Abstract
H/ACA small nucleolar RNAs (snoRNAs) guide pseudouridylation as part of a small nucleolar ribonucleoprotein complex (snoRNP). Disruption of H/ACA snoRNA levels in stem cells impairs pluripotency, yet it remains unclear how H/ACA snoRNAs contribute to differentiation. To determine if H/ACA snoRNA levels are dynamic during differentiation, we comprehensively profiled H/ACA snoRNA abundance in multiple murine cell types and during differentiation in three cellular models, including mouse embryonic stem cells and mouse myoblasts. We determined that the profiles of H/ACA snoRNA abundance are cell-type specific, and we identified a subset of snoRNAs that are specifically regulated during differentiation. Additionally, we demonstrated that a decrease in Snora27 abundance upon differentiation corresponds to a decrease in pseudouridylation of its target site within the E-site transfer RNA (tRNA) binding region of the 28S ribosomal RNA (rRNA) in the large ribosomal subunit. Together, these data point toward a potential model in which H/ACA snoRNAs are specifically regulated during differentiation to alter pseudouridylation and fine tune ribosome function.
INTRODUCTION
Pseudouridine, which results from the isomerization of a uridine base, is an essential RNA modification that regulates gene expression through diverse mechanisms including translation efficiency and fidelity (1–3). Pseudouridine is the most abundant RNA modification (4,5). In mouse or human rRNA, 7–8% of uridines are pseudouridines (6). They are clustered in functionally important regions of the ribosome, including the peptidyl transferase center, tRNA and messenger RNA (mRNA) binding sites, and the subunit interface (7–12). Pseudouridines are present in all major types of cellular RNAs, and pseudouridylation of mRNA, rRNA and noncoding RNAs is inducible in response to cell stress or cell signaling (13–17). Additionally, snoRNA-mediated changes in pseudouridylation are developmentally regulated during two life cycle stages of Trypanosoma brucei (18). As a result, dynamic pseudouridylation may fine-tune ribosome function and gene expression in response to changes in the cellular environment or during development.
Pseudouridylation is catalyzed by enzymes called pseudouridine synthases. Pseudouridine synthases are either RNA-independent, achieving substrate specificity by recognizing specific sequences or structures within their target RNA, or are RNA-dependent, forming box H/ACA snoRNPs and relying upon box H/ACA snoRNAs to guide them to their target RNAs (1,19,20). Here, we focus on the box H/ACA snoRNPs, which are composed of one of many H/ACA snoRNAs and four core proteins: DKC1, the pseudouridine synthase; NOP10; NHP2 and GAR1 (21–26). The H/ACA snoRNAs impart target specificity by base-pairing with substrate RNAs ranging from rRNAs to small nuclear RNAs (snRNAs) to mRNAs and directing the box H/ACA snoRNP to convert the target uridine to pseudouridine (Figure 1A; 27–29).
Figure 1.
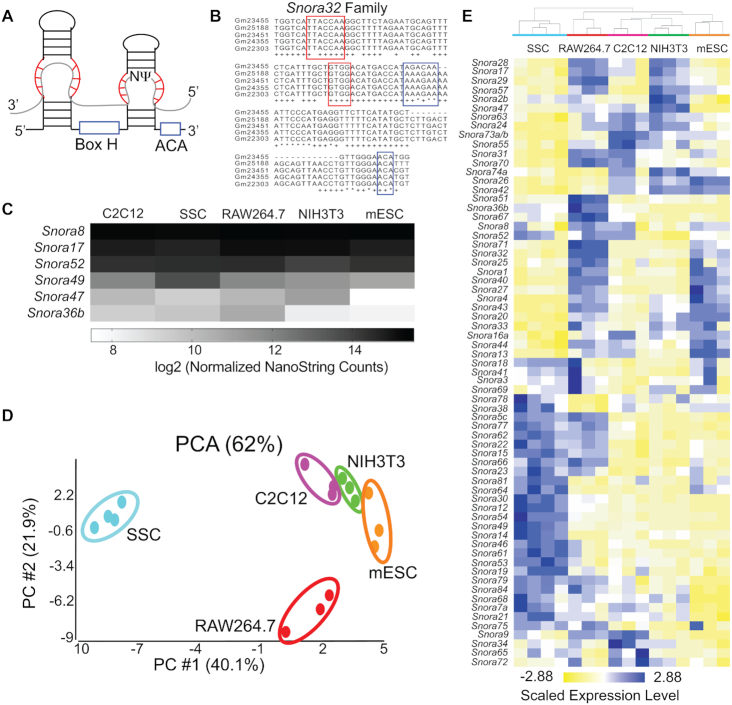
H/ACA snoRNA expression is heterogeneous. (A) Diagram of H/ACA snoRNA structure. The conserved box H and ACA sequence motifs are outlined in blue and the regions of the snoRNA that base pair with target RNAs (gray) are outlined in red. (B) Multiple-sequence alignment of members of the Snora32family in Mus musculus.Sequences of the five Snora32 family members that were detected by small-RNA sequencing of mESCs are shown.The remaining 11 members of the SNORA32 family are either expressed in other tissue or cell types or are pseudogenes and are not expressed at all. The regions that base pair with target RNA(s) are outlined in red. The box H and ACA sequence motifs are outlined in blue. Nucleotides that are identical in all 16 Snora32 family members are denoted with an * and nucleotides that are conserved in ≥75% of family members are denoted with a +. The lengths of Snora32 family members vary from 79–133 nucleotides, and three family members appear to be truncated at the 5′ end. (C) Heat map of the log2 transformed average, normalized snoRNA levels as quantified by NanoString nCounter from C2C12 cells, SSCs, RAW264.7 cells, NIH3T3 cells and mESCs. The most stable and most variable snoRNAs are plotted. (D) PCA plot of the variance in H/ACA snoRNA levels as quantified by NanoString nCounter between the mouse cell types tested: SSCs (light blue), RAW264.7 cells (red), NIH3T3 cells (green), C2C12 cells (pink) and mESCs (orange). (E) Hierarchical clustering of H/ACA snoRNA levels as quantified by NanoString nCounter standardized to a mean of 0 and scaled to a standard deviation of 1. Clustering was performed using Euclidean distance and average linkage in Partek Genomics Suite (Partek Incorporated).
Alterations in the abundance of specific H/ACA snoRNAs are hallmarks of cancer and other human diseases. Multiple H/ACA snoRNAs have been found to be differentially expressed in lung cancer, colorectal cancer, and hepatocellular carcinoma (30–36). As such, they are potential diagnostic biomarkers or therapeutic targets (34,37,38). Recently, McMahon et al., demonstrated that a single H/ACA snoRNA, SNORA24, acts as an oncogene and promotes development of hepatocellular carcinoma. In the absence of SNORA24, ribosomes lack the SNORA24-guided pseudouridine within the 18S rRNA and are prone to miscoding and stop codon readthrough (39). H/ACA snoRNA dysregulation is not limited to cancer. For example, SNORA12 was found to be downregulated in T-cells from patients with lupus, and lower levels of SNORA12 are proposed to contribute to disease pathogenesis (40). Similarly, the levels of multiple snoRNAs were found to be significantly changed in the myocardium of infants born with the congenital heart defect, tetralogy of Fallot (41).
Dysregulation of specific H/ACA snoRNAs in human diseases ranging from congenital heart defects to cancer suggests that snoRNA abundance is regulated during development and cellular differentiation. However, only a few studies have addressed box H/ACA snoRNP function and H/ACA snoRNA abundance in pluripotent versus differentiated cells. It has been shown that expression of box H/ACA snoRNP components is downregulated when mouse embryonic stem cells (mESCs) are differentiated with retinoic acid (RA), and DKC1 knockdown leads to decreased expression of pluripotency genes (42). In the case of the bone-marrow failure disorder, X-linked dyskeratosis congenita (X-DC), mutations in human DKC1 result in reduced levels of specific H/ACA snoRNAs, a loss of pseudouridylation of their target nucleotides within the rRNA, and ultimately defective hematopoietic differentiation (43). In undifferentiated human umbilical cord mesenchymal stem cells, SNORA7A is highly abundant (44). Overexpression of SNORA7A promotes self-renewal and inhibition suppresses proliferation. Likewise, snoRNA abundance is developmental- and lineage-specific during human hematopoiesis (45). Taken together, this suggests that regulation of H/ACA snoRNA abundance is likely to be critical for differentiation.
Here, we generated global profiles of murine H/ACA snoRNA abundance across many cell types and during differentiation in multiple model systems. Through targeted gene expression profiling, we identified cell-type specific patterns of H/ACA snoRNA abundance, determined that snoRNA abundance was consistently regulated during differentiation in multiple models, and demonstrated that the observed changes in snoRNA abundance were largely independent of expression of their host genes. Furthermore, we demonstrated that changes in Snora27 abundance during mESC differentiation correlated with changes in pseudouridylation of its target nucleotide within the E-site tRNA binding region of the 28S rRNA. We propose a model wherein regulated changes in snoRNA abundance during cellular differentiation contribute to the production of heterogeneous ribosomes that are differentially pseudouridylated and may have specialized or cell-type specific functions.
MATERIALS AND METHODS
Animals
All animal procedures were approved by the Institutional Animal Care and Use Committee of the National Institute of Environmental Health Sciences (NIEHS) and were performed in accordance with the approved NIEHS animal study proposal (ASP 2014-0004).
Cell culture
Mouse NIH3T3 cells were kindly provided by Dr Karen Adelman and were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Gibco), 2 mM l-glutamine (Gibco), 100 U/ml penicillin and 100 μg/ml streptomycin (Sigma) at 37°C in 5% CO2. Murine macrophage RAW 264.7 cells were kindly provided by Dr Perry Blackshear and were cultured in DMEM supplemented with 10% fetal bovine serum (Gibco), 2 mM l-glutamine (Gibco), 100 U/ml penicillin and 100 μg/ml streptomycin (Sigma) at 37°C in 5% CO2.
Mouse SSCs were isolated from 6- to 10-day-old DBA/2J mouse testes as described previously (46). Briefly, a single cell suspension was generated by mechanically separating the detunicated testes and incubating the tubules in DNaseI and 0.25% trypsin before filtering the cells through a 40 μM strainer. Spermatogonia were enriched through a 30% Percoll gradient and plated onto SIM mouse embryo-derived thioguanine- and ouabain-resistant (STO) feeder cells treated with mitomycin C and were cultured in mouse SSC serum-free medium (mSFM; MEM alpha (Gibco), 0.2% BSA, 5 μg/ml insulin (Sigma), 10 μg/ml Transferrin (Sigma), 30 nM Na2SeO3 (Sigma), 60 μM putrescine (Sigma), 2 mM l-glutamine (Gibco), 50 μM β-mercaptoethanol (Sigma), 2.36 μM palmitic acid (Sigma), 0.21 μM palmitoleic acid (Sigma), 0.88 μM stearic acid (Sigma), 1.02 μM oleic acid (Sigma), 2.71 μM linoleic acid (Sigma), 0.43 μM linolenic acid (Sigma), 10 mM HEPES (Gibco), 50 U/ml penicillin and 50 μg/ml streptomycin (Sigma) supplemented with 1 ng/ml recombinant human basic Fibroblast Growth Factor (bFGF; BD Biosciences) and 20 ng/ml recombinant human glial-derived neurotrophic factor (hGDNF; PeproTech). SSCs were cultured at 37°C in 5% CO2 and were subcultured onto new feeder cells weekly.
