Abstract
DNA synthesis is a fundamental requirement for cell proliferation and DNA repair, but no single method can identify the location, direction and speed of replication forks with high resolution. Mammalian cells have the ability to incorporate thymidine analogs along with the natural A, T, G and C bases during DNA synthesis, which allows for labeling of replicating or repaired DNA. Here, we demonstrate the use of the Oxford Nanopore Technologies MinION to detect 11 different thymidine analogs including CldU, BrdU, IdU as well as EdU alone or coupled to Biotin and other bulky adducts in synthetic DNA templates. We also show that the large adduct Biotin can be distinguished from the smaller analog IdU, which opens the possibility of using analog combinations to identify the location and direction of DNA synthesis. Furthermore, we detect IdU label on single DNA molecules in the genome of mouse pluripotent stem cells and using CRISPR/Cas9-mediated enrichment, determine replication rates using newly synthesized DNA strands in human mitochondrial DNA. We conclude that this novel method, termed Replipore sequencing, has the potential for on target examination of DNA replication in a wide range of biological contexts.
INTRODUCTION
DNA replication is a fundamental requirement for the development of an organism and the life-long maintenance of organ function. An estimated 2–3 × 1011 cell divisions occur in the human body each day to replenish lost cells and repair tissue damage. While DNA is replicated just once during a cell cycle, the pattern by which this occurs varies between cell types (1), and is different in malignancy (2). During the neural differentiation of mouse embryonic stem cells, for example, as much as 20% of the genome switches replication timing (3). Mammalian development induces replication program changes, which affect at least 50% of the genome (4). The generation of induced pluripotent stem cells is associated with changes in replication timing, which reset the replication pattern to resemble the one in embryonic stem cells (5). Importantly, cancer cells acquire replication programs, which differ from the ones in their normal counterparts. For instance, allelic replication asynchrony at the p53 and 21q22 loci has been described in invasive carcinomas (6). Delayed replication of one allele of a tumor suppressor can interfere with gene expression and create a situation similar to loss of heterozygosity, which is associated with malignancy (6). Chromosome-wide delays in DNA replication have been shown to delay chromosome condensation and contribute to the chromosomal instability of various tumor cell lines (2). Therefore, in addition to studying differential gene expression, there is a strong rationale to study differences in DNA replication patterns in development and disease.
Methods that are currently available to analyze the progression of DNA replication include a pulse of BrdU and immunoprecipitation for labeled DNA (7). Replication patterns can also be inferred from the sequencing of Okazaki fragments (8) or from counting the number of reads in next generation sequencing data, which allows the identification of early and late replicating regions as well as of replication origins (9). However, next generation sequencing requires amplification during library preparation and averages the signal from different cells and DNA strands. To identify the progression of replication on a single DNA strand, a commonly used technique is DNA combing, which relies on sequential pulses with two modified nucleotides—IdU and CldU, fiber stretching on glass slides and staining with specific antibodies. While this technique can track replisome progression, stalling and restart in a genome-wide manner, it does not provide any sequence information. To study replication dynamics at a locus of interest, fiber analysis requires combination with fluorescent in situ hybridization (10). This method, however, is very laborious and not scalable to the level of a complete eukaryotic genome. Thus, to the best of our knowledge, there is currently no technique that can capture location, direction and speed of replication fork progression on single molecules at a genome-wide level with high resolution.
Nanopore technology has recently emerged as a powerful third-generation sequencing technique, primarily utilized for genome assembly due to its ability to produce ultra-long reads (11). The system transports DNA through a collection of nanopores and bases are identified by measuring changes in the electric current across the surface of the pores (12). In reality, signal shift is produced not by a single base, but is influenced by 5–6 neighboring nucleotides, making contact with the pore and termed a k-mer. Computational algorithms are then used to process the signal and determine the identity of the bases passing through. Since the nanopore platform does not require DNA amplification for library preparation, it can sequence cellular DNA directly and thereby avoid averaging signals from different DNA strands. Because nanopore technology does not rely on base-pairing to determine DNA sequence, it may be used to distinguish not only the four canonical nucleotides (A, T, G, C), but potentially other types of bases, as well. Indeed, the system is able to detect methylated bases (13), which suggests that the sequencing of nucleotide analogs, incorporated during DNA replication may also be possible.
Here, we present the use of nanopore sequencing on the Oxford Nanopore Technologies (ONT) MinION to detect thymidine analogs that can be directly incorporated during DNA replication or generated in click cycloaddition reactions. Using a series of templates, we show that the MinION can distinguish between thymidine and all 11 analogs with the greatest signal to noise ratio recorded for IdU, CldU and biotin-dU and that the two analogs—IdU and biotin can be distinguished from each other. We also demonstrate the detection of IdU on single strands of substituted genomic mammalian DNA and show that our method can be applied to determine replication rates of the human mitochondrial genome.
MATERIALS AND METHODS
Training template assembly
For training sequence assembly, the pCALNL-GFP vector (Addgene plasmid#13770) was digested with AgeI and NotI to release the E-GFP insert. A double stranded DNA oligomer with 5 nb.BbvCI sites was prepared by hybridizing two single-stranded DNA oligos (top clone and bottom clone) and ligated into the purified empty vector, as described in Luzzietti et al. (14). The product was then transformed into Escherichia coli. Nicking and substitution reactions were performed as outlined in (14) with oligos, purchased from Baseclick, Germany. Prior to MinION library prep, all templates were linearized with SpeI-HF (NEB R3133S).
Top clone:
5′Phos-CCGGGCCTCAGCTTGCCACGACCTCAGCGTCAATTGTCCTCAGCTCAGATGACCTCAGCAGATTGTAGCCTCAGC-3′
Bottom clone:
5′Phos-GGCCGCTGAGGCTACAATCTGCTGAGGTCATCTGAGCTGAGGACAATTGACGCTGAGGTCGTGGCAAGCTGAGGC-3′
Substitute oligo single:
5′Phos-TGAGGCTACAATCTGCTGAGGTCATCTGAGCXGAGGACAATTGACGCTGAGGTCGTGGCAAGC-3′
X = EdU, CldU, BrdU, IdU
Substitute oligo multi:
5′Phos-XGAGGCXACAAXCXGAGGXCAXCXGAGCXGAGGACAAXXGACGCXGAGGXCGXGGCAAGC-3′
X = EdU, CldU, IdU
Click cycloaddition reactions
Click cyclo-addition was performed on single stranded EdU replace oligos prior to the substitution reaction. Briefly, 1 ug phosphorylated oligo was incubated with reagents from the CuAAC Biomolecule Reaction Buffer kit (Jena Bioscience CLK-072) and 250 uM azide for 1 h at 37°C. Separate reactions were carried out with the following azides: sodium azide (Sigma S2002), nicotinoyl azide (Sigma CDS006775), 6-azido-6-deoxy-d-glucose (Sigma 712760) Biotin azide (Thermo Fisher B10184), AF488 azide (Thermo Fisher A10266), AF647 azide (Thermo Fisher A10277) and a ssDNA oligo purchased from Integrated DNA Technologies: 5′-GGATAGCCTC/3AzideN/-3′. All click products were purified by precipitation.
Dot blots
For dot blots, 100–200 ng DNA was denatured with 20 mM NaOH for 5 min at 99°C, cooled on ice and neutralized with 0.6 M ammonium acetate. Spotting was performed manually on a 0.45 uM nylon membrane (Thermo Fisher 77016). Once fully dried, the membrane was baked in a microwave for 2.5 min. Prior to imaging, Biotin-containing blots were stained with A647-Streptavidin (Thermo Fisher S32357). For this procedure, the baked membrane was incubated with 3% BSA in TBST for 15 min, followed by A647 Streptavidin 1:500 (Thermo Fisher S32357) for 45 min at room temperature and three washes with TBST. To detect CldU or IdU-containing DNA, 500 ng–1 ug of sample was spotted on the membrane and stained with rat BrdU/CldU 1:500 (Bio Rad OBT0030S) or mouse BrdU/IdU 1:500 (BD 347580) in 3% BSA in TBST for 1h at room temperature. Staining with mouse ssDNA antibody 1:500 (Millipore MAB3034) was used as a loading control. To visualize the signal, the membrane was incubated with Alexa Fluor 647 and Alexa Fluor 488 secondary antibodies at 1:500. Following washes, the membranes were imaged with a Biorad ChemiDoc MP Imaging System.
