Abstract
The glucocorticoid receptor is an important immunosuppressive drug target and metabolic regulator that acts as a ligand-gated transcription factor. Generally, GR’s anti-inflammatory effects are attributed to the silencing of inflammatory genes, while its adverse effects are ascribed to the upregulation of metabolic targets. GR binding directly to DNA is proposed to activate, whereas GR tethering to pro-inflammatory transcription factors is thought to repress transcription. Using mice with a point mutation in GR’s zinc finger, that still tether via protein–protein interactions while being unable to recognize DNA, we demonstrate that DNA binding is essential for both transcriptional activation and repression. Performing ChIP-Seq, RNA-Seq and proteomics under inflammatory conditions, we show that DNA recognition is required for the assembly of a functional co-regulator complex to mediate glucocorticoid responses. Our findings may contribute to the development of safer immunomodulators with fewer side effects.
INTRODUCTION
Steroids such as glucocorticoids are widely used immunosuppressants with potent effects on mammalian physiology. Upon ligand binding, the glucocorticoid receptor (GR) enters the nucleus to both activate and repress gene expression and thereby control diverse biological processes, including development, metabolism and inflammation (1–3). However, the mechanisms determining transcriptional activation versus repression remain incompletely understood. Generally, GR is known to dimerize and bind palindromic consensus DNA sequences termed glucocorticoid response elements (GREs), which is largely thought to activate target genes. On the other hand, ‘tethering’ of GR indirectly to chromatin (potentially as a monomer), for example by binding AP-1 or NF-κB proteins instead of DNA sequences, is proposed to mediate negative regulation (4–6).
Many mechanistic studies on the suppression of inflammatory responses by GR have therefore focused on protein–protein interactions with transcription factors such as AP-1 or NF-κB (4,7–9). However, when we profiled GR-occupied promoters and enhancers associated with repressed inflammatory target genes by ChIP-Seq and reporter screening, we unexpectedly found classical GREs to be significantly enriched. This opened up the possibility that DNA binding by the GR may also be involved in the negative regulation of inflammatory target genes (10,11).
Further corroborating this hypothesis, we identified GRE half site motifs in GR ChIP-sequences obtained from dimerization-defective GRdim mutant macrophages, which implicates DNA contacts (12). The GRdim point mutant mouse line was originally designed to separate dimeric DNA binding from monomeric protein–protein interactions. Analyses of GRdim mutants initially supported the tethering model, but in the meantime were found to actually bind DNA, calling for reinterpretation and re-analysis (8,13). Concordantly, recent studies have shown GR to bind directly to response elements embedded in the DNA sequence of AP-1 and NF-κB motifs (14,15).
To gain mechanistic insight into potentially polarizing scenarios, we have now created a mouse model which selectively interferes with DNA recognition while maintaining protein–protein interactions. Here we show that essentially all glucocorticoid responses in inflammatory and metabolic cells and tissues require direct DNA binding of the receptor, and that tethering alone is not sufficient to explain GR’s anti-inflammatory actions.
MATERIALS AND METHODS
Animals
The GRΔZn line was generated by Cyagen US Inc. via homologous recombination in mouse ES cells. The C437G (TGC to GGC) mutation was introduced into exon 3 of Nr3c1 via the 5′ homology arm, with a LoxP flanked Neomycin cassette in the next intron. After breeding to ROSA26 Cre deleters for Neo removal, GRΔZn mice were kept on a C57BL/6 background. Please see Supplementary Figure S1 for verification of the mutation by PCR, digestion and sequencing.
Mice were housed in a controlled SPF facility with a 12 h light/dark cycle at 23°C with constant humidity and fed ad libitum. Genotyping primers are listed in Supplementary Table S1.
GR null mice were generated by crossing GRflox mice (a generous gift from J. Tuckermann, Ulm, Germany) with ROSA26 Cre deleters.