Mouse C2C12 cells were obtained from the ATCC and were cultured in DMEM supplemented with 10% fetal bovine serum (Gibco), 1% sodium pyruvate (Sigma), 100 U/ml penicillin and 100 μg/ml streptomycin (Sigma) at 37°C in 5% CO2. For myotube formation experiments, C2C12 cells were cultured until ∼90% confluent and then the medium was exchanged for a differentiation medium comprising DMEM supplemented with 2% horse serum (Gibco), 1% sodium pyruvate, 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in 5% CO2.
E14Tg2a mESCs were obtained from the Mutant Mouse Resource and Research Center. mESCs were maintained on gelatin-coated plates in ESGRO Complete PLUS Clonal Grade medium (Millipore) at 37°C in 5% CO2. For all experiments, mESCs were cultured on gelatin-coated plates in M15 medium (DMEM supplemented with 15% Embryonic Stem Cell FBS (Gibco), 10 μM β-mercaptoethanol (Sigma), 1× non-essential amino acids (Invitrogen), 1× EmbryoMax nucleosides (Millipore), 1000 U/ml mouse LIF (Gemini Biosciences)) at 37°C in 5% CO2. For RA differentiation experiments, mESCs were cultured on gelatin-coated plates in M15 medium for 48 h prior to the initiation of differentiation. After 48 h, mESCs were cultured on gelatin-coated plates in M15-LIF medium (DMEM supplemented with 15% Embryonic Stem Cell FBS (Gibco), 10 μM β-mercaptoethanol (Sigma), 1× non-essential amino acids (Invitrogen), 1× EmbryoMax nucleosides (Millipore)) supplemented with 1 μM RA (Sigma) at 37°C in 5% CO2.
To generate transgenic cells, E14Tg2a mESCs were transfected with αMHC-puro Rex-neo (AddGene Plasmid #21230; 47) using Lipofectamine 2000 (Invitrogen). Transfected clones were selected by growth in the presence of 200 μg/ml G418 (Invitrogen) for 12 days. The mESCs were maintained in an undifferentiated state by culturing on gelatin-coated plates in ESGRO Complete PLUS Clonal Grade medium (Millipore) at 37°C in 5% CO2. To induce cardiomyocyte differentiation, αMHC-puro Rex-neo E14Tg2a cells were first cultured in M15 (DMEM supplemented with 15% Embryonic Stem Cell FBS (Gibco), 10 μM β-mercaptoethanol (Sigma), 1× non-essential amino acids (Invitrogen), 1× EmbryoMax nucleosides (Millipore), 1000 U/ml mouse LIF (Gemini Biosciences)).
After 2 days, cardiomyocyte differentiation was induced following the hanging drop protocol as described previously (48). Cardiomyocytes were selected by growth in the presence of 1 μg/ml puromycin for 8 days beginning one day after plating embryoid bodies on gelatin-coated dishes.
snoRNA NanoString nCounter analysis
RNA was extracted with TRIzol Reagent according to the manufacturer's recommendations, DNase-treated and further purified using RNeasy mini spin columns (Qiagen). RNA concentration, purity, and quality were analyzed on a Qubit 3 Fluorometer, a NanoDrop One spectrophotometer and an Advanced Analytical Fragment Analyzer. H/ACA snoRNAs were analyzed using a custom NanoString nCounter probe set (Supplementary Table S1; NanoString Technologies, Inc., Seattle, WA, USA). Raw data were normalized to the geometric mean of the positive control probes and to the geometric mean of snoRNAs that remain unchanged as determined by percent coefficient of variation (%CV) <20 using nSolver Analysis Software 3.0 (NanoString Technologies, Inc.). Experiments were performed in triplicate. The snoRNA gene expression nCounter data has been submitted to GEO (Gene Expression Omnibus) under accession number GSE140623.
Statistical analyses
Principal component analysis (PCA), ANOVA analysis, and hierarchical clustering were performed using the Partek Genomic Suite (Partek Incorporated, St. Louis, MO, USA). Differentially-expressed snoRNAs had a fold-change > |2| and a false discovery rate-corrected P-value <0.05.
Small RNA sequencing and bioinformatics
Total RNA was isolated using TRIzol Reagent, DNase-treated and further purified using RNeasy mini spin columns (Qiagen). Small RNAs (>17 nt, <200 nt) were purified using the mirVana miRNA isolation kit (Invitrogen). The 5.8S and 5S ribosomal RNAs were removed using the RiboMinus Eukaryote System v2 (Invitrogen). The purified small RNAs were treated with T4 Polynucleotide Kinase (NEB), purified using the Directzol microprep kit (Zymo) and their concentrations were determined using a Qubit 3 Fluorometer. For RNA-sequencing library construction, TGIRT template-switching reverse transcription reactions and adapter ligations were performed as described previously (49,50) using 50 ng of RNA. The ligated cDNA products were purified using a MinElute column (Qiagen) and PCR amplified using Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific) and 200 nM of both Illumina multiplex primer and Illumina barcode primer (Supplementary Table S2). PCR was performed for 17 cycles, as described previously (49), and PCR products were purified using Agencourt AMPure XP beads (Beckman Coulter). Libraries were sequenced on an Illumina NextSeq 500 instrument to obtain 75-nt paired-end reads. Sequenced reads were assigned to an index of mouse H/ACA snoRNA sequences using Salmon 0.12.0 with default parameters (51). Read counts were combined by snoRNA family designation (Supplementary Table S3). Differentially-expressed snoRNAs were identified using DESeq2 with default parameters (52). Proportions of reads by transcript type were determined through a separate assignment with Salmon to an index of all Ensembl mouse transcripts (version 96). The small RNA-seq data have been submitted to GEO (Gene Expression Omnibus) under accession number GSE140623.
RNA isolation and quantitative real-time RT-PCR
All parameters for quantitative real-time RT-PCR are reported according to the MIQE guidelines in Supplementary Table S4 (53). Total RNA was isolated from cells and DNase-treated using RNeasy mini kits (Qiagen). RNA concentration and quality were analyzed on a Qubit 3 Fluorometer and an Agilent 4200 TapeStation. For reverse transcription (RT) prior to gene expression analysis, cDNAs were generated using Superscript III (Invitrogen) according to manufacturer's instructions. Purified RNA (1 μg), 50 ng random hexamers, and 50 μM oligo(dT)20 were combined in a 10 μl reaction, denatured for 5 min at 65°C. Reactions were immediately cooled on ice and 10 μl extension mix (2 μl 10× RT buffer (Invitrogen), 4 μl 25 mM MgCl2, 2 μl 0.1 M DTT, 1 μl Superscript III (Invitrogen), 1 μl RNase Out (Invitrogen)) was added to each. Reactions were incubated at 25°C for 10 min, 50°C for 50 min and 85°C for 5 min. Each reaction was then treated with 1 μl RNase H (Invitrogen) for 20 min at 37°C. As negative controls for each primer set, mock ‘no RT’ reactions and no-template reactions were performed using identical conditions except that either the reverse transcriptase (Superscript III) was omitted (no RT) or the RNA was omitted (no template).
All oligonucleotides for quantitative real-time PCR and their specifications, including primer sequences, amplicon sizes and amplification efficiencies are provided in Supplementary Table S4. Quantitative real-time PCR was performed using 2 ng of cDNA, 5 μM forward and 5 μM reverse oligonucleotides, and SsoAdvanced Universal SYBR Green Supermix (BioRad) using a CFX96 Real-Time PCR system thermocycler (BioRad) with the following cycling conditions: (i) 95°C for 30 s, (ii) 95°C for 5 s, (iii) 59°C for 10 s, (iv) repeat steps 2–3 for 40 cycles. Melt curves were generated with a range of 65–95°C, at increments of 0.5°C. Analyses were completed using the comparative CT method (ΔΔCT) where Actin served as a reference gene. Three technical replicates were performed for each measurement. Three biological replicates were performed for each sample.
RNA sequencing and bioinformatics
Total RNA was isolated from C2C12 myoblasts before and after differentiation into myotubes using TRIzol Reagent, DNase-treated, and further purified using RNeasy mini spin columns (Qiagen). RNA concentration and quality were analyzed on a Qubit 3 Fluorometer and an Agilent Bioanalyzer. Libraries for RNA-seq analysis were prepared using the TruSeq Stranded Total RNA Gold Library Prep Kit (Illumina) following manufacturer's instructions. Libraries were sequenced on an Illumina NovaSeq instrument to obtain 75-nt paired-end reads. Read pairs were filtered to remove those where mean base qualities of one or both mates was below 20, then aligned to the mm10 reference genome using STAR version 2.5.1b, specifying –outMultimapperOrder random, –outFilterType BySJout and –outFilterIntronMotifs RemovNoncanonical, but otherwise using default parameters (54). Counts of fragments mapped to each gene defined in the GENCODE M24 annotation set were determined using featureCounts v1.5.1 in a strand-specific manner, setting a minimum mapping quality of 10 and excluding chimeric read pairs (-Q 10 -p -C -s 2) (55). Counts for all samples were imported into DESeq2 1.18.1 running under R 3.4.0 and time points were compared using the default differential expression analysis workflow (52). The sequencing data have been submitted to GEO under accession number GSE140623.
Molecular visualizations
The atomic coordinates for a cryo-electron microscopy structure of an 80S eukaryotic ribosome (PDB ID: 6SGC) were obtained from the Protein Data Base (PDB), and visualized using PyMol molecular visualization software (56).
CMC treatment and primer extension
Total RNA was isolated using TRIzol as per the manufacturer's instructions, DNase-treated and further purified using RNeasy mini spin columns (Qiagen). For CMC treatment, 20 μg of RNA in a total volume of 20 μl was denatured in 5 mM EDTA for 3 min at 80°C. The denatured RNA was mixed with either 100 μl 0.3 M CMC in BEU buffer (+CMC; 50 mM Bicine pH 8.5, 4 mM EDTA pH 8.0, 7 M urea) or with 100 μl BEU buffer (+CMC) and incubated at 37°C for 30 min. The RNA was precipitated and resuspended in 30 μl freshly prepared buffer (50 mM Na2CO3 pH 10.4, 2 mM EDTA pH 8.0) and incubated at 50°C for 2 h. The RNA was precipitated and resuspended in 7 μl dH2O. The concentration was determined using a Qubit 3 Fluorometer.
For primer extension reactions, 2 μg of +CMC RNA or −CMC RNA was hybridized with 2 pmol FAMN-labeled oligo (Supplementary Table S2) in a 6.5 μl reaction containing 1.5 μl 4× hybridization buffer (200 mM Tris pH 8.3, 240 mM NaCl, 40 mM DTT) for 10 min at 65°C. For sequencing reactions, 5 μg of total RNA was hybridized with 2 pmol oligo in a 6.5 μl reaction containing 1.5 μl 4× hybridization buffer for 10 min at 65°C. Reactions were moved immediately to ice and 4.5 μl of extension mix (2 μl 5× Superscript II buffer (Invitrogen), 0.5 μl 0.1 M DTT, 0.5 μl RNase OUT (Invitrogen), 1 μl 10 mM dNTPs, 0.5 μl Superscript II (Invitrogen)) was added to +CMC and −CMC reactions. For sequencing reactions, 4.5 μl of sequencing extension mix (2 μl 5× Superscript II buffer (Invitrogen), 0.5 μl 0.1 M DTT, 0.5 μl RNase OUT (Invitrogen), 1 μl 2.5 mM dNTPs, 0.5 μl Superscript II (Invitrogen), 1 μl 5 mM ddNTP) was added. Reactions were incubated at 42°C for 30 min and 8 μl of stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol) was added to each reaction. Reactions were run on 10% denaturing acrylamide gels, visualized on a Typhoon FLA 9500 (GE Healthcare), and quantified using ImageQuant TL (GE HealthCare). The pseudouridine ratio was defined as the intensity of primer extension stops at a specific pseudouridylated nucleotide in the +CMC lane minus that of the −CMC control. Three biological replicates were performed for each sample.