MinION runs on synthetic templates
All samples were sequenced on MinION R9 flow cells. Sequencing libraries were prepared with Ligation Sequencing Kit 1D (ONT SQK-LSK108), following manufacturer's instructions. Library preparation for the training template with two modifications (biotin and IdU) was carried out with Ligation Sequencing Kit 1D (ONT SQK-LSK109). Each run was performed with 250–500 ng purified DNA. Typically, a single flow cell was used for multiple samples with washes in between, carried out with the Flow Cell Wash Kit (ONT EXP-WSH002). Fast5 data files from each run were analyzed with NanoMod version 0.2.1, an updated version of the NanoMod software, described in Liu et al. (15).
MinION runs with genomic DNA
For genomic DNA runs, mouse embryonic stem cells from a pure C57BL/6J strain (Jackson LAB Strain # 000664) were incubated with 25 uM IdU for 24 h. Cells were then harvested and genomic DNA was extracted with a High Pure PCR Template Preparation Kit (Roche 11796828001). MinION runs were performed on R9 flow cells with 400–500 ng purified DNA per sample following library preparation with Ligation Sequencing Kit 1D (ONT SQK-LSK108). For each run, unlabeled DNA was loaded first and sequenced for 5–6 h. This was followed by a wash step with the ONT wash kit (ONT EXP-WSH002), priming and loading of IdU-substituted gDNA, which was run until the flow cell expired, typically from 8 to 20 h without voltage adjustments. Analysis was performed with data from two combined runs. The combined control data set included 719 295 long reads, covering 1.2G bases, while the IdU sample was represented by 140 667 long reads across 209M bases.
Computational simulations with NanoMod to determine the coverage required for the detection of single and multi-modified reads
We used NanoMod version 0.2.1(15) for the analysis of MinION sequencing data in this study. In NanoMod version 0.2.1, we implemented k-mer-based analysis for low coverage nanopore data for a large genome. This was accomplished by incorporating signals from different genomic positions, which represent the same k-mer to increase the coverage of individual k-mers in the context of low whole genome coverage. In addition, we used a window-based method (i.e. 60 bp) to identify a region with multiple modifications. This was done by assigning the top percentile (top 10% or top 15%) P-value calculated for individual Ts in the 60-bp window to the entire region. Finally, we implemented computational simulations on single modification and multiple modifications data to determine the coverage required to detect reads with modification rates of <20%.
Detection of single modified reads of 2.0 kb or more in mammalian genomic DNA
Long reads from control and IdU-substituted genomic DNA were basecalled with Albacore v2.3.1 and aligned with the mouse reference genome (GRCm38, also known as mm10). The next steps of the analysis considered only k-mers present in the IdU-substituted genomic DNA dataset. To identify individual modified DNA molecules, we randomly selected 70% of the reads from the control data and calculated the signal mean and standard deviation for each of the central five bases (centered 5-mer) in all T-containing 9-mers. Next, we set a read length threshold (500, 800, 1000, 1500, 2000 bp) and calculated the combined P-value for any T in the reads, which crossed the length threshold. This was accomplished by using the signal mean and standard deviation to calculate the P-value for each base in a centered 5-mer. This generated a set of P-values from different T-containing 9-mers along the read. The P-value in the smallest percentile was used as a P-value for the read. This process was applied to the remaining 30% reads in the control sample and to the IdU-substituted data set.
CRISPR/Cas9-mediated enrichment and sequencing of human mitochondrial DNA
Enrichment for human mitochondrial DNA for MinION sequencing was performed on two samples of unlabeled and 8h IdU-labeled (30uM) genomic DNA from C9012 human embryonic stem cells, grown in StemFlex media. High molecular weight DNA from ∼20 × 106 cells/sample was prepared with a phenol–chloroform extraction method, as outlined in Giesselmann et al. (16). Enrichment for mitochondrial DNA was carried out by incubation of high molecular weight gDNA with Alt-R® S.p. HiFi Cas9 Nuclease V3 (IDT 1081060) and the gRNA:
mC*mA*mC*rUrUrUrCrArCrCrGrCrUrArCrArCrGrArCrGrUrUrUrUrArGrArGrCrUrArGrArArArUrArGrCrArArGrUrUrArArArArUrArArGrGrCrUrArGrUrCrCrGrUrUrArUrCrArArCrUrUrGrArArArArArGrUrGrGrCrArCrCrGrArGrUrCrGrGrUrGrCmU*mU*mU*rU
which introduces a cut between positions 8146 and 8147 in the human mitochondrial genome (gRNA target sequence CACTTTCACCGCTACACGAC), thereby linearizing it. Following enrichment, library preparation proceeded with the Ligation Sequencing Kit 1D (ONT SQK-LSK109), as in Giesselmann et al. (16). The two samples (unlabeled and 8h IdU-labeled) were sequenced on a single flow cell (FLO-MIN106D R9 SpotON) with a wash step in-between, performed with the ONT EXP-WSH003 kit. The order of sequencing was as follows: unlabeled control (630 ng DNA loaded on the flow cell), which was sequenced for 10h and 8h-IdU labeled sample (485 ng DNA loaded on the flow cell), sequenced for ∼20 h until the flow cell expired.
Analysis of human mitochondrial DNA replication
Sequencing reads from the two samples-unlabeled control (Sample 1) and 8h IdU (Sample 2) were first basecalled with Albacore v2.3.4. Long reads were then mapped against hg38 and NanoMod was used to anchor signals, according to their alignment against hg38. We found 15,119 forward-aligned long reads and 1503 reverse-aligned long reads in the unlabeled control as well as 9201 forward-aligned long reads and 668 reverse-aligned long reads in the sample, incubated for 8h with IdU. Since not all long reads in the IdU-incubated sample contain the label, we designed a strategy to separate labeled from unlabeled reads. To do this, we (i) split long reads from the control unlabeled sample into two groups: one group with long reads, basecalled from odd-numbered folders (Sample1_odd), and another with long reads, basecalled from even-numbered folders (Sample 1_even). We then (ii) obtained signals for each strand-specific reference position in the mitochondrial genome according to the alignment of long reads from Sample1_odd. Each position was associated with two groups of signals: one for reverse-aligned long reads and the other for forward-aligned long reads. In the next step, we (iii) calculated signal mean and standard deviation for each strand-specific reference position in the mitochondrial genome and used this result to calculate a P-value for each thymidine in long reads from Sample1_odd, Sample1_even and Sample 2 (8h IdU). For this calculation, we assumed that a base Ti in a long read with signal Si is mapped against a strand-specific reference base Tj with the signal means mj and dj and used the value of (Si- mj)/ dj to calculate a P-value for Ti under a normal distribution with the mean mj and the standard deviation dj. We, then used Stouffer's method to aggregate the P-values of Ti-2, Ti-1, Ti, Ti+1 and Ti+2, thereby taking into account the neighborhood base effect. In the next step, we (iv) obtained P-values for all thymidines in a long read in an ascending order, and used the P-value at the 10% percentile in the list as the P-value of the long read. Then, for each group of Sample1_odd, Sample1_even or Sample 2, we (v) plotted violin and boxplots as shown in Figure 4C and used the P-value at the 25% percentile of the list of long-read P-values in Sample1_ odd as the threshold to categorize the long reads in Sample 2 into labeled long reads with P-value less than the threshold and unlabeled long reads with P-value above the threshold. Finally, (vi) we used NanoMod to calculate P-values for all thymidines in the reference mitochondrial genome by comparing Sample1_even against labeled long reads in Sample 2 or by comparing Sample1_even against unlabeled long reads in Sample 2. This comparison is strand-specific and also grouped by the length of mapped long reads: in one comparison, we only considered long reads of 7–9 kb length (labeled 8 kb long reads), and in the other- long reads of 15-16.6 kb length (labeled 16 kb long reads). In NanoMod, to avoid bias caused by the different coverage of the light and heavy strands in the mitochondrial genome, reads for the reference position of the light strand were downsampled to 500, and reads for the reference position of the heavy strand to 50. This process was repeated 100 times for each position and the smallest 25% quantile P-value was used as the P-value of each reference position.
