen T.S., Gu H., Gnirke A., Regev A., Meissner A.. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012; 484:339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Surani M.A. Reprogramming of genome function through epigenetic inheritance. Nature. 2001; 414:122–128. [DOI] [PubMed] [Google Scholar]
- 67. Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002; 3:662–673. [DOI] [PubMed] [Google Scholar]
- 68. Liu H., Kim J.M., Aoki F.. Regulation of histone H3 lysine 9 methylation in oocytes and early pre-implantation embryos. Development. 2004; 131:2269–2280. [DOI] [PubMed] [Google Scholar]
- 69. Liu S., Brind’Amour J., Karimi M.M., Shirane K., Bogutz A., Lefebvre L., Sasaki H., Shinkai Y., Lorincz M.C.. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev. 2014; 28:2041–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kang Y.K. Surveillance of retroelement expression and nucleic-acid immunity by histone methyltransferase SETDB1. Bioessays. 2018; 40:e1800058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data for DELP-seq and RNA-seq were deposited under GSE111266:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE111266
UCSC Genome Browser session link for DELP-seq and MBD-seq:
http://genome.ucsc.edu/s/mbk0asis/DELPseq_MBDseq_Min_et_al_NAR