Northern blotting
Total RNA was isolated using TRIzol as per the manufacturer's instructions. Northern blotting was performed as described previously (57). Briefly, 3 μg of RNA was run on a 1% agarose/1.25% formaldehyde gel and transferred to a Hybond-XL membrane (GE Healthcare RPN303 S).
Membranes were hybridized with radiolabeled oligonucleotide probes (Supplementary Table S2) to detect mature rRNAs and pre-rRNA species (58). Northern blots were imaged on a Typhoon FLA 9500 (GE Healthcare) and quantified using ImageQuant TL (GE Healthcare). Ratio Analysis of Multiple Precursors (RAMP) was performed as described previously (58). Three biological replicates were performed for each sample.
RESULTS
H/ACA snoRNA levels are cell-type specific
To systematically and comprehensively interrogate H/ACA snoRNA abundance, we used NanoString nCounter technology, a target-directed approach. By examining existing snoRNA and genome databases (59–61), we identified 357 unique, putative H/ACA snoRNAs in the mouse genome (Supplementary Table S3). All 357 snoRNAs contain the canonical H box (ANANNA) and ACA box (ACA) sequences and are predicted to form the characteristic hairpin-hinge-hairpin-tail structure (Figure 1A, B; 62). These snoRNAs were further classified into 72 distinct families based on sequence homology, particularly within the regions that recognize and bind their target RNAs (Figure 1B). We selected one snoRNA from each family to target in our custom NanoString nCounter assay (Supplementary Table S1). However, due to the high degree of sequence homology between family members (Figure 1B), the NanoString nCounter probes were predicted to efficiently target all family members, thereby providing a measure of snoRNA abundance at the family level.
Our NanoString nCounter data indicate H/ACA snoRNA abundance varies widely across the different families. H/ACA snoRNA abundance was measured in multiple cell types: E14Tg2a mESCs, NIH3T3 mouse embryonic fibroblast cells, RAW264.7 mouse macrophage cells, C2C12 mouse myoblast cells and mouse spermatogonial stem cells (SSCs) (Supplementary Table S5). In mESCs, NIH3T3 cells and RAW264.7 cells, the same five H/ACA snoRNAs (Snora76, Snora11, Snora35, Snora50 and Snora58) were either not expressed or were not sufficiently captured by the NanoString nCounter probes, so we excluded them from further analyses. We examined H/ACA snoRNA abundance across all five cell types and found that some H/ACA snoRNA families showed very little variability in abundance across cell types, while others were much more varied. For example, Snora8, Snora17 and Snora52 levels were relatively stable across cell types, but the levels of Snora47, Snora49 and Snora36b differed 6–10-fold depending on the cell line (Figure 1C, Supplementary Figure S1 and Supplementary Table S6).
We found that the overall profiles of H/ACA snoRNA abundance are cell-type specific. Principal component analysis (PCA) of the normalized nCounter data (Supplementary Table S5) revealed that the patterns of snoRNA abundance are unique for each cell type, as the biological replicates from each cell type clustered together (Figure 1D). Additionally, hierarchical clustering of H/ACA snoRNA abundance data resulted in the clustering of all replicates from each cell type into discrete groups, where each cell type has a distinct pattern of H/ACA snoRNA abundance (Figure 1E). The PCA plot revealed that the SSCs and the RAW264.7 cells were the most dissimilar from the other cell types. Comparing snoRNA abundance between these cell types and others revealed many differentially expressed snoRNAs (Figure 1E and Supplementary Figure S2A–G). In contrast, C2C12 and NIH3T3 cells were most similar with only Snora57 differentially expressed (Supplementary Figure S2H). mESCs are similar to C2C12 and NIH3T3 cells. Few snoRNAs were differentially expressed comparing mESCs to these cell types (Supplementary Figure S2I, J).
H/ACA snoRNA abundance is regulated during embryonic stem cell differentiation
H/ACA snoRNA abundance varies across multiple cell types, in cancer cells versus normal cells, and has been shown to change during the trypanosome lifecycle (Figure 1; 18,30–36,39,45). Thus, we hypothesized that H/ACA snoRNAs would be differentially expressed during mESC differentiation. To test this hypothesis, we analyzed H/ACA snoRNA profiles before and after differentiation of mESCs with retinoic acid (RA), a metabolite of vitamin A that promotes differentiation into ectodermal cells (63,64). We used two orthogonal approaches: NanoString nCounter assay and small RNA sequencing (small RNA-seq, Figure 2A). The NanoString nCounter assay is advantageous for two main reasons: (i) the RNA sample requires less processing: it does not need to be enriched for small RNAs or depleted of highly abundant rRNAs, and no library preparation is necessary; (ii) no potential amplification bias is introduced because the sample is not PCR amplified during preparation. Small RNA-seq is advantageous as an untargeted approach: it provides information regarding abundance of individual H/ACA snoRNAs within each family and how they contribute to the family level analysis. It also detects H/ACA snoRNAs that were not captured by the NanoString nCounter codeset.
Figure 2.
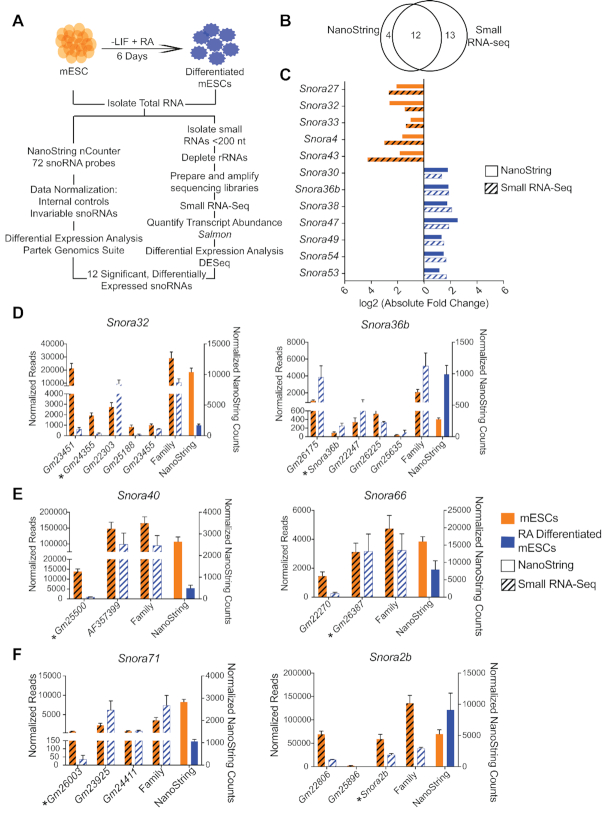
H/ACA snoRNA abundance is regulated during mESC differentiation. (A) Schematic of the experimental systems used to measure H/ACA snoRNA abundance before and after RA-induced of mESCs. (B) Venn diagrams showing the overlap in significantly differentially-expressed H/ACA snoRNAs detected by the two methodologies. (C) The levels of 12 H/ACA snoRNAs are significantly changed upon mESC differentiation with RA. The bar graph compares the log2 fold changes observed in the NanoString nCounter (solid colors) and small RNA-seq (hash lines) data sets. The color indicates the cell type in which the snoRNA is most abundant. Orange = mESCs. Blue = RA-differentiated mESCs. (D, E) The bar graphs plot the normalized sequencing reads for individual snoRNAs and the total for the entire family before and after differentiation of mESCs with RA and the normalized NanoString nCounter counts before (orange) and after (blue) differentiation. (D) The levels of the individual members of the Snora32 and Snora36b families were highly variable and had patterns of abundance distinct from the family-level pattern. (E) Snora40 and Snora66 were significantly differentially expressed by NanoString nCounter analysis but missed the significance cutoff in the small RNA-seq analysis. (F) The Snora71 and Snora2b families showed opposite trends in abundance in the two methods. Asterisks denote the family member that was targeted by the NanoString nCounter probes.
Using these two complementary methodologies, we identified H/ACA snoRNA families that were significantly differentially expressed during mESC differentiation with RA (Figure 2A, B). Using the NanoString nCounter assay, we found that 16 of the 72 H/ACA snoRNA families (∼22%) were differentially expressed: eight were more abundant in mESCs and eight were more abundant in RA-differentiated cells (Figure 2B and Supplementary Table S7). In parallel, small RNA-seq libraries from 17–200 nt, rRNA-depleted RNA isolated from mESCs and from RA-differentiated cells were constructed and sequenced (Figure 2A). Transcript abundance was determined using Salmon (51). On average, ∼72% of the assigned reads were ncRNAs and ∼27% of the ncRNA reads were H/ACA snoRNAs (Table 1). Similar to small-RNA sequencing analyses of human cell lines (65), ∼20 H/ACA snoRNAs account for the vast majority (∼80%) of H/ACA snoRNA reads in both mESCs and RA-differentiatied cells (Supplementary Figure S3A). Interestingly, five highly abundant H/ACA snoRNAs are shared between all three datasets suggesting that these snoRNAs may have a conserved, essential function (Supplementary Figure S3B). Small RNA-seq analyses confirmed the absence of Snora35 and Snora50 expression in mESCs, two snoRNAs that were not detected by the NanoString nCounter assay. It also detected Snora11, Snora76 and Snora58, which were not previously detected by the NanoString nCounter assay. In total, we found that 25 of the 72 H/ACA snoRNA families (∼37.5%) were differentially expressed upon treatment of mESCs with RA: 12 were more abundant in mESCs and 13 were more abundant upon treatment with RA (Figure 2B and Supplementary Table S7). Importantly, we identified 12 H/ACA snoRNA families that were significantly differentially expressed in both analyses and showed similar fold changes in abundance (Figure 2A–C and Table 2). We therefore conclude that H/ACA snoRNA levels are dynamic during mESC differentiation.
Table 1.
Summary of small RNA-sequencing statistics
| mESCs rep. 1 | mESCs rep. 2 | mESCs rep. 3 | RA-differentiated mESCs rep 1 | RA-differentiated mESCs rep 2 | RA-differentiated mESCs rep 3 | |
|---|---|---|---|---|---|---|
| Raw reads | 25 664 849 | 16 421 212 | 18 386 473 | 24 517 078 | 15 140 722 | 14 632 744 |
| Assigned reads | 14 042 942 | 11 358 769 | 12 144 174 | 13 199 225 | 7 216 843 | 7 622 604 |
| % Assigned | 54.7% | 69.2% | 66% | 53.8% | 47.7% | 52.1% |
| rRNAs | 31 809 | 21 788 | 19 663 | 50 918 | 2302 | 5677 |
| ncRNAs | 10 409 217 | 8 991 257 | 8 399 714 | 10 448 667 | 4 519 994 | 5 291 673 |
| % ncRNAs | 74.1% | 79.2% | 69.2% | 79.2% | 62.6% | 69.4% |
| snoRNAs | 4 826 535 | 4 366 464 | 4 939 639 | 4 679 234 | 2 307 155 | 2 899 401 |
| C/D snoRNAs | 1 672 941 | 1 710 320 | 1 901 676 | 2 653 251 | 1 112 356 | 1 856 683 |
| H/ACA snoRNAs | 3 153 594 | 2 656 144 | 3 037 963 | 2 025 983 | 1 194 799 | 1 042 718 |
| % H/ACA snoRNAs of ncRNAs | 30.3% | 29.54% | 36.17% | 19.39% | 26.43% | 19.7% |
Table 2.