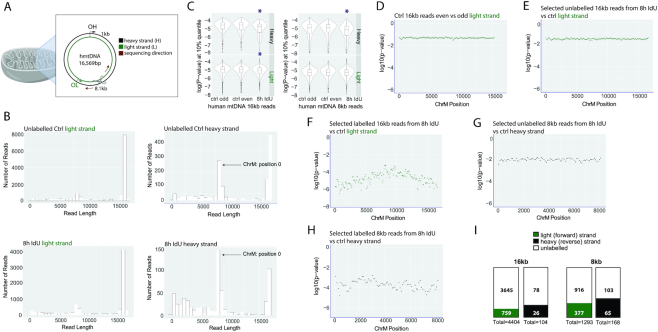
Figure 4.
Detection of newly-synthesized DNA in the human mitochondrial genome. (A) A schematic of mitochondria and their genome. The light (forward) and heavy (reverse) strands are shown in green and black, respectively. The Cas9-induced cut to linearize the mitochondrial genome occurs approximately between positions 8146 and 8147. The MiniION sequencing direction is indicated with a dark red arrow. The light and heavy strand replication origins, OL and OH, are shown in green and black, respectively. (B) Read length distribution for the light and heavy strands of the unlabeled control and the 8h IdU-labeled sample. (C) P-values in the 10% percentile for 16kb and 8kb reads for the light and heavy strands of the unlabeled control and the 8h IdU-labeled sample. Purple stars indicate significant differences. (D). P-values in the 25% percentile for the comparison between 16kb reads, basecalled from the even and the odd-numbered data folders of the unlabeled control. P-values in the 25% percentile from a comparison of (E) unlabeled or (F) labeled 16kb reads from the 8h IdU-sample and 16kb reads from the unlabeled control. (G). P-values in the 25% percentile for the comparison between unlabeled or (H). labeled 8kb reads from the 8h IdU sample and 8kb reads from the unlabeled control. The x-axis indicates position on the mitochondrial genome and the y-axis shows the log10(P-value). (I). Numbers of labeled 16kb and 8kb long reads from the light and heavy strands of the 8h IdU-labeled sample. The filled and empty portions of each box contain the numbers of labeled and unlabeled reads, respectively.
RESULTS
Detection of single-base DNA modifications with nanopore sequencing
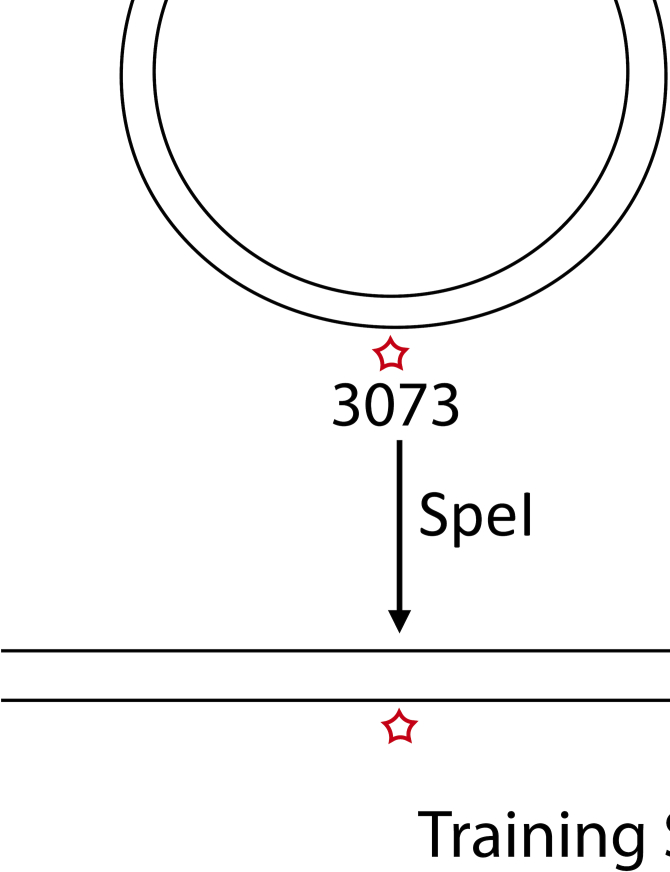
To determine the ability of the ONT MinION sequencer to detect modified bases, we designed synthetic templates containing a single modification in a defined location. Template assembly involved a vector nicking step with endonuclease nb.BbvCI, which generated a 63 bp region of ssDNA on the minus strand, followed by the ligation of an oligo with one modified thymidine base (Figure 1A). This approach to replace segments within a plasmid with a labeled oligo has been previously used for FRET studies and reported to result in >75% substitution (14). We used oligos without modification, or containing five different modifications, EdU, CldU, BrdU, IdU or biotin for the substitution reaction (Figure 1B). In addition, we included an oligo with two different analogs-Biotin and IdU (Figure 1B). Also, we modified EdU with a series of compounds of increasing molecular weight: azide, nicotine, glucose, AF488, AF647 and a 10-bp ssDNA oligo through click cycloaddition reactions (17) (Supplementary Figure S1A). This method allows the attachment of any azide group-containing moiety to alkyne-substituted DNA and has been reported to be nearly 100% efficient (18). Dot blots for AF647, AF488, BrdU and biotin modified plasmids showed efficient labeling (Figure 1C). Following the replace reaction, the vector was linearized with the restriction enzyme SpeI to generate a 6184-bp double-stranded training template with one modified thymidine on the minus strand at position 3073, which was sequenced on the MinION (Figure 1E). The modifications on the template with two analogs were located at 3049 (biotin) and 3098 (IdU).
Figure 1.
Design and assembly of training templates with single and multiple modifications at defined locations. (A) A schematic of the two-step assembly process of modified training templates. The first step is nicking the vector with the restriction enzyme nb.BbvCI and the second- ligation of a ssDNA oligo which carries 1, 2 or 14 modifications. The vector is then linearized with SpeI-HF to generate a 6184bp modified template, sequenced on the MinION. (B) Sequence of the single, double and multiple modification oligos used for ligation. (C) Dot blots with ligation products for single modification templates, carrying AF647, AF488, Biotin or a BrdU moiety, demonstrating efficient replacement with the modified oligo. (D) Dot blots with ligation products for multi-modified templates, carrying 14 CldU or 14 IdU modifications. (E) A schematic illustrating the sequencing of a training template with one or more modifications through a nanopore.