Formal approval for animal experiments was obtained from the district government of Upper Bavaria (55.2-1-54-2532-33-14 and ROB-55.2-2532.Vet_02-19-43) in accordance with HMGU guidelines for the care and use of animals.
Primary cell cultures
Mouse embryonic fibroblasts MEFs
MEFs were generated from E13.5 embryos after 12 min trypsinization (0.25% Trypsin–EDTA) at 37°C and homogenization following standard protocols. Non-immortalized cells up to passage 4 were cultured in high glucose DMEM with 10% FBS and 1% Pen/Strep at 37°C and 5% CO2. Dialyzed FBS was used for ligand-free samples.
Fetal liver macrophages
Livers from E13.5 embryos were washed twice with cold PBS and then homogenized. Red blood cells were lysed with AKC lysis buffer (155 mM NH4Cl, 10 mM KHCO3 and 0.1 mM EDTA) and remaining cells were kept in macrophage differentiation medium (30% L929 conditioned medium, 20% FBS, 1% Pen/Strep and high glucose DMEM supplemented with M-CSF (315-02, PeproTech)). Macrophages were differentiated during 7 days on non-coated plates and cellular identity was validated by qRT-PCR (Supplementary Figure S4b).
Cell lines
The male CV-1 fibroblast cell line was used for luciferase measurements and was cultured in high glucose DMEM with 10% FBS and 1% Pen/Strep at 37°C with 5% CO2. Dialyzed FBS was used for ligand-free samples.
ChIP coupled to mass spectrometry (ChIP-MS)
ChIP-MS was performed in wild type and homozygous GRΔZn MEFs treated with 1 μM Dex overnight and 100 ng/μl LPS for 3 h. ChIP-MS was carried out as described previously with minor modifications (11): cells were lysed in IP-buffer (50 mM Tris–HCl pH 8, 100 mM NaCl, 5 mM EDTA, 0.3% SDS, 1.7% Triton X-100) and chromatin was sonicated to an average size of 200 bp. After overnight immunoprecipitation with rabbit α-GR (24050-1-AP, Proteintech) or rabbit IgG (2729, Cell Signaling), antibody-bait complexes were captured by Protein A coupled Sepharose beads (CL-4B, GE healthcare), washed, eluted and analyzed by mass spectrometry.
Peptide separation, chromatography and mass acquisition was performed as described with the following changes: we used an EASY‐nLC 1200 ultra‐high‐pressure system, a gradient over 100 min, 10 data‐dependent MS/MS scans (15 K resolution, 60 ms max. injection time, AGC targets 1e5), an isolation window of 1.4, normalized collision energy of 27 and 30 s exclusion for multiple sequencing of peptides.
ChIP-MS data analysis
Raw mass spectrometry data were analyzed with MaxQuant (v1.5.3.54) and Perseus (v.1.6.0.2078) as previously reported (11). Protein entries referring to contaminants, identified via matches to the reverse database, or identified only via modified sites, were removed, LFQ values were log2 transformed. Significant outliers were defined by permutation-controlled Student's t-test (FDR < 0.01, s0 = 1) comparing triplicate ChIP-MS samples for each antibody, requiring at least two valid values in the GR replicates. Functional enrichment annotation was carried out using GOrilla (16).
We first defined this set of putative GR interactors with the FDR cutoff described above (over IgG), and then determined differential enrichment between genotypes by Student's t-test P-value (cutoff P < 0.01, fold change > 2).
The mass spectrometry proteomics data have been deposited at the PRIDE ProteomeXchange Consortium repository (17) with the dataset identifier PXD013772. Please see List S3 for a list of peptides.
ChIP-Seq and data analysis
ChIP-Seq was performed on 15 million MEFs per biological replicate, treated with 1 μM Dex overnight or with 1 μM Dex overnight plus 100 ng/μl LPS for 3 h. Cells were fixed with disuccinimidyl glutarate (DSG) for 30 min and 1% formaldehyde for 10 min as previously described (10), with the addition of 1 h pre-clearing with