Comparison of NanoString nCounter and small RNA-sequencing analyses
| SnoRNA | NanoString fold change | NanoString P-value | Small RNA- Seq fold change | Small RNA-Seq P-value | Expression trend upon RA differentiation |
|---|---|---|---|---|---|
| Differentially expressed snoRNAs in both analyses | |||||
| Snora27 | 0.24 | 0.028 | 0.14 | 6.26 × 10−6 | Down |
| Snora32 | 0.16 | 0.00022 | 0.36 | 0.0076 | Down |
| Snora33 | 0.49 | 0.023 | 0.35 | 0.0004 | Down |
| Snora4 | 0.32 | 0.014 | 0.11 | 5.69 × 10−7 | Down |
| Snora43 | 0.28 | 6.71 × 10−7 | 0.05 | 1.77 × 10−44 | Down |
| Snora30 | 3.45 | 0.0012 | 2.38 | 0.0009 | Up |
| Snora36b | 3.57 | 0.0076 | 2.50 | 0.031 | Up |
| Snora38 | 3.37 | 2.67 × 10−5 | 4.35 | 0.023 | Up |
| Snora47 | 5.88 | 0.00066 | 3.45 | 3.07 × 10−6 | Up |
| Snora49 | 2.50 | 0.0012 | 2.63 | 0.015 | Up |
| Snora53 | 2.22 | 0.037 | 3.23 | 0.01 | Up |
| Snora54 | 2.78 | 0.0024 | 2.94 | 0.0066 | Up |
| Differentially expressed snoRNAs in the NanoString analysis only | |||||
| Snora40 | 0.18 | 0.0011 | 0.57 | 0.205 | Down |
| Snora66 | 0.50 | 0.049 | 0.68 | 0.5 | Down |
| Snora28 | 2.27 | 0.0023 | 1.89 | 0.05 | Up |
| Snora71 | 0.38 | 0.00028 | 2.08 | 0.16 | |
| Differentially expressed snoRNAs in the small RNA-seq analysis only | |||||
| Snora1 | 0.51 | 0.087 | 0.13 | 5.86 × 10−11 | Down |
| Snora15 | 0.73 | 0.21 | 0.31 | 0.005 | Down |
| Snora26 | 0.90 | 0.04 | 0.11 | 9.66 × 10−7 | Down |
| Snora44 | 0.64 | 0.003 | 0.16 | 4.82 × 10−12 | Down |
| Snora57 | 0.65 | 0.00013 | 0.27 | 0.0018 | Down |
| Snora67 | 0.71 | 0.0047 | 0.19 | 4.56 × 10−9 | Down |
| Snora14 | 1.54 | 0.006 | 4.76 | 0.014 | Up |
| Snora46 | 2.63 | 0.16 | 2.50 | 0.0025 | Up |
| Snora55 | 1.32 | 0.035 | 2.63 | 0.042 | Up |
| Snora68 | 1.45 | 0.007 | 2.08 | 0.04 | Up |
| Snora58 | n.d. | n.d. | 2.04 | 0.0099 | Up |
| Snora64 | 1.05 | 0.91 | 2.44 | 0.033 | Up |
| Snora2b | 1.49 | 0.38 | 0.28 | 2.53 × 10−7 | |
The changes we observed in snoRNA levels in the small RNA-seq analysis largely recapitulated the trends we observed in the NanoString nCounter data set. Most differentially expressed snoRNAs that were identified by only one method showed similar changes in abundance using the other method but the degree of change either did not meet the 2-fold change in abundance threshold or statistical significance (Table 2). One snoRNA, Snora58, was not detected by NanoString nCounter assay, however, small RNA-seq analysis identified Snora58 as a differentially expressed snoRNA that was upregulated ∼2-fold upon treatment with RA (Table 2). Only two snoRNAs showed opposite trends in change in abundance upon differentiation using the two methods: Snora71 and Snora2b.
We examined the small RNA-seq data in greater depth to determine how levels of individual members of snoRNA families contribute to the total measured abundance of the family. We found that the levels of individual family members can vary considerably, and changes in individual snoRNAs may be distinct from the overall family-level changes (Table 2, Figure 2D, Supplementary Table S8). For example, Snora32 levels decrease overall upon RA-induced differentiation, yet the individual family member Gm22303 increases upon differentiation. Similarly, Snora36b levels increase upon differentiation while family member Gm26225 decreases. Although NanoString nCounter analysis captures snoRNA abundances at the family-level, small RNA-seq provides additional, informative details.
The extra details afforded by small RNA-seq explained a few differences between the NanoString nCounter assay and small RNA-seq data. For example, the Snora40 and Snora66 families were significantly more abundant in mESCs than RA-differentiated cells in the NanoString nCounter analysis but were not significantly differentially expressed in the small RNA-seq dataset (Table 2). The small RNA-seq data for the individual snoRNAs in these families revealed that one member in each of the Snora40 and Snora66 families recapitulated the change in abundance captured by the NanoString nCounter probes. However, these family members were the least abundant and their contributions to the family-level changes were diluted by the more abundant members (Figure 2E and Supplementary Table S8). For Snora40, the change in abundance of the individual snoRNA that was the direct target of the NanoString nCounter probe matched the NanoString result. Thus, it appears that the NanoString probes could favor the direct target detection and may not always capture all family members equally, as we predicted based on sequence similarity.
We also analyzed the changes in abundance of the individual members of the Snora71 and Snora2b families, which showed opposite trends upon differentiation using the two methods (Table 2). For these two snoRNA families, levels of the individual members varied widely (Figure 2F and Supplementary Table S8). Although it appears that the Snora71 NanoString probes are not capturing all family members equally, we were not able to discern why there was such a discrepancy between the NanoString nCounter analysis and the small RNA-seq analyses for the Snora2b family.
In sum, our NanoString nCounter and small RNA-seq analyses indicate that at least 12 H/ACA snoRNAs are significantly regulated in response to RA-induced differentiation of mESCs (Figure 2B, C). The snoRNA families that were identified in both analyses are considered a high confidence set. While the snoRNAs that were identified in only one analysis may be regulated, they are classified as a lower confidence set. Moreover, our small RNA-seq analysis validates the use of the NanoString nCounter assay as a method for assessing relative snoRNA levels and also provides detailed information about individual family member abundance. We opted to use the NanoString nCounter assay for subsequent analyses of snoRNA levels due to its advantage of streamlined sample preparation, which outweighed the need for information on individual family members.
Regulation of H/ACA snoRNA abundance is a hallmark of cellular differentiation
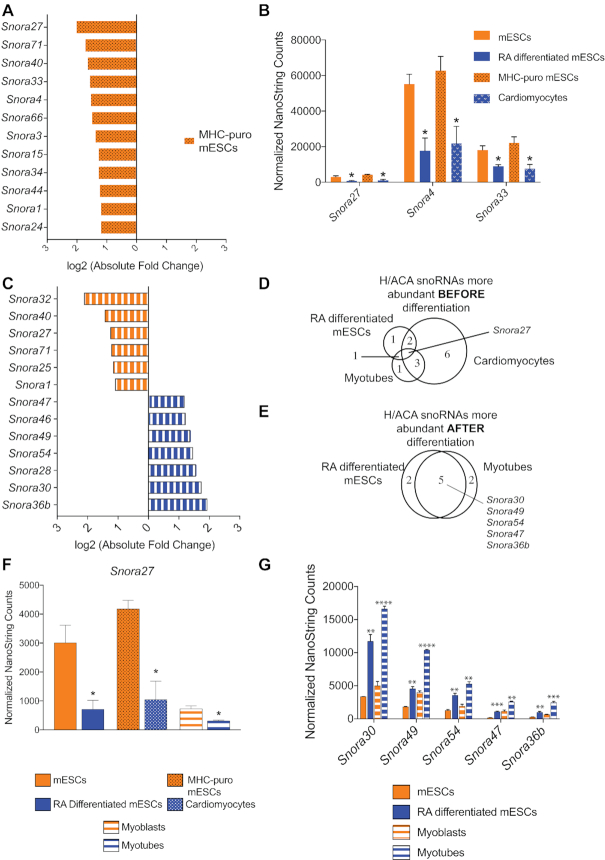
We next examined H/ACA snoRNA abundance upon mESC differentiation into cardiomyocytes, which are mesodermal. By using this alternative pathway of differentiation, we could identify dynamic changes in H/ACA snoRNA levels toward a different cell fate. We could also determine whether the changes in snoRNA levels we observed upon RA treatment were due to differentiation or were a response to RA-induced signaling. We generated a transgenic mESC line in which the puromycin resistance gene was under the control of the cardiomyocyte-specific a-myosin promoter (aMHC-Puro). After inducing cardiomyocyte differentiation of this cell line using an established method (48), we could enrich for cardiomyocytes by puromycin selection. We analyzed H/ACA snoRNA levels before and after mESC differentiation into cardiomyocytes using our NanoString nCounter assay. We identified 12 H/ACA snoRNAs that were significantly downregulated upon differentiation into cardiomyocytes (Figure 3A). Three of these snoRNAs were also downregulated upon mESC differentiation with RA (Figure 3B). Importantly, the abundance of Snora27, Snora4 and Snora33 before and after differentiation is similar in both differentiation model systems, suggesting that the observed changes are neither random nor a response to a specific signal; rather they are a regulated response to cellular differentiation (Figure 3B and Supplementary Figure S4).
Figure 3.
H/ACA snoRNA abundance is regulated consistently across multiple models of differentiation. (A) Twelve H/ACA snoRNAs are more abundant in mESCs than they are in the differentiated cardiomyocytes. The bar graph plots the log2 fold changes in expression as determined by NanoString nCounter analysis. (B) Three H/ACA snoRNAs are consistently and significantly downregulated after both RA-induced mESC differentiation and differentiation into cardiomyocytes. The bar graph plots the normalized NanoString nCounter counts before (orange) and after (blue) differentiation for all three snoRNAs. (C) Expression of 13 H/ACA snoRNAs is significantly changed upon myoblast differentiation into myotubes. The bar graph plots the log2 fold-changes in expression as determined by nCounter analysis. The color indicates the cell type in which the snoRNA is most abundant. Orange = myoblasts. Blue = myotubes. (D, E). Venn diagrams showing the overlap in H/ACA snoRNAs that are downregulated upon differentiation (D), or upregulated upon differentiation (E). (F) Snora27 is consistently and significantly downregulated after differentiation. The bar graph plots the normalized counts before (orange) and after (blue) differentiation. (G) Five H/ACA snoRNAs are consistently and significantly upregulated after differentiation. The bar graph plots the normalized counts before (orange) and after (blue) differentiation for all three snoRNAs. Data are represented as mean ± SEM. *P ≤ 0.05, **P ≤ 0.01, *** P ≤ 0.001. **** P ≤ 0.0001.
We found that snoRNA abundance is also dynamically regulated during mouse myogenesis. To probe whether regulation of H/ACA snoRNA abundance is unique to pluripotent mESC cells or if it is a common feature of differentiation in pluripotent and multipotent cells, we interrogated H/ACA snoRNA abundance during myogenesis. We used C2C12 myoblast cells that readily differentiate in vitro into myotubes, thereby mimicking skeletal muscle development (Supplementary Figure S5; 66). We quantified H/ACA snoRNA levels before and after myoblast differentiation into myotubes using our NanoString nCounter assay. Like mESC differentiation, 13 H/ACA snoRNAs were significantly differentially expressed during differentiation: six were more abundant in myoblasts and seven were more abundant in myotubes (Figure 3C).