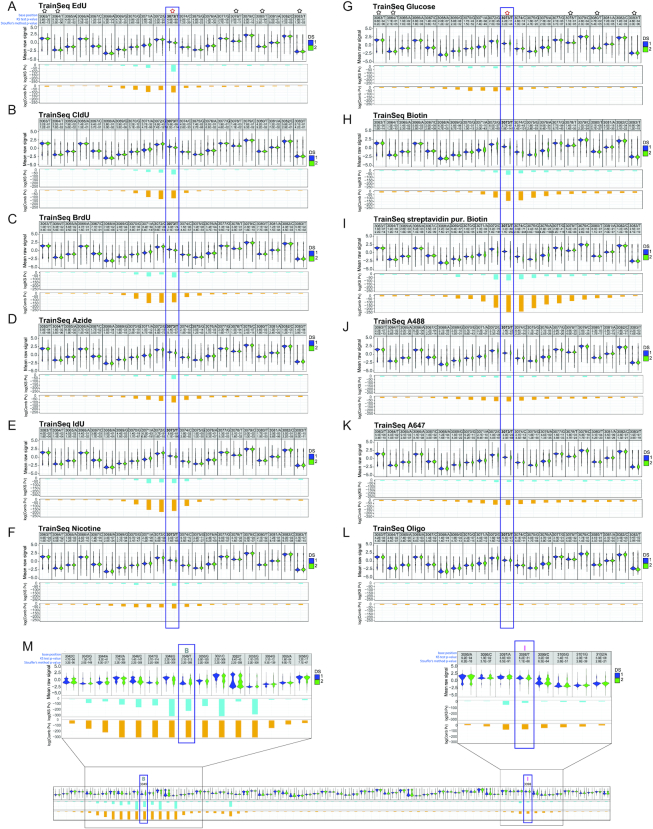
To identify modified nucleotides, we developed NanoMod, an analysis tool which calls DNA modifications directly from electrical signals (15). NanoMod uses Albacore for base calling and performs indel error correction by aligning signals to a reference sequence. The input to NanoMod is a dataset with two groups of samples-a modified template and a sequence-matched control, while the output is the ranked list of regions with modifications. Signals from control and substituted samples are then compared using two statistical assays-Komogorov–Smirnov test and Stouffer's method for calculating combined P values. We used NanoMod to score individual nucleotides in 5-bp sliding windows to detect the presence of thymidine analogs. All 11 modifications (Supplementary Figure S1A) in the dataset caused signal shifts at base 3073, as well as, at neighboring positions (Figure 2A–L). The analogs EdU, CldU, BrdU and IdU caused signal shifts spreading as far as 3–4 bases upstream and 2 bases downstream of the modified position (Figure 2A, B, C, E), consistent with the MinION detecting signals from a 5–6 bp k-mer. Therefore, the signal of the modified T is present in neighboring bases. The magnitude of the signal change was proportional to the size of the moiety with one of the largest unclicked analogs IdU (MW = 354.1 g/mol) generating the most significant change in pore current, log(combined P-value) = –175 (Figure 2E). The smallest modification, EdU (MW = 252.23 g/mol) was detectable, as well at log(combined P-value) = –125 (Figure 2A). A highly significant change in signal at 3073 was also recorded for the bulkier analog—biotin, with a log(combined P-value) = –150 (Figure 2H). Purification of the plasmid with streptavidin beads after the substitution reaction, followed by sequencing further increased the significance of the change, with a log(combined P-value) = –250 (Figure 2I). Therefore, modification at 75% (14), was sufficient to call the modified base, while 100% modification further magnified signal change. Biotin influenced electrical signals further away from the modification than EdU, CldU, BrdU or IdU. The increased number of surrounding bases affected- as many as 3–4 nucleotides upstream and about 10 downstream of the modified position (Figure 2H, I) indicates that the effect of bulky modifications can spread beyond a single k-mer.
Figure 2.
Detecting a series of modifications with increasing molecular weight through MinION sequencing. A violin plot of the known modification site and the surrounding bases for (A) EdU, (B) CldU, (C) BrdU, (D) azide, (E) IdU, (F) nicotine, (G) glucose, (H) biotin, (I) streptavidin purified biotin, (J) AF488, (K) AF647, (L) 10 bp ssDNA oligo, attached to position 3073 by click cycloaddition and (M) dual-labeled template with Biotin at 3049 and IdU at 3098. In each panel, the first line denotes position, followed by the base, the P-value for a Kolmogorov–Smirnov test and the P-value for Stouffer's method. The second line shows a violin plot of mean normalized signal from the control and modified sample, the third line contains the logarithm of the P-value (pv) for Kolmogorov–Smirnov test, and the fourth line displays the logarithm of the combined P-value, calculated using Stouffer's method. ‘DS 1’ in blue represents the non-modified oligo sample, while ‘DS 2’ in green stands for the modified template; ‘- strand’ denotes reverse strand. Black stars represent unmodified T’s, while a modified position is denoted by a red star. B = biotin and I = IdU.
We also compared modifications attached to the training template by click cycloaddition—an azide, nicotine, glucose, AF488 and AF647. In regions outside the modified base, we did not observe significant changes in signals, indicating that the click reaction did not affect the ability to sequence DNA on the MinION (e.g. compare position T3063 and T3083 on the control and experimental templates in Figure 2A and F). At the modified base, nicotine showed a significant change with a log(combined P-value) = –74 (Figure 2F). Bulky adducts caused a wider spread in signal, which generally affected 5–6 bases preceding the modification and about 2–4 bases downstream (Figure 2D, F, G). Clicked moieties AF488 and AF647 caused a more extensive downstream current shift, spreading as far as 8–10 bases beyond the modified position (Figure 2J, K). The mean raw signal at the modified base was visibly different when compared to control, though the change did not translate into lower P-values in the NanoMod statistical analysis. The largest clicked modification—a ssDNA oligo, expected to generate a branched structure, caused the smallest change in signal, which spread over 10 bases upstream and 2–3 positions downstream of 3073 (Figure 2L). In this particular case, reduced progression through the pore or signal integration from bases on the training template and the clicked oligo might have contributed to the low shift in current, recorded for the modified position.
These observations point to a relationship between adduct size and the magnitude of signal shift at the modified position, as well as, the neighboring bases. In general, bulkier moieties caused more pronounced changes in the raw signal at the modified position, and affected more bases in its vicinity. The differences in signal spread between adducts of different molecular weight show that pairs of analogs, such as IdU and biotin, can be distinguished from each other based on the width of the window affected by nanopore current shifts (Figure 2E and I). To test this experimentally, we prepared a training sequence with two different thymidine analogs—biotin at position 3049 and IdU at position 3098 on the same strand of the template (Figure 1B). Following purification with streptavidin beads, the training sequence was run on the MinION. Analysis with NanoMod showed that both modifications could be clearly detected at the expected positions (Figure 2M and Supplementary Figure S1B). The signal at Biotin 3049 had a log(combined P-value) = –300, spreading over 7–9 base pairs upstream and downstream of the modification (Figure 2M and Supplementary Figure S1B), similar to the log(combined P-value) of –250, recorded for the single Biotin modification at 3073 on Figure 2I. The IdU modification at 3098 had a log(combined P-value) of –200 and affected 3–4 neighboring bases, similarly to the single IdU at 3073 for which the log(combined P-value) was –175 (Figure 2E). The clear differences in signal magnitude and the extent of signal spread over the neighboring bases allowed us to distinguish the biotin analog from IdU in a blinded experimental set up.
These observations collectively demonstrate that various thymidine analogs, sequenced on the MinION platform, can be detected with NanoMod analysis. Furthermore, bulkier moieties, such as biotin can be distinguished from smaller analogs, such as IdU, which is required for dual color labeling and conclusive identification of DNA synthesis directionality.
Detection of multiple DNA modifications with nanopore sequencing
To mimic the scenario of pulsing replicating cells with nucleotide analogs, which are incorporated at multiple positions into newly synthetized DNA, we repaired the nb.BbvCI nicks on the minus strand of the training sequence by ligating an oligo with 14 thymidine modifications between positions 3042 and 3104. Based on the results with a single defined position, we selected three analogs—EdU, CldU and IdU (Figure 1B). Efficient replacement was confirmed by dot blots for CldU and IdU (Figure 1D). Templates with replacements were sequenced and analysis was carried out by dividing the reference sequence into ∼400 regions of 60 bp each with a 30-bp overlap between adjacent windows. For all analogs tested, NanoMod correctly identified the modified region between 3042 and 3104, as shown by the P-values for Kolmogorov-Smirnov test and Stouffer's method (Supplementary Figure S1C–E). The most significant shift of signal was centered around a modified base, gradually abated with increasing distance from the analog and raised again in the vicinity of the next modification on the template. Within each template, some modified thymidines generated stronger signals than others (Supplementary Figure S1C–E). As in templates with single modifications, the magnitude of signal shift in sequences with multiple analogs was determined by the size of the adducts with IdU generating the greatest signal to noise ratio (Supplementary Figure S1E). This dataset on multiple modifications demonstrates that MinION sequencing and analysis with NanoMod can be successfully used to identify short genomic regions with modified bases in DNA.