Several of the snoRNAs that were regulated during mESC differentiation were also regulated during myogenesis. Comparison of the differentially expressed H/ACA snoRNAs revealed that although the model systems were distinct, several snoRNAs were regulated similarly (Figure 3D, E). For example, Snora27 was consistently downregulated during differentiation in all three model systems (Figure 3D, F). Likewise, Snora30, Snora49, Snora54, Snora47 and Snora36b, which were upregulated during RA-induced differentiation of mESCs, were also upregulated upon myotube formation (Figure 3E, G).
Regulation of snoRNA abundance can be independent of its host gene
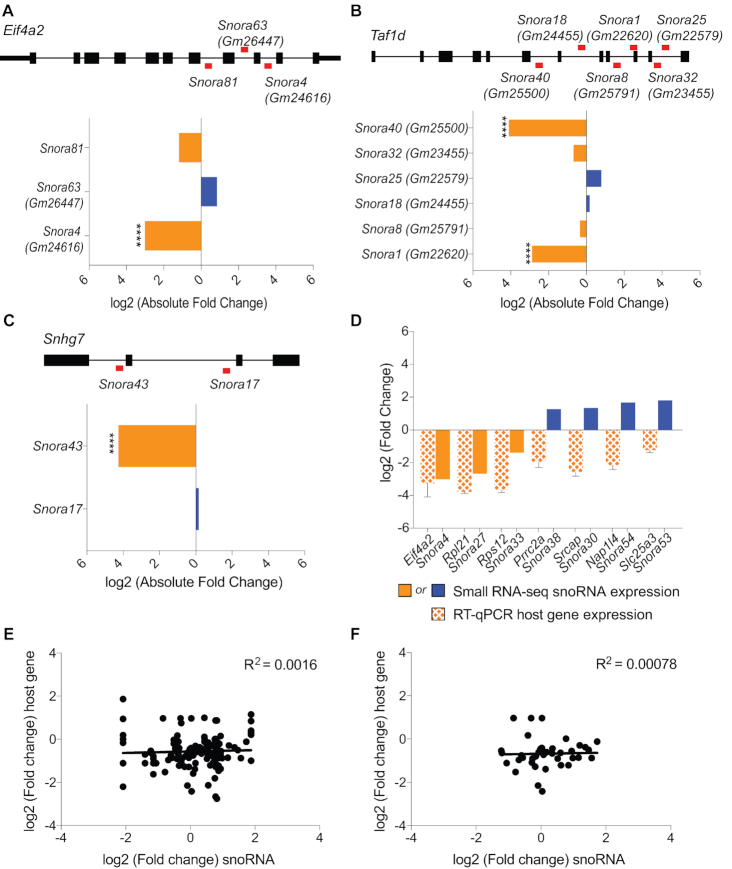
While snoRNA genes sometimes exist as independent transcripts or within genomic clusters, the majority of mammalian snoRNAs are embedded within introns of protein-coding host genes (67–69). As such, we investigated whether the observed changes in snoRNA abundance are tied to changes in host gene expression during cellular differentiation. We first compared the small RNA-sequencing profiles of multiple snoRNAs that are embedded within the same host gene to determine whether or not their expression is coordinated (Figure 4A–C). Two protein-coding genes and one long noncoding RNA (lncRNA) encode multiple H/ACA snoRNAs whose levels were regulated in our study. In all three cases, levels of the snoRNAs derived from a single host gene were not coordinated upon RA-differentiation of mESCs (Figure 4A–C). For example, Snora81, Snora63 (Gm26447) and Snora4 (Gm24616) are all embedded within introns of the protein-coding gene Eif4a2. While Snora4 levels were significantly downregulated, Snora81 levels were only mildly reduced, and Snora63 levels were slightly increased upon differentiation with RA (Figure 4A). Similarly, the protein-coding gene Taf1d is the host of Snora40 (Gm25500), Snora32 (Gm23455), Snora25 (Gm22579), Snora18 (Gm24455), Snora8 (Gm25791) and Snora1 (Gm22620) (70). Upon RA-differentiation of mESCs, levels of Snora40 and Snora1 were significantly downregulated whereas levels of Snora32, Snora8, Snora25, and Snora18 were not significantly changed (Figure 4B). In addition, Snora43 and Snora17 are encoded within the introns of the lncRNA Snhg7 (Figure 4C). Although Snora17 levels did not significantly change upon mESC differentiation with RA, Snora43 levels were significantly downregulated (Figure 4C). These three examples of snoRNAs linked to the same host gene suggest that changes in snoRNA abundance are independent of host gene expression changes.
Figure 4.
SnoRNA levels are uncoupled from expression of its host gene. (A–C) SnoRNAs expressed from the same host gene are independently regulated in response to mESC differentiation with RA. The locations of snoRNAs within host genes Eif4a2 (A), Taf1d (B) and Snhg7 (C) are indicated by red bars. The bar graphs plot the log2 fold change in abundance of snoRNAs expressed from the same host genes upon differentiation of mESCs with RA. Orange bars denote snoRNAs that are downregulated upon differentiation. Blue bars denote snoRNAs that are upregulated upon differentiation. (D) Host gene expression and snoRNA abundance are not always coordinated. The bar graph plots the log2 fold change in snoRNA abundance as quantified by Small RNA-seq and the log2 fold change in host gene expression as determined by quantitative real-time RT-PCR. Host gene expression is normalized to Actin and represented as mean ± SEM. (E, F) Scatter plots comparing the log2 fold changes in the corresponding host gene expression during myogenesis as quantified by RNA-seq and the log2 fold change in snoRNA abundance as quantified by our NanoString nCounter assay. We plotted the data for all snoRNAs and all potential host genes (E) or for snoRNAs that are expressed from one genomic locus and their respective host genes (F).
We next directly investigated the association between snoRNA abundance and host gene expression during differentiation and found that snoRNA abundance can change irrespective of host gene expression variations in both mESC and myoblast differentiation systems. To analyze the relationship between the expression of snoRNA and host gene pairs in mESCs, we identified host genes that were the sole genomic locus of expression for a snoRNA that was significantly differentially expressed in our small RNA-seq dataset (Supplementary Table S8). We monitored expression of seven host genes before and after mESC differentiation with RA using real-time quantitative PCR and compared the observed changes in expression of the host gene to the changes in abundance of the corresponding snoRNA detected by small RNA-seq. Expression of the seven host genes was downregulated upon differentiation (Figure 4D). Similarly, levels of Snora4, Snora27 and Snora33 decreased, in accordance with the observed changes in their respective host genes. In contrast, levels of Snora38, Snora30, Snora54 and Snora53 increased upon differentiation, opposite to the changes in their respective host genes (Figure 4D). We also examined the relationship between host gene and snoRNA expression during myoblast differentiation. We compared changes in snoRNA abundance as quantified by NanoString nCounter analysis to changes in host gene expression as detected by RNA-seq (Figure 4E, F and Supplementary Table S9). In an initial comparison we included all possible snoRNA host genes to encompass all individual family members that contribute to the NanoString nCounter data (Figure 4E). To focus our analysis on corresponding pairs of host genes and individual snoRNAs, we then restricted our comparison to snoRNAs that are known to be expressed from a single genomic locus (Figure 4F). In both analyses, changes in snoRNA levels did not correlate with changes in host gene expression (r2 = 0.0016 and r2 = 0.00078). Similar to what we determined for mESC differentiation, expression of the majority of snoRNA host genes was downregulated upon differentiation of myoblasts into myotubes, but the corresponding snoRNA levels increased, decreased, or remained unchanged (Figure 4E, F). Taken together, our data are consistent with previous findings that regulation of snoRNA abundance is largely independent of its host gene expression (45,68,71).
Snora27 abundance correlates with levels of ribosomal RNA pseudouridylation
SnoRNAs guide pseudouridylation of target RNAs as part of the box H/ACA snoRNP. We hypothesized that regulation of snoRNA abundance during differentiation is a mechanism for dynamically pseudouridylating target nucleotides within the rRNA. We focused on the mammalian-specific snoRNAs that are regulated during mESC differentiation with RA. These snoRNAs target nucleotides within functionally important regions of the ribosome, including the E-site tRNA binding region (Snora27), the A-site finger (Snora47), the subunit interface (Snora54) and the region bound by ribosomal protein RPL3 (Snora33, Snora43, Snora30 and Snora54; Supplementary Figure S6 and Table 3). To test our hypothesis, we investigated pseudouridylation of the known rRNA target of Snora27, which is the sole snoRNA targeting nucleotide 3371 within the E-site tRNA binding region where pseudouridylation could impact translation fidelity based on the known functions of the E-site in translation (72).
Table 3.
Summary of mammalian-specific modified nucleotides of differentially expressed snoRNAs
| snoRNA | Modified nucleotide (mouse) | Region of the ribosome |
|---|---|---|
| Snora27 | 28S: 3371 | In E-tRNA acceptor stem binding region |
| Snora33 | 28S: 4648 | Near Rpl3 binding site |
| Snora43 | 28S: 4620 | Near Rpl3 binding site |
| Snora30 | 28S: 4325 | Near Rpl3 binding site |
| Snora36 | 18S: 105 | Within helix 7 of the body of the small subunit |
| Snora38 | Unknown | N/A |
| Snora47 | 28S: 1776 | A-site finger |
| Snora49 | Unknown | N/A |
| Snora53 | Unknown | N/A |
| Snora54 | 28S: 3478 | Near subunit interface |
| Snora54 | 28S: 4221 | Part of domain 0, near Rpl3 extension binding region |
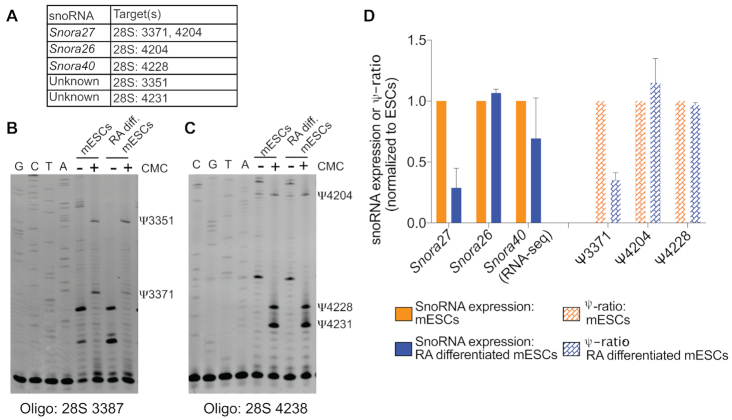
We interrogated pseudouridine (Ψ) modification using N-cyclohexyl N′-(2-morpholinoethyl) carbodiimide (CMC) modification and primer extension (Figure 5). By chemically treating RNA with CMC, the pseudouridines retain a chemical adduct that blocks elongation by the reverse transcriptase, resulting in a truncated product during primer extension (73,74). Snora27 targets two nucleotides within the mouse 28S rRNA: 3371, in the E-site, as noted above and 4204, adjacent to the peptidyl transferase center (59). Unlike site 3371, which is targeted solely by Snora27, site 4204 is also targeted by Snora26 (Figure 5A; 59).
Figure 5.
Changes in 28S rRNA pseudouridylation coincide with altered Snora27 levels in RA-differentiated mESCs. (A) SnoRNAs and their known RNA target sites that were detected in the primer extension assays. (B) Pseudouridylation of the 28S rRNA at nucleotides 3351 and 3371, a target of Snora27, before and after mESC differentiation with RA was assessed using a CMC-based primer extension assay. Control lanes (−) were not treated with CMC and (+) lanes were treated with CMC. Sequencing lanes are shown on the left. (C) Pseudouridylation of the 28S rRNA at nucleotides 4204, a target of Snora27 and Snora26; 4228, a target of Snora40; and 4231 before and after mESC differentiation with RA was assessed using a CMC-based primer extension assay. Control lanes (−) were not treated with CMC and (+) lanes were treated with CMC. Sequencing lanes are shown on the left. (D). Bar graph plotting both snoRNA expression and the ψ-ratio (intensity of ψ in +CMC condition minus the intensity of ψ in −CMC) normalized to the undifferentiated (mESCs) state. Data are represented as mean ± SEM.
Using an oligonucleotide complementary to a sequence downstream of position 3371 of the 28S rRNA (Oligo 28S 3387), we detected pseudouridines at position 3371 and at position 3351 (Figure 5A, B). Consistent with our hypothesis, the reduction in pseudouridine at position 3371 correlated with the reduction in Snora27 abundance in RA-differentiated mESCs (Figure 5B, D). This correlation was specific to Ψ3371, which is only targeted by Snora27. We used a second oligonucleotide to detect Snora27- and Snora26-guided pseudouridylation at position 4204 (Oligo 28S 4238). This oligonucleotide also detected Ψ4228, which is guided by Snora40, and Ψ4231, whose guide RNA is unknown (Figure 5A, C). Unlike Snora27 abundance that is regulated during differentiation, Snora26 abundance as quantified by NanoString is stable during differentiation (Figure 5D). Consistent with stable Snora26 abundance, Ψ4204 was relatively unchanged in RA-differentiated mESCs compared to mESCs (Figure 5C, D), and the reduction of Snora27 was not sufficient to affect this shared site. Ψ4228 was also unchanged in RA-differentiated mESCs compared to undifferentiated mESCs, which correlated with the relatively stable levels of Snora40 as quantified by RNA-seq (Figure 5D). As discussed above, the NanoString nCounter probes do not capture all members of the Snora40 family equally (Figure 2E). As such, the small RNA-seq dataset provides the most accurate measure of the changes in expression. Thus, for Snora27, pseudouridylation of its target nucleotide in the E-site tRNA-binding region correlated with snoRNA abundance. The correlation between Snora27 abundance and modification suggests that changes in snoRNA abundance during differentiation may be a mechanism for fine-tuning ribosome function through changes in pseudouridylation of important functional regions.
Pre-rRNA processing is altered during differentiation
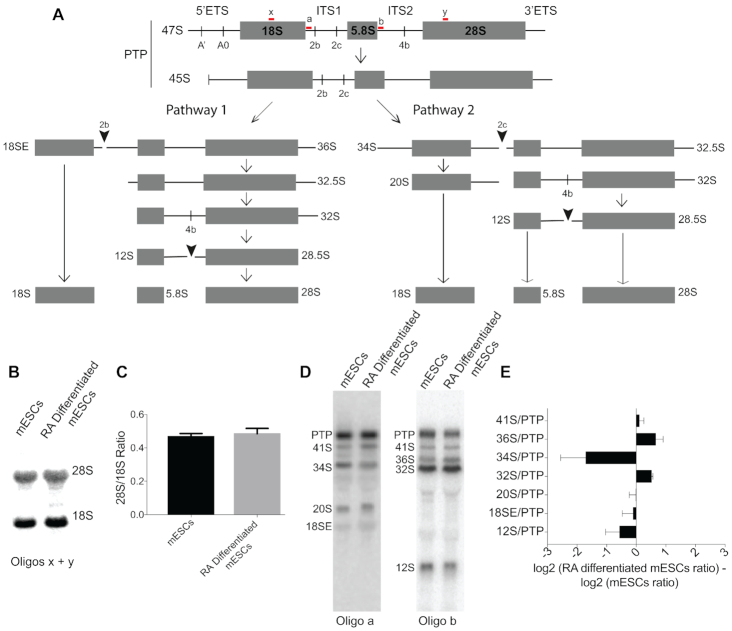
Since the most well documented targets of H/ACA snoRNAs are nucleotides within the rRNAs and because pseudouridylation of the rRNA occurs concurrent with processing of the pre-rRNA, we hypothesized that pre-rRNA processing may also be changed during differentiation. We therefore interrogated pre-rRNA processing in mESCs and RA-differentiated mESCs through Northern blot analysis. In mammalian cells, ribosome assembly initiates in the nucleolus with the transcription of a 47S primary transcript plus (PTP) that encodes the mature 18S, 5.8S, and 28S rRNAs as well as two external transcribed spacers (5′ ETS and 3′ ETS) and two internal transcribed spacers (ITS1 and ITS2) that must be removed through a series of endo- and exonucleolytic cleavage steps (Figure 6A). The primary transcript is processed through two parallel pathways, depending upon whether cleavage in ITS1 occurs at one of two positions, site 2b or site 2c (58). For each cleavage site, distinct pre-rRNA intermediates are generated. By using oligonucleotide probes complementary to regions within ITS1 (oligo a) and ITS2 (oligo b), we can detect these unique intermediates and ascertain the favored pathway for pre-rRNA processing (58).
Figure 6.
Pre-rRNA processing is altered upon differentiation. (A) Schematic of mouse pre-rRNA processing. PTP: Primary transcript plus. The oligonucleotide probes used for Northern blot analysis are shown in red. (B) Northern blot of the mature 28S and 18S rRNAs using oligos x and y shown in panel A of RNA from mESCs and mESCs differentiated with RA. (C) Ratio of 28S to 18S rRNAs quantified from the Northern blot in panel B. (D) Northern blots of RNA from mESCs and mESCs differentiated with RA using the oligonucleotide probes shown in panel A to detect pre-rRNA intermediates. (E) Profile comparing the ratios of each pre-rRNA to the PTP from mESCs and mESCs differentiated with RA. A positive log2 ratio indicates there is more of the pre-rRNA intermediate relative to the PTP in mESCs compared with mESCs differentiated with RA whereas a negative log2 ratio indicates there is less of the pre-rRNA intermediate relative to the PTP in mESCs compared with mESCs differentiated with RA.
Our analysis indicated that the pathway of pre-rRNA processing changes during differentiation of mESCs, although differences are not detected at the levels of mature 28S and 18S rRNAs (Figure 6B, C). Northern blot analysis of pre-rRNA intermediates revealed that upon differentiation of mESCs by RA treatment, Pathway 1 is used more frequently than Pathway 2 (Figure 6A), as indicated by an increase in 36S pre-rRNA levels relative to the primary transcript produced by site 2b cleavage and a corresponding decrease in 34S pre-rRNA levels produced by site 2c cleavage (Figure 6D, E). It has been proposed that the preference for Pathway 1 indicates that conditions are more favorable for small ribosomal subunit assembly than large ribosomal subunit assembly (58). In agreement with this proposal, we also detected an accumulation of 32S pre-rRNA and a decrease in 12S pre-rRNA, a direct product of 32S processing, relative to the primary transcript in RA-differentiated mESCs, which indicates less favorable processing of large subunit rRNAs.
DISCUSSION
To gain insight into how H/ACA snoRNAs, critical mediators of multiple cellular functions, are expressed and regulated during differentiation, we developed a custom NanoString nCounter assay to quantify their abundance. Our analyses revealed that H/ACA snoRNA levels are cell-type specific and are regulated during cellular differentiation in three murine model systems. Strikingly, we identified a core subset of snoRNAs whose levels are consistently regulated during differentiation of mESCs and during myogenesis. Additionally, we determined that the observed changes in snoRNA levels are largely independent of changes in their host genes, suggesting the snoRNA levels are regulated through mechanisms beyond transcription of the host gene. Furthermore, the changes we observed in Snora27 expression during mESC differentiation correlated with a change in pseudouridylation of its target nucleotide within the E-site tRNA binding region of the 28S rRNA, where it has the potential to influence translation fidelity. Taken together, our findings suggest that H/ACA snoRNA levels are key determinants of cell identity, and specific regulation of snoRNAs during differentiation and development has the potential to fine tune ribosome function through changes in rRNA pseudouridylation.
As H/ACA snoRNAs levels are changed upon stem cell differentiation, it raises the question, are altered H/ACA snoRNA levels a cause or consequence of differentiation? Our data do not differentiate between these options, but they add to the accumulating evidence supporting the idea that regulation of H/ACA snoRNA levels contributes to cell fate specification. For example, knockdown of the catalytic component of the box H/ACA snoRNP, Dkc1, results in differentiation of mESCs (42). This result suggests that changes in snoRNA levels or corresponding pseudouridylation may cause differentiation, since Dkc1 is essential for snoRNA stability with mutations in Dkc1 resulting in a reduction of specific snoRNA levels (43,75). The most direct evidence that a snoRNA may influence cell fate specification is the demonstration that SNORA7A, which is highly abundant in undifferentiated human umbilical cord mesenchymal stem cells, promotes proliferation and self-renewal while suppressing differentiation (44). In addition to our results demonstrating that snoRNA levels are cell-type specific and differentially expressed across multiple differentiation models, snoRNA levels are lineage-specific and developmentally regulated during human hematopoiesis (45). Differentially-expressed snoRNAs may play a role in promoting the exit of self-renewal, loss of pluripotency, or maintenance of the differentiated state. H/ACA snoRNAs likely promote differentiation and cell fate specification through multiple mechanisms, which may include modulating ribosome function, ribosome assembly, or modification of other cellular RNAs.
Changes in snoRNA abundance and pseudouridylation have been shown to influence ribosome function, which may be utilized to alter gene expression in stem cell differentiation. For example, in yeast, depletion of snoRNAs that guide pseudouridylation in functionally important regions of the ribosome leads to defects in translation, including stop codon readthrough, frameshifting, and reduced amino acid incorporation rates (8,10–12). Importantly, a recent publication detailing the role of SNORA24 in hepatocellular carcinoma revealed that ribosomes lacking SNORA24-guided pseudouridine modifications made more miscoding errors during translation (39). Moreover, mutations in DKC1, such as those associated with in X-linked dyskeratosis congentia, result in a decrease in H/ACA snoRNA abundance and impaired translation of internal ribosome entry site containing mRNAs (11,43,76). Strikingly, we identified that Snora27 levels are regulated during differentiation in multiple cellular models (Figure 3F), and we demonstrated that changes in Snora27 levels correlated with the degree of pseudouridylation of its target nucleotide within the E-site of the ribosome (Figure 5D and Supplementary Figure S6B). The E-site is critical for maintaining the translational reading frame and for preventing harmful misincorporation errors (72). Pseudouridylation changes within the E-site have the potential to affect the kinetics of E-site tRNA release, thereby altering the fidelity of translation. However, the mechanisms relating E-site pseudouridylation, alterations in E-site functionality and cellular differentiation remain to be determined. Moreover, Snora27 is one of several snoRNAs that target key functional regions of the ribosome and were differentially expressed during cellular differentiation (Table 3 and Supplementary Figure S6). Thus, snoRNAs have the potential to fine-tune ribosome function through regulated pseudouridylation of multiple important functional regions to control translation and affect gene expression during differentiation and development.