A comparative analysis of nucleotide analogs for gDNA modifications
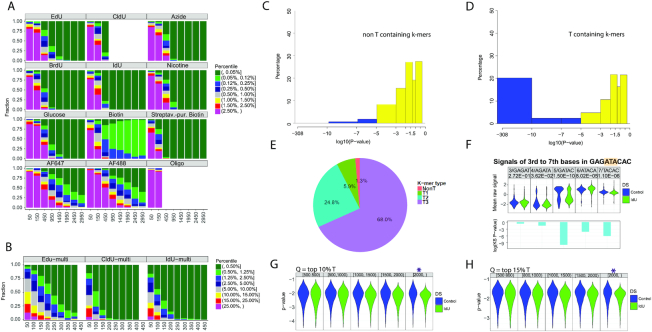
To determine if pore sequencing and the analysis method NanoMod, developed here, can be applied to sequencing base analogs, incorporated by replicating cells in genomic DNA, we performed computational simulations. We first determined the level of analog substitution in vivo by exposing primary human fibroblasts to EdU for 24h, or a complete cell cycle. Purified, EdU-containing genomic DNA was then labeled with AF647 in a click cycloaddition reaction and the level of EdU-AF647 substitution was determined by measuring sample fluorescence against a standard curve of PCR products with known percentage substitutions of EdU, clicked with AF647. This analysis revealed that 20% of thymidine bases in genomic DNA were substituted with EdU (Supplementary Figure S2A, B). This is the expected percent modification for replication tracts pulsed with thymidine analogs (19). Based on this result, we performed computational simulations with 20% modified and 80% unmodified reads to determine what coverage would be required to detect single (Figure 3A) and multiple (Figure 3B) modifications in genomic DNA. Since the fraction of modified reads without purification is approximately 75% (14), the proportion of modified reads in the simulation is ∼15%. IdU and CldU were easiest to detect in the group of analogs that can be directly incorporated during DNA synthesis, requiring 450 reads of a modified site to rank strands with a single modification in the 0.05 percentile or less (Figure 3A). The calling of BrdU and the calling of single EdU-modified strands also obtained the highest 0.05 percentile rank with as low as 450 or 950 reads, respectively. Similarly, Biotin modifications were ranked in the 0.05 percentile range with only 450 reads. The clicked analogs-an azide group, nicotine and glucose required 950 reads, almost twice the number noted for IdU and CldU, to achieve the 0.05 percentile rank. The clicked fluorophores AF488 and AF647 required more extensive coverage—1950 and 2450 reads, respectively for more than 75% of the strands to be ranked in the 0.05 percentile or between percentiles 0.05 and 0.12. This result is consistent with the wide-spread signals of lower significance, described for AF488, AF647 and the ssDNA oligo in Figure 2. This result corroborates the observation that thymidine analogs of higher molecular weight—IdU, and biotin are readily detected on the MinION platform.
Figure 3.
Detection of IdU, incorporated into replicating mammalian DNA. Computational modeling to determine the sequencing depth, required for calling base analogs in mixed samples of modified and unmodified reads, using (A) single modified base templates, (B) multi-modified templates. The X axis represents the number of reads used for the analysis, and the Y axis shows the fraction a modified site can be ranked by percentile among all sites for each sample. Analysis was performed using Stouffer's combined statistic. IdU and CldU modifications cause the strongest signal shifts and require the least number of reads for reliable detection. (C) combined P-values calculated with Stouffer's method for non-T containing 9-mers in the mouse genome. The combined P-value includes P-values for the central base of the 9-mer, as well as, 2 bases upstream and downstream. (D) Combined P-values for T-containing k-mers in the mouse genome. All T-containing 9-mers were considered, including ones with multiple Ts. (E) Pie chart showing the percentage of 9-mers with P-values less than 10−7. Non-T, 1T, 2T and 3T-containing k-mers are shown in red, green, blue and purple, respectively. (F). Signal changes as a selected 9-mer from IdU-substituted mouse genomic DNA with the sequence GAGATACAC was separated in five different k-mers. The k-mers were covered by 200–400 reads. (G) Detection of single modified reads. The ranges of read lengths are indicated at the top of the panel. The purple star indicates the read length required for the detection of individual IdU-substituted genomic DNA reads when integrating signals from k-mers with P-values in the 10% and the H). 15% percentile. Q = quantile.
To determine the number of reads required to identify a region with multiple modifications, we performed the same simulation with 14 thymidine analogs in a single template (Figure 3B). Multiple EdU modifications required 250 reads for reliable detection (percentile 0.05 or between the 0.05 and 0.12 percentiles). For other analogs CldU and IdU, only 100 reads proved enough for correct calling (percentile 0.05 or between the 0.05 and 0.12 percentiles). This analysis showed that the presence of multiple modified bases in a DNA segment facilitates detection with NanoMod, requiring a smaller number of reads compared to a single modification. IdU was the modification that was most readily detectable in the modeling of both single and multiple modifications.
IdU incorporated in replicating mammalian DNA can be detected on single DNA strands
To test if the MinION platform in conjunction with NanoMod can detect nucleotide analogs, incorporated in replicating DNA, we incubated mouse embryonic stem (ES) cells from a pure C57BL/6J strain with 25uM IdU for 24h. Genomic DNA from unlabeled and IdU-incubated samples was then harvested, sheared in 6–8 kb fragments and prepared for 1D sequencing on the MinION system. Error correction and signal annotation were performed with NanoMod. Base calling was accomplished with Albacore v2.3.1 and long reads were aligned with the GRCm38 (mm10) mouse reference genome. We aligned all reads from the IdU sample to the mouse reference genome and used a set of IdU-covered reads and control reads in the subsequent analysis. These reads were from different, rather than the same genomic location. The reference genome was split into 9-mers with no thymidines (XVVVVVVVX) where V = AGC and X is any base, 1 thymidine in the center of the k-mer (XVVVTVVVX) or two and more thymidines at different positions on the plus strand of the k-mer. This classification included 335 non-T k-mers and 668 T-containing k-mers of different sequence composition with coverage of >500 reads per k-mer. Current signals were collected from the central T, as well as, two nucleotides upstream and two downstream of the base and used in a Kolmogorov–Smirnov test to calculate P-values. Stouffer's method was then applied to calculate a combined P-value for each 9-mer. The procedure was repeated 50 times and the median of the resulting 50 P-values was plotted on a logarithmic scale. Comparing IdU labeled samples with unlabeled controls revealed that 20% of the T-containing k-mers in the IdU sample had combined log10P-values less than –10, while none of the non-T k-mers had a P-value in this range, showing that IdU substitution in T-containing k-mers generates a detectable signal shift (Figure 3C, D). Comparison of k-mers containing either 1T or 2T resulted in greater significance of detected differences: 25% of 1T and almost 40% of 2T-contaning 9-mers had combined log10P-values less than –7 (Supplementary Figure S2C, D). Analysing k-mers with P-values below 10−7 showed that only 1.3% of non-T k-mers have P-values in this range, while 5.9% of the 1T, 24.8% of the 2T and 68% of the 3T containing k-mers had signals with P-values <10−7 (Figure 3E). This result demonstrates that the method employed here has low background and can detect incremental signal differences generated by the presence of increasing numbers of IdU-substituted thymidines. To determine whether signals from a k-mer of a specific sequence could be used to distinguish IdU from thymidine, two selected 9-mers GAGATACAC and GAGTTACAC were divided into five different k-mers. Electrical signals from control and IdU-labeled samples of the same k-mer were plotted and compared using NanoMod. A significant shift was seen in the IdU containing k-mers, with the shift depending on the position of the T within the k-mer (Figure 3F and Supplementary Figure S2E). This analysis used 427IdU/229 control reads for GAGATACAC and 310IdU/211 control for GAGTTACAC to identify an IdU containing k-mer in cellular DNA. This analysis shows that thymidine analogs, incorporated by replicating mammalian cells, can be called with nanopore sequencing, and that T containing k-mers from different locations of the genome can be combined for the calling of base modifications. The combination of ∼200 different alignments per k-mer should provide a tool to identify replicated DNA from long nanopore reads.
To determine if we can detect single IdU-substituted DNA molecules with NanoMod, we randomly selected 70% of the reads in the control genomic DNA sample with T-containing k-mers from different genomic locations. We calculated the signal mean and standard deviation for the central 5 bases (central T as well as 2 bases upstream and 2 bases downstream) of each 9-mer. These results were used to calculate a combined P-value for each T in the remaining 30% of the T-containing k-mers in the control genomic DNA sample and the IdU-substituted sample with the following thresholds for read length: 500, 800, 1000, 1500 and 2000 bp. This analysis showed that IdU-substituted single reads of length 2.0 kb or more can be detected with NanoMod in the 10% and 15% percentile (Figure 3G, H). These results demonstrate that IdU, incorporated in newly-synthesized mammalian DNA can be detected on individual DNA molecules with MinION sequencing and NanoMod analysis.
Quantifying mitochondrial replication rates with Replipore Sequencing
To analyze DNA replication in mammalian cells, we focused on the mitochondrial genome. This allows for high coverage without requiring a large number of flow cells as for a complete mammalian genome. Human mitochondria contain a genome of 16,569 bp, which can be labeled with nucleotide analogs (20). We adapted a CRISPR/Cas9-based enrichment approach to increase the coverage of human mitochondrial DNA (hmtDNA). We induced a Cas9-mediated cut to linearize the mitochondrial genome at position 8146–8147 and used the 5′ phosphate at the cut site for the ligation of the MinION sequencing adapter (Figure 4A). We determined the levels of enrichment for hmtDNA in two high molecular weight gDNA samples from unlabeled and 8 h-IdU-labeled human embryonic stem cells. Both samples were run on the same flow cell with an average sequencing depth of the human genome of 0.54× in the control and 0.33× in the IdU-labeled sample (Supplementary Figure S2F). The coverage of the mitochondrial genome in both samples was enriched by a factor of 7000× in the control and 6500× in the sample, incubated with IdU.
The mitochondrial genome has a light (CA-rich) and a heavy (GT-rich) strand, which were sequenced as the forward and reverse strands on the MinION, respectively (Figure 4A). The light strand was predominantly represented by 16kb long reads, while the heavy strand showed two read pools- 8kb and 16kb in both the control and IdU-labeled sample (Figure 4B). The ∼8kb reads in the heavy strand occur due to the position of the Cas9 cut at 8.1kb and read direction clockwise toward the OH (Figure 4A and Supplementary Figure S2H) on DNA molecules that are not yet ligated at the initiation site and form a nicked genome (21). In addition, the heavy strand is underrepresented relative to the light strand: up to 7000 16kb long reads covered the light, while only 500 covered the heavy strand in the control sample (Figure 4B). A similar result was recorded for the 8h-IdU labeled sample: over 4000 16kb long reads for the light strand, compared to about 100 for the heavy strand (Figure 4B). This difference is consistent with the RITOLS (RNA Incorporation Throughout the Lagging Strand) model of mitochondrial DNA replication, which describes base-pairing between the parental heavy strand and RNA (22). Efficient Cas9-mediated enrichment requires dsDNA; RNA/DNA hybrids, converted to ssDNA as a result of RNAse A treatment (23) during sample preparation depress the coverage of the heavy strand as S. pyogenes Cas9 does not cleave ssDNA efficiently (24) (Supplementary Figure S2H).
We next computed P-values in the 10% percentile for 16kb long reads and found significant differences when reads from the IdU-labeled sample were compared to the unlabeled control for both the heavy and light strands (Figure 4C and Supplementary Figure S2G). A significant result was also seen in the analysis of 8kb reads from the heavy strand (Figure 4C). The 8kb reads from the light strand showed differences, though they were not significant. This analysis defines the range of signal from unlabeled reads.
To distinguish and segregate labeled reads of length 16kb and 8kb from unlabeled ones, we next calculated P-values in sliding windows of 200 bp in the IdU-incubated sample. To set the baseline for IdU detection, we compared reads, basecalled from even and odd data folders from the unlabeled control sample to each other (Figure 4D). We then compared unlabeled 16kb reads from the IdU-incubated sample (P-values above the 25% percentile) to reads from the control, without detecting any signal (Figure 4E). Next, we compared selected 16kb labeled reads (P-values below the 25% percentile) from the IdU-incubated sample to reads from the control which resulted in log(10) P-values between −6 and −3, clearly indicating the presence of IdU label (Figure 4F). Since the heavy strand was represented by two pools of reads- 16kb and 8kb, we decided to extend the same analysis to 8kb reads from the heavy strand. Thus, we selected unlabeled 8kb reads (P-value above the 25% percentile) and labeled 8kb reads (P-value below the 25% percentile) and compared them to reads from the unlabeled control (Figure 4G, H). While the comparison of 8kb unlabeled reads from the IdU-incubated sample to control remained close to baseline (Figure 4G), the log(10) P-values from the comparison of 8kb labeled reads from the IdU-treated sample to the control dropped to a range between −5 and −3 (Figure 4H), demonstrating the detection of IdU label in newly synthesized DNA strands. This analysis led to the identification of 759/4404 (17%) labeled light strand and 26/104 (25%) labeled heavy strand reads in the 16kb read pool (Figure 4I). The number of 8kb labeled reads was 377/1293 (29%) for the light strand and 65/168 (39%) for the heavy strand (Figure 4I). The greater percentage of labeled heavy strands in both the 16kb and 8kb read pools is consistent with the initiation of DNA replication from OH prior to OL.
The total number of labeled 16kb and 8kb reads was 1227/5969 (21%). Because DNA replication is semiconservative and at most half of the DNA strands are expected to be labeled within a complete cell cycle, we conclude that in a timeframe of 8h, about 42% of mtDNA has replicated. This replication rate allows mtDNA homeostasis in human embryonic (hES) stem cells, which have a cell cycle of about 15–20 h (25). This analysis shows that our tools in combination with the MinION sequencing platform can be used for identifying and counting the number of IdU-labeled strands in human mitochondrial DNA.
DISCUSSION
Analysis of DNA replication and repair at a single molecule level with a readily amenable sequencing platform would greatly increase our understanding of DNA replication and genetic stability in development and disease. Established methods, such as fiber analysis and in situ hybridization are limited in resolution and scalability. Towards this aim, we show here that Replipore sequencing can be used to detect a panel of thymidine modifications in synthetic templates as well as IdU, incorporated in the DNA of replicating mammalian cells. Compounds, which are chemically alike, but with different molecular weights or different structure, can generate distinct signals when passing through a pore (26). Here, we present increased signal changes with increasing molecular weight from EdU, CldU and BrdU to IdU with a signal spread affecting 2–4 neighboring bases on each side of the analog. Structurally bulkier moieties of high molecular weight, such as biotin, generated large signal shifts spreading as far as 10 nucleotides in the neighborhood of the modified base. Larger polymers have been reported to have increased pore residence times compared to shorter molecules (27) and thus the extensive signal spread with biotin-modified DNA may be due to longer dwell times in the pore. The difference in signal spread between small analogs and bulky adducts can be leveraged for identification purposes. In this study, we demonstrate that NanoMod analysis can be used to distinguish between Biotin and IdU when present on the same DNA strand. Studies on the progression of DNA replication may use the sequential application of IdU and EdU with the intent to modify the latter further with Biotin and form a larger base. While we show proof of principle of dual color labeling and sequencing using a plasmid, identifying alternating tracks of IdU and Biotin in large mammalian genomes will require further development of the current algorithms. The use of two analogs is routinely applied in studies of fork progression through DNA combing, which is not associated with sequence information. The ability to add information on location in the genome on single DNA strands through pore sequencing would greatly facilitate our understanding of the pattern of DNA synthesis in normal and abnormal cells. To this end, we provide a proof of principle that noncanonical bases that can be incorporated into cells during DNA replication can be detected with nanopore sequencing and that bulky adducts can be distinguished from smaller analogs.