SnoRNAs may also promote cell fate specification by altering ribosome assembly. Here, we focus on the major function of snoRNAs to direct pseudouridylation, but snoRNAs have also been shown to modify pre-rRNA processing directly (18,77). The rRNA is pseudouridylated very early during ribosome biogenesis, and the resulting modifications are thought to play important roles in the assembly of ribosomal proteins and the formation of key RNA structures (78–80). Importantly, we identified several developmentally-regulated snoRNAs that target nucleotides in the region of the 28S rRNA that binds ribosomal protein RPL3 (Table 3 and Supplementary Figure S6D). RPL3 is essential for assembly of the large ribosomal subunit and for the function of the peptidyl transferase center (81,82). In yeast, loss of Rpl3p leads to defects in processing of the large rRNA precursors (83). Thus, snoRNA-mediated changes in pseudouridylation in the RPL3 binding region could influence the timing of recruitment of RPL3 or its ability to associate with and stabilize the assembling pre-rRNA, leading to changes in the cleavage patterns of the pre-rRNA. In line with this hypothesis, we observed a change in the processing of the large rRNA precursors upon RA-differentiation of mESCs (Figure 6). Further studies aimed at determining the importance of snoRNA-mediated pseudouridylation in recruiting ribosomal proteins such as RPL3 to the assembling ribosome will be pivotal for elucidating the potential mechanistic connections between regulated snoRNA expression, rRNA modifications, and ribosome assembly.
SnoRNAs may also contribute to cell fate specification by guiding pseudouridylation of RNA targets beyond the rRNA, leading to changes in the function or stability of those RNAs. Indeed, H/ACA snoRNA-guided pseudouridines have been identified in mRNAs, snoRNAs, and snRNAs (13,15,17). Snora49, which is upregulated upon RA differentiation of mESCs and myoblast differentiation (Figure 3E), is a leading H/ACA snoRNA candidate for mRNA targeting as it is not known to target the rRNA. The presence of a pseudouridine in an mRNA may alter its coding potential or fate. For example, pseudouridines within a stop codon have been shown to cause stop codon readthrough in vitro and in vivo (84). This indicates that pseudouridines can alter how the ribosome recognizes and decodes codons containing pseudouridine. Pseudouridines within the 5′ or 3′ untranslated regions (UTRs) could alter stability or translation of the mRNA (1). Moreover, box H/ACA snoRNP-mediated pseudouridine sites have been detected in the 5′UTR of the mRNA that encodes ribosomal protein RPL15 in human cells (13,15). The snoRNA(s) responsible for guiding these modifications have not been identified, yet the presence of pseudouridines within an mRNA encoding a ribosomal protein suggests that H/ACA snoRNAs could also regulate ribosome assembly by modulating the stability or translation of ribosomal protein mRNAs.
Our data raise another important question: what are the mechanisms regulating snoRNA abundance during differentiation? We found that snoRNA abundance is uncoupled from host gene expression, therefore, snoRNA levels are likely controlled through post-transcriptional mechanisms including their assembly into and association with the snoRNP, processing into snoRNA-derived small RNAs, and snoRNA turnover. Compared to mRNAs, little is known about how snoRNA levels are regulated. Interestingly, it has been shown, some snoRNAs can be capped with NAD+, which targets them for degradation (85). NAD+ is important for maintaining ‘stemness,’ as a reduction in cellular NAD+ levels in human embryonic stem cells leads to spontaneous differentiation (86). It is interesting to postulate that snoRNAs that promote differentiation, such as Snora30 or Snora49 (Figure 3E), are selectively capped with NAD+ and targeted for differentiation in stem cell populations. Deciphering how snoRNA levels are normally controlled during differentiation is imperative. It is likely that these mechanisms fail during disease pathogenesis, as multiple snoRNAs have been found to be dysregulated in cancer, lupus and hematological disorders (30–38,40).
H/ACA snoRNAs, like the ribosome, are not static. They are dynamic, noncoding RNAs with underappreciated regulatory potential during development and in disease. Our data point toward a potential model in which H/ACA snoRNAs are specifically regulated during differentiation to alter rRNA pseudouridylation and fine tune ribosome production or function. We note that the majority of the snoRNAs that were differentially expressed upon mESC treatment with RA (Table 3) target the large subunit rRNAs. However, further analyses will be needed to elucidate how these changes in snoRNA abundance influence pre-rRNA processing and ribosome funcition.
DATA AVAILABILITY
The snoRNA gene expression nCounter data and the small RNA-seq data have been submitted to GEO (Gene Expression Omnibus) under accession number GSE140623.
Supplementary Material
ACKNOWLEDGEMENTS
We thank K. Gerrish and R. Fannin of the NIEHS Molecular Genomics Core Facility for assistance with NanoString nCounter data collection. We thank G. Solomon and the staff of the NIEHS Epigenetics Core Facility for assistance with small RNA-sequencing and the staff of the NIEHS Integrative Bioinformatics Support Group. We thank our colleagues, P. Wade, M. Pillon and Y.H. Lo for stimulating discussion and critical comments.
Author contributions: Conceptualization, K.L.M. and T.M.T.H.; Methodology, K.L.M., A.B.B., T.M.T.H.; Investigation, K.L.M., S.L.K, A.B.B., B.T.P.; Writing – original draft, K.L.M., T.M.T.H.; Writing – Review & Editing; K.L.M., S.L.K., B.T.P, A.B.B., T.M.T.H; Funding Acquisition, T.M.T.H.
Notes
Present address: Sanam L. Kavari, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA.
Present address: Bart T. Phillips, Research Square, Durham, NC 27701, USA.
Contributor Information
Kathleen L McCann, Epigenetics and Stem Cell Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA.
Sanam L Kavari, Epigenetics and Stem Cell Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA.
Adam B Burkholder, Epigenetics and Stem Cell Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA.
Bart T Phillips, Epigenetics and Stem Cell Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA.
Traci M Tanaka Hall, Epigenetics and Stem Cell Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institutes of Health; National Institute of Environmental Health Sciences (to T.M.T.H.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Adachi H., De Zoysa M.D., Yu Y.T.. Post-transcriptional pseudouridylation in mRNA as well as in some major types of noncoding RNAs. Biochim. Biophys. Acta - Gene Regul. Mech. 2019; 1862:230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Henras A.K., Plisson-Chastang C., Humbert O., Romeo Y., Henry Y.. Synthesis, function, and heterogeneity of snoRNA-guided posttranscriptional nucleoside modifications in eukaryotic ribosomal RNAs. Enzymes. 2017; 41:169–213. [DOI] [PubMed] [Google Scholar]
- 3. Penzo M., Montanaro L.. Turning uridines around: Role of rRNA pseudouridylation in ribosome biogenesis and ribosomal function. Biomolecules. 2018; 8:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohn W.E., Volkin E.. Nucleoside-5′-phosphates from ribonucleic acid. Nature. 1951; 167:483–484. [Google Scholar]
- 5. Cantara W.A., Crain P.F., Rozenski J., McCloskey J.A., Harris K.A., Zhang X., Vendeix F.A.P., Fabris D., Agris P.F.. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011; 39:D195–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ofengand J. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett. 2002; 514:17–25. [DOI] [PubMed] [Google Scholar]
- 7. Decatur W.A., Fournier M.J.. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002; 27:344–351. [DOI] [PubMed] [Google Scholar]
- 8. Liang X. hai, Liu Q., Fournier M.J.. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007; 28:965–977. [DOI] [PubMed] [Google Scholar]
- 9. Piekna-Przybylska D., Przybylski P., Baudin-Baillieu A., Rousset J.P., Fournier M.J.. Ribosome performance is enhanced by a rich cluster of pseudouridines in the A-site finger region of the large subunit. J. Biol. Chem. 2008; 283:26026–26036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baudin-baillieu A., Fabret C., Liang X.H., Piekna-Przybylska D., Fournier M.J., Rousset J.P.. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 2009; 37:7665–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jack K., Bellodi C., Landry D.M., Niederer R.O., Meskauskas A., Musalgaonkar S., Kopmar N., Krasnykh O., Dean A.M., Thompson S.R. et al.. RRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell. 2011; 44:660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. King T.H., Liu B., McCully R.R., Fournier M.J.. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell. 2003; 11:425–435. [DOI] [PubMed] [Google Scholar]
- 13. Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M., Gilbert W. V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014; 515:143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Courtes F.C., Gu C., Wong N.S.C., Dedon P.C., Yap M.G.S., Lee D.Y.. 28S rRNA is inducibly pseudouridylated by the mTOR pathway translational control in CHO cell cultures. J. Biotechnol. 2014; 174:16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li X., Zhu P., Ma S., Song J., Bai J., Sun F., Yi C.. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 2015; 11:592–597. [DOI] [PubMed] [Google Scholar]
- 16. Lovejoy A.F., Riordan D.P., Brown P.O.. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One. 2014; 9:e110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schwartz S., Bernstein D.A., Mumbach M.R., Jovanovic M., Herbst R.H., León-Ricardo B.X., Engreitz J.M., Guttman M., Satija R., Lander E.S. et al.. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014; 159:148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chikne V., Doniger T., Rajan K.S., Bartok O., Eliaz D., Cohen-Chalamish S., Tschudi C., Unger R., Hashem Y., Kadener S. et al.. A pseudouridylation switch in rRNA is implicated in ribosome function during the life cycle of Trypanosoma brucei. Sci. Rep. 2016; 6:25296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hamma T., Ferré-D’Amaré A.R.. Pseudouridine synthases. Chem. Biol. 2006; 13:1125–1135. [DOI] [PubMed] [Google Scholar]
- 20. Kiss T., Fayet-Lebaron E., Jády B.E.. Box H/ACA small ribonucleoproteins. Mol. Cell. 2010; 37:597–606. [DOI] [PubMed] [Google Scholar]
- 21. Watkins N.J., Gottschalk A., Neubauer G., Kastner B., Fabrizio P., Mann M., Lührmann R.. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA- binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998; 4:1549–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lafontaine D.L.J., Bousquet-Antonelli C., Henry Y., Caizergues-Ferrer M., Tollervey D.. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998; 12:527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bousquet-Antonelli C., Henry Y., Gélugne J.P., Caizergues-Ferrer M., Kiss T.. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J. 1997; 16:4770–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dragon F., Pogacic V., Filipowicz W.. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 2000; 20:3037–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Henras A., Henry Y., Bousquet-Antonelli C., Noaillac-Depeyre J., Gélugne J.P., Caizergues-Ferrer M.. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998; 17:7078–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang C., Meier U.T.. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004; 23:1857–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Balakin A.G., Smith L., Fournier M.J.. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996; 86:823–834. [DOI] [PubMed] [Google Scholar]
- 28. Ganot P., Bortolin M.L., Kiss T.. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997; 89:799–809. [DOI] [PubMed] [Google Scholar]
- 29. Ni J., Tien A.L., Fournier M.J.. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997; 89:565–573. [DOI] [PubMed] [Google Scholar]