In this study, we also demonstrate that click cycloaddition reactions can be used to generate any desired chemical change after incorporation of a nucleotide with a reactive group. These analogs could be detected without further modification, and the clicking of bulky adducts again showed a wider spread in signals. Though click chemistry with copper catalysts can introduce DNA damage, novel copper-free click is now available and will likely be more suitable for this application (28). For example, AmdU is an azide group-containing analog, which is incorporated during DNA replication and reacts with alkynes in copper-free reactions, allowing the formation of a bulky adduct while protecting DNA from damage (29, 30). A base with a vinyl-group, such as VdU, also allows the introduction of further modifications via copper-free alkene-tetrazine ligation (31).
We chose IdU to establish detection of nucleotide analogs in mammalian cellular DNA, as it performed well in modeling to detect a modified nucleotide when only 20% of T’s are substituted. By applying IdU label to human embryonic stem cells through a complete cell cycle, we demonstrate that IdU-substituted genomic DNA can be distinguished from control DNA. A coverage of 200–300 reads per 9-mer was sufficient to observe significant differences. As these k-mers are from different locations of the genome, k-mers from the same read may be combined for the analysis of a labeled DNA strand, thereby amplifying the signal and reducing the coverage requirement. We used this strategy to detect IdU-substitutions on individual DNA molecules and showed that single IdU-containing reads of 2.0 kb can be called with NanoMod when sampling from the top 10% lowest P-values of T-containing k-mers along a single read.
To apply our tools to a biological problem, we chose to study and detect replication of the human mitochondrial genome. Mitochondria are organelles involved in cellular respiration and defects in mitochondrial DNA replication are associated with a wide spectrum of disorders, such as Alpers syndrome, ataxia-neuropathy syndrome, epilepsy and others, (reviewed in Milone et. al (32)). We demonstrated that CRISPR/Cas9-based enrichment for human mitochondrial DNA results in increase of coverage of up to 7000× on a single MinION flow cell. Most of the reads in our pool were from the light strand of the mitochondrial genome. The reduced coverage of the heavy strand is likely due to the presence of RNA/DNA hybrids of the parental heavy strand, as predicted by the RITOLS model (22), which are not captured after RNAse A treatment and Cas9-mediated target enrichment. The heavy strand was represented in two read populations, 8kb and 16kb, due to a nick at OH in replicated, but not yet ligated mtDNA molecules. Our analysis with NanoMod revealed that IdU-labeled strands can be identified and counted as a fraction of all sequenced reads, showing that about 42% of mtDNA molecules replicate within a period of 8h in human embryonic stem cells, consistent with the replication rates required to maintain mtDNA homeostasis. Therefore, this method provides a path to studying mitochondrial DNA replication dynamics. Such information is valuable for the analysis of differential replication when two different mtDNA haplotypes are present in the same cell after mitochondrial replacement. Competition between two mtDNA haplotypes determines the efficacy of mitochondrial replacement therapy (33), intended to prevent the genetic transmission of mitochondrial disease.
The MinION has been successfully employed for the assembly of eukaryotic genomes, such as yeast (34), C. elegans (35) and the human genome with a median coverage of 26×, though it required 43 flow cells (36). Therefore, nanopore sequencing is an accessible technology that can readily be applied to organisms with small genomes. Recently, the MinION has also been used for the analysis of DNA replication of the smaller yeast genome using the BrdU analog (37). For the larger mammalian genomes, the approach of CRISPR/Cas9-mediated enrichment, as used here, is currently more practical and will be useful for on target sequencing of specific genomic regions. An important question for future studies is whether the MinION can be applied to the study DNA replication progression at repetitive DNA regions, such as the insulin ILPR associated with type 1 diabetes, or the C9ORF72 repeats causing frontotemporal dementia. A similar approach has recently been used for the study of DNA methylation at repetitive loci (16). CRISPR/Cas9-mediated enrichment may be multiplexed, such that multiple regions are analyzed in a single sequencing run, providing information on replication progression that thus far requires the specialized skills of fiber isolation, combing and in situ hybridization (38).
We conclude that sequencing DNA replication using nanopores, termed here Replipore sequencing, is a novel technology with significant potential in research and diagnostics. DNA replication is the fundamental requirement of cell proliferation and regeneration, and occurs in a cell type and locus-specific manner. Abnormalities in DNA replication can result in genome instability, cellular senescence or apoptosis, and the increase of toolsets to study replication is therefore highly relevant to regenerative medicine. Insights about polymerase progression, derived from sequencing modified nucleotides through nanopores, will likely prove critical to improving our understanding and ultimately treatment of diseases, characterized by abnormalities in DNA replication.
DATA AVAILABILITY
NanoMod is an open source software, available at https://github.com/WGLab/NanoMod.
The datasets generated and analyzed in this study have been deposited in the Sequence Read Archive (SRA) under study accession PRJNA563770 (biological samples: SAMN12687803, SAMN12692999, SAMN14479298 and SAMN14479297).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Marcus Stoiber from the Lawrence Berkeley National Laboratory of Environmental Genomics and Systems Biology in Berkeley, CA for assistance in initial nanopore modification detection.
Contributor Information
Daniela Georgieva, Integrated Program in Cellular, Molecular, and Biomedical Studies, Columbia University, New York, NY 10032, USA; Naomi Berrie Diabetes Center, Columbia University, New York NY 10032, USA; Columbia Stem Cell Initiative, Columbia University, New York, NY 10032, USA.
Qian Liu, Raymond G. Perelman Center for Cellular and Molecular Therapeutics, Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA.
Kai Wang, Raymond G. Perelman Center for Cellular and Molecular Therapeutics, Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA; Department of Pathology and Laboratory Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Dieter Egli, Naomi Berrie Diabetes Center, Columbia University, New York NY 10032, USA; Columbia Stem Cell Initiative, Columbia University, New York, NY 10032, USA; Department of Pediatrics and Department of Obstetrics and Gynecology, Columbia University, New York, NY 10032, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Naomi Berrie Diabetes Center of Columbia University; NIH [R21 HG010165-01A1]; CHOP Research Institute (to K.W.); D.E. personal funds. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hansen R.S., Thomas S., Sandstrom R., Canfield T.K., Thurman R.E., Weaver M., Dorschner M.O., Gartler S.M., Stamatoyannopoulos J.A.. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith L., Plug A., Thayer M.. Delayed replication timing leads to delayed mitotic chromosome condensation and chromosomal instability of chromosome translocations. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:13300–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hiratani I., Ryba T., Itoh M., Yokochi T., Schwaiger M., Chang C.-.W., Lyou Y., Townes T.M., Schübeler D., Gilbert D.M. et al.. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS. Biol. 2008; 6:e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pope B.D., Ryba T., Dileep V., Yue F., Wu W., Denas O., Vera D.L., Wang Y., Hansen R.S., Canfield T.K. et al.. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014; 515:402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu J., Li H., Hu M., Sasaki T., Baccei A., Gilbert D.M., Liu J.S., Collins J.J., Lerou P.H.. The distribution of genomic variations in human iPSCs is related to replication-timing reorganization during reprogramming. Cell Rep. 2014; 7:70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Amiel A., Kolodizner T., Fishman A., Gaber E., Klein Z., Beyth Y., Fejgin M.D.. Replication pattern of the p53 and 21q22 loci in the premalignant and malignant stages of carcinoma of the cervix. Cancer. 1998; 83:1966–1971. [PubMed] [Google Scholar]