- 30. Mannoor K., Shen J., Liao J., Liu Z., Jiang F.. Small nucleolar RNA signatures of lung tumor-initiating cells. Mol. Cancer. 2014; 13:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mei Y.P., Liao J.P., Shen J., Yu L., Liu B.L., Liu L., Li R.Y., Ji L., Dorsey S.G., Jiang Z.R. et al.. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 2012; 31:2794–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Okugawa Y., Toiyama Y., Toden S., Mitoma H., Nagasaka T., Tanaka K., Inoue Y., Kusunoki M., Boland C.R., Goel A.. Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer. Gut. 2017; 66:107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang X., Li Y., Li L., Liu J., Wu M., Ye M.. SnoRNAs are involved in the progression of ulcerative colitis and colorectal cancer. Dig. Liver Dis. 2017; 49:545–551. [DOI] [PubMed] [Google Scholar]
- 34. Yoshida K., Toden S., Weng W., Shigeyasu K., Miyoshi J., Turner J., Nagasaka T., Ma Y., Takayama T., Fujiwara T. et al.. SNORA21 – an oncogenic small nucleolar RNA, with a prognostic biomarker potential in human colorectal cancer. EBioMedicine. 2017; 22:68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nogueira Jorge N.A., Wajnberg G., Ferreira C.G., de Sa Carvalho B., Passetti F.. snoRNA and piRNA expression levels modified by tobacco use in women with lung adenocarcinoma. PLoS One. 2017; 12:e0183410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu L., Zheng J., Chen P., Liu Q., Yuan Y.. Small nucleolar RNA ACA11 promotes proliferation, migration and invasion in hepatocellular carcinoma by targeting the PI3K/AKT signaling pathway. Biomed. Pharmacother. 2017; 90:705–712. [DOI] [PubMed] [Google Scholar]
- 37. Ronchetti D., Mosca L., Cutrona G., Tuana G., Gentile M., Fabris S., Agnelli L., Ciceri G., Matis S., Massucco C. et al.. Small nucleolar RNAs as new biomarkers in chronic lymphocytic leukemia. BMC Med. Genomics. 2013; 6:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Valleron W., Ysebaert L., Berquet L., Fataccioli V., Quelen C., Martin A., Parrens M., Lamant L., De Leval L., Gisselbrecht C. et al.. Small nucleolar RNA expression profiling identifies potential prognostic markers in peripheral T-cell lymphoma. Blood. 2012; 120:3997–4005. [DOI] [PubMed] [Google Scholar]
- 39. McMahon M., Contreras A., Holm M., Uechi T., Forester C.M., Pang X., Jackson C., Calvert M.E., Chen B., Quigley D.A. et al.. A single H/ACA small nucleolar RNA mediates tumor suppression downstream of oncogenic RAS. Elife. 2019; 8:e48847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lai N.-S., Yu H.-C., Huang K.-Y., Tung C.-H., Huang H.-B., Lu M.-C.. Decreased T cell expression of H/ACA box small nucleolar RNA 12 promotes lupus pathogenesis in patients with systemic lupus erythematosus. Lupus. 2018; 27:1499–1508. [DOI] [PubMed] [Google Scholar]
- 41. O’Brien J.E., Kibiryeva N., Zhou X.G., Marshall J.A., Lofland G.K., Artman M., Chen J., Bittel D.C.. Noncoding RNA expression in myocardium from infants with tetralogy of fallot. Circ. Cardiovasc. Genet. 2012; 5:279–286. [DOI] [PubMed] [Google Scholar]
- 42. Fong Y.W., Ho J.J., Inouye C., Tjian R.. The dyskerin ribonucleoprotein complex as an OCT4/SOX2 coactivator in embryonic stem cells. Elife. 2014; 3:e03573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bellodi C., McMahon M., Contreras A., Juliano D., Kopmar N., Nakamura T., Maltby D., Burlingame A., Savage S., Shimamura A. et al.. H/ACA small RNA dysfunctions in disease reveal key roles for noncoding RNA modifications in hematopoietic stem cell differentiation. Cell Rep. 2013; 3:1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang Y., Xu C., Gu D., Wu M., Yan B., Xu Z., Wang Y., Liu H.. H/ACA box small nucleolar RNA 7A promotes the self-renewal of human umbilical cord mesenchymal stem cells. Stem Cells. 2017; 35:222–235. [DOI] [PubMed] [Google Scholar]
- 45. Warner W.A., Spencer D.H., Trissal M., White B.S., Helton N., Ley T.J., Link D.C.. Expression profiling of snoRNAs in normal hematopoiesis and AML. Blood Adv. 2018; 2:151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Oatley J.M., Brinster R.L.. Spermatogonial stem cells. Methods Enzymol. 2006; 419:259–282. [DOI] [PubMed] [Google Scholar]
- 47. Kita-Matsuo H., Barcova M., Prigozhina N., Salomonis N., Wei K., Jacot J.G., Nelson B., Spiering S., Haverslag R., Kim C. et al.. Lentiviral vectors and protocols for creation of stable hESC lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLoS One. 2009; 4:e5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fuegemann C.J., Samraj A.K., Walsh S., Fleischmann B.K., Jovinge S., Breitbach M.. Differentiation of mouse embryonic stem cells into cardiomyocytes via the hanging-drop and mass culture methods. Curr. Protoc. Stem Cell Biol. 2010; doi:10.1002/9780470151808.sc01f11s15. [DOI] [PubMed] [Google Scholar]
- 49. Shurtleff M.J., Yao J., Qin Y., Nottingham R.M., Temoche-Diaz M.M., Schekman R., Lambowitz A.M.. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E8987–E8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Qin Y., Yao J., Wu D.C., Nottingham R.M., Mohr S., Hunicke-Smith S., Lambowitz A.M.. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA. 2016; 22:111–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C.. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017; 14:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. et al.. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009; 55:611–622. [DOI] [PubMed] [Google Scholar]
- 54. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liao Y., Smyth G.K., Shi W.. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 56. Chandrasekaran V., Juszkiewicz S., Choi J., Puglisi J.D., Brown A., Shao S., Ramakrishnan V., Hegde R.S.. Mechanism of ribosome stalling during translation of a poly(A) tail. Nat. Struct. Mol. Biol. 2019; 26:1132–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pestov D.G., Lapik Y.R., Lau L.F.. Assays for ribosomal RNA processing and ribosome assembly. Curr. Protoc. Cell Biol. 2008; doi:10.1002/0471143030.cb2211s39. [DOI] [PubMed] [Google Scholar]
- 58. Wang M., Anikin L., Pestov D.G.. Two orthogonal cleavages separate subunit RNAs in mouse ribosome biogenesis. Nucleic Acids Res. 2014; 42:11180–11191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yoshihama M., Nakao A., Kenmochi N.. SnOPY: a small nucleolar RNA orthological gene database. BMC Res. Notes. 2013; 6:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lestrade L., Weber M.J.. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006; 34:D158–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pignatelli M., Vilella A.J., Muffato M., Gordon L., White S., Flicek P., Herrero J.. NcRNA orthologies in the vertebrate lineage. Database. 2016; 2016:bav127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kalvari I., Argasinska J., Quinones-Olvera N., Nawrocki E.P., Rivas E., Eddy S.R., Bateman A., Finn R.D., Petrov A.I.. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018; 46:D335–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rochette-Egly C. Retinoic acid signaling and mouse embryonic stem cell differentiation: cross talk between genomic and non-genomic effects of RA. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids. 2015; 1851:66–75. [DOI] [PubMed] [Google Scholar]
- 64. Zhang J., Gao Y., Yu M., Wu H., Ai Z., Wu Y., Liu H., Du J., Guo Z., Zhang Y.. Retinoic acid induces embryonic stem cell differentiation by altering both encoding RNA and microRNA expression. PLoS One. 2015; 10:e0132566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jorjani H., Kehr S., Jedlinski D.J., Gumienny R., Hertel J., Stadler P.F., Zavolan M., Gruber A.R.. An updated human snoRNAome. Nucleic Acids Res. 2016; 44:5068–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Burattini S., Ferri R., Battistelli M., Curci R., Luchetti F., Falcieri E.. C2C12 murine myoblasts as a model of skeletal muscle development: morpho-functional characterization. Eur. J. Histochem. 2004; 48:223–233. [PubMed] [Google Scholar]
- 67. Dieci G., Preti M., Montanini B.. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009; 94:83–88. [DOI] [PubMed] [Google Scholar]
- 68. Boivin V., Deschamps-Francoeur G., Couture S., Nottingham R.M., Bouchard-Bourelle P., Lambowitz A.M., Scott M.S., Abou-Elela S.. Simultaneous sequencing of coding and noncoding RNA reveals a human transcriptome dominated by a small number of highly expressed noncoding genes. RNA. 2018; 24:950–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Maxwell E., Fournier M.. The small nucleolar RNAs. Annu. Rev. Biochem. 1995; 64:897–934. [DOI] [PubMed] [Google Scholar]
- 70. Agarwala R., Barrett T., Beck J., Benson D.A., Bollin C., Bolton E., Bourexis D., Brister J.R., Bryant S.H., Canese K. et al.. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2018; 47:D23–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lykke-Andersen S., Chen Y., Ardal B.R., Lilje B., Waage J., Sandelin A., Jensen T.H.. Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev. 2014; 28:2498–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Márquez V., Wilson D.N., Tate W.P., Triana-Alonso F., Nierhaus K.H.. Maintaining the ribosomal reading frame: The influence of the E site during translational regulation of release factor 2. Cell. 2004; 118:45–55. [DOI] [PubMed] [Google Scholar]
- 73. Bakin A., Ofengand J.. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry. 1993; 32:9754–9762. [DOI] [PubMed] [Google Scholar]
- 74. Ho N.W.Y., Gilhamt P.T.. Reaction of pseudouridine and inosine with N-cyclohexyl-N’-γ- (4-methylmorpholinium) ethylcarbodiimide. Biochemistry. 1971; 10:3651–3657. [PubMed] [Google Scholar]
- 75. Mochizuki Y., He J., Kulkarni S., Bessler M., Mason P.J.. Mouse dyskerin mutations affect accumulation of telomerase RNA and small nucleolar RNA, telomerase activity, and ribosomal RNA processing. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:10756–10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yoon A., Peng G., Brandenburg Y., Zollo O., Xu W., Rego E., Ruggero D.. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006; 312:902–906. [DOI] [PubMed] [Google Scholar]
- 77. Enright C.A., Stuart Maxwell E., Sollner-Webb B.. 5′ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3. RNA. 1996; 2:1094–1099. [PMC free article] [PubMed] [Google Scholar]
- 78. Maden B.E.H., Forbes J.. Standard and non standard products in combined T1 plus pancreatic RNAase fingerprints of HeLa cell rRNA and its precursors. FEBS Lett. 1972; 28:289–292. [DOI] [PubMed] [Google Scholar]
- 79. Maden B.E.H. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1990; 39:241–303. [DOI] [PubMed] [Google Scholar]
- 80. Bertrand E., Fournier M.. The snoRNPs and related machines: ancient devices that mediate maturation of rRNA and other RNAs. Madame Curie Bioscience Database. 2000-2013); Landes Bioscience. [Google Scholar]
- 81. Schulze H., Nierhaus K.H.. Minimal set of ribosomal components for reconstitution of the peptidyltransferase activity. EMBO J. 1982; 1:609–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nam H.G., Fried H.M.. Effects of progressive depletion of TCM1 or CYH2 mRNA on Saccharomyces cerevisiae ribosomal protein accumulation. Mol. Cell. Biol. 1986; 6:1535–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rosado I. V., Kressler D., de la Cruz J.. Functional analysis of Saccharomyces cerevisiae ribosomal protein Rpl3p in ribosome synthesis. Nucleic Acids Res. 2007; 35:4203–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Karijolich J., Yu Y.T.. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011; 474:395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jiao X., Doamekpor S.K., Bird J.G., Nickels B.E., Tong L., Hart R.P., Kiledjian M.. 5′ end nicotinamide adenine dinucleotide cap in human cells promotes RNA decay through DXO-mediated deNADding. Cell. 2017; 168:1015–1027.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Son M.J., Son M.Y., Seol B., Kim M.J., Yoo C.H., Han M.K., Cho Y.S.. Nicotinamide overcomes pluripotency deficits and reprogramming barriers. Stem Cells. 2013; 31:1121–1135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The snoRNA gene expression nCounter data and the small RNA-seq data have been submitted to GEO (Gene Expression Omnibus) under accession number GSE140623.