- 7. Haye-Bertolozzi J.A., Oscar M.. Quantitative bromodeoxyuridine immunoprecipitation analyzed by high-throughput sequencing (qBrdU-Seq or QBU). In: Muzi-Falconi M., Brown G. (eds) Genome instability. Methods Mol. Biol. 2018; 1672:209–225. [DOI] [PubMed] [Google Scholar]
- 8. McGuffee S.R., Smith D.J., Whitehouse I.. Quantitative, genome-wide analysis of eukaryotic replication initiation and termination. Mol. Cell. 2013; 50:123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koren A., Handsaker R.E., Kamitaki N., Karlić R., Ghosh S., Polak P., Eggan K., McCarroll SA.. Genetic variation in human DNA replication timing. Cell. 2014; 159:1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Norio P., Kosiyatrakul S., Yang Q., Guan Z., Brown N.M., Thomas S., Riblet R., Schildkraut C.L.. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol. Cell. 2005; 20:575–587. [DOI] [PubMed] [Google Scholar]
- 11. Jenjaroenpun P., Wongsurawat T., Pereira R., Patumcharoenpol P., Ussery D.W., Nielsen J., Nookaew I.. Complete genomic and transcriptional landscape analysis using third-generation sequencing: a case study of Saccharomyces cerevisiae CEN.PK113-7D. Nucleic Acids Res. 2018; 46:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goodwin S., Gurtowski J., Ethe-Sayers S., Deshpande P., Schatz M.C., McCombie WR.. Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res. 2015; 25:1750–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rand A.C., Jain M., Eizenga J.M., Musselman-Brown A., Olsen H.E., Akeson M., Paten B.. Mapping DNA methylation with high-throughput nanopore sequencing. Nat. Methods. 2017; 14:411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Luzzietti N., Knappe S., Richter I., Seidel R.. Nicking enzyme-based internal labeling of DNA at multiple loci. Nat. Protoc. 2012; 7:643–653. [DOI] [PubMed] [Google Scholar]
- 15. Liu Q., Georgieva D.C., Egli D., Wang K.. NanoMod: a computational tool to detect DNA modifications using Nanopore long-read sequencing data. BMC Genomics. 2019; 20:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giesselmann P., Brändl B., Raimondeau E., Bowen R., Rohrandt C., Tandon R., Kretzmer H., Assum G., Galonska C., Siebert R. et al.. Analysis of short tandem repeat expansions and their methylation state with nanopore sequencing. Nat. Biotechnol. 2019; 37:1478–1481. [DOI] [PubMed] [Google Scholar]
- 17. Liang L., Astruc D.. The copper(I)-catalyzed alkyne-azide cycloaddition (CuAAC) ‘click’ reaction and its applications. An overview. Coord. Chem. Rev. 2011; 255:2933–2945. [Google Scholar]
- 18. Hong V., Presolski S.I., Ma C., Finn M.G.. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew. Chem. Int. Ed. Engl. 2009; 48:9879–9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Russev G.C.T., Tsanev R.G.. Continuous labeling of mammalian DNA in vivo. Analyt. Biochem. 1973; 54:115–119. [DOI] [PubMed] [Google Scholar]
- 20. Phillips A.F., Millet A.R., Tigano M., Dubois S.M., Crimmins H., Babin L., Charpentier M., Piganeau M., Brunet E., Sfeir A. et al.. Single-molecule analysis of mtDNA replication uncovers the basis of the common deletion. Mol. Cell. 2017; 65:527–538. [DOI] [PubMed] [Google Scholar]
- 21. Macao B., Uhler J.P., Siibak T., Zhu X., Shi Y., Sheng W., Olsson M., Stewart J.B., Gustafsson C.M., Falkenberg M. et al.. The exonuclease activity of DNA polymerase gamma is required for ligation during mitochondrial DNA replication. Nat. Commun. 2015; 6:7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yasukawa T., Reyes A., Cluett T.J., Yang M.Y., Bowmaker M., Jacobs H.T., Holt I.J.. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO. J. 2006; 25:5358–5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barbieri C.L., Li T.K., Guo S., Wang G., Shallop A.J., Pan W., Yang G., Gaffney B.L., Jones R.A., Pilch D.S.. Aminoglycoside complexation with a DNA RNA hybrid duplex: the thermodynamics of recognition and inhibition of RNA processing enzymes. Am. Chem. Soc. 2003; 125:6469–6477. [DOI] [PubMed] [Google Scholar]
- 24. Ma E., Harrington L.B., O’Connell M.R., Zhou K., Doudna J.A.. Single-stranded DNA cleavage by divergent CRISPR-Cas9 enzymes. Mol. Cell. 2015; 60:398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Becker K.A., Ghule P.N., Therrien J.A., Lian J.B., Stein J.L., van Wijnen A.J., Stein G.S.. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J. Cell. Physiol. 2006; 209:883–893. [DOI] [PubMed] [Google Scholar]
- 26. Li-Qun G.U., Braha O., Conlan S., Cheley S., Bayley H.. Stochastic sensingof organic analytesbyapore-forming protein containing amolecularadapter. Nature. 1999; 398:686–690. [DOI] [PubMed] [Google Scholar]
- 27. Joseph W.F, Robertson C.G.R., Stanford V.M., Rubinson K.A., Krasilnikov O.V., Kasianowicz J.J.. Single-molecule mass spectrometry in solution using a solitary nanopore. PNAS. 2007; 104:8207–8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abel G.R. Jr, Calabrese Z.A., Ayco J., Hein J.E., Ye T.. Measuring and suppressing the oxidative damage to DNA during Cu(I)-catalyzed azide-alkyne cycloaddition. Bioconjug. Chem. 2016; 27:698–704. [DOI] [PubMed] [Google Scholar]
- 29. Tera M., Glasauer S.M.K, Luedtke N.W.. In vivo incorporation of azide groups into DNA by using membrane-permeable nucleotide triesters. Chembiochem. 2018; 19:1939–1943. [DOI] [PubMed] [Google Scholar]
- 30. Neef A.B., Luedtke N.W.. An azide-modified nucleoside for metabolic labeling of DNA. Chembiochem. 2014; 15:789–793. [DOI] [PubMed] [Google Scholar]
- 31. Rieder U., Luedtke N.W.. Alkene-tetrazine ligation for imaging cellular DNA. Angew. Chem. Int. Ed. Engl. 2014; 53:9168–9172. [DOI] [PubMed] [Google Scholar]
- 32. Milone M., Massie R.. Polymerase gamma 1 mutations: clinical correlations. Neurologist. 2010; 16:84–91. [DOI] [PubMed] [Google Scholar]
- 33. Yamada M., Emmanuele V., Sanchez-Quintero M.J., Sun B., Lallos G., Paull D., Zimmer M., Pagett S., Prosser R.W., Sauer M.V. et al.. Genetic drift can compromise mitochondrial replacement by nuclear transfer in human oocytes. Cell. Stem. Cell. 2016; 18:749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Istace B., Friedrich A., d’Agata L., Faye S., Payen E., Beluche O., Caradec C., Davidas S., Cruaud C., Liti G. et al.. de novo assembly and population genomic survey of natural yeast isolates with the Oxford Nanopore MinION sequencer. Gigascience. 2017; 6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tyson J.R., O’Neil N.J., Jain M., Olsen H.E., Hieter P., Snutch T.P.. MinION-based long-read sequencing and assembly extends the Caenorhabditis elegans reference genome. Genome Res. 2018; 28:266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A., Tyson J.R., Beggs A.D., Dilthey A.T., Fiddes I.T. et al.. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018; 36:338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muller C.A., Boemo M.A., Spingardi P., Kessler B.M., Kriaucionis S., Simpson J.T., Nieduszynski C.A.. Capturing the dynamics of genome replication on individual ultra-long nanopore sequence reads. Nat. Methods. 2019; 16:429–436. [DOI] [PubMed] [Google Scholar]
- 38. Gerhardt J., Tomishima M.J., Zaninovic N., Colak D., Yan Z., Zhan Q., Rosenwaks Z., Jaffrey S.R., Schildkraut C.L.. The DNA replication program is altered at the FMR1 locus in fragile X embryonic stem cells. Mol. Cell. 2014; 53:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NanoMod is an open source software, available at https://github.com/WGLab/NanoMod.
The datasets generated and analyzed in this study have been deposited in the Sequence Read Archive (SRA) under study accession PRJNA563770 (biological samples: SAMN12687803, SAMN12692999, SAMN14479298 and SAMN14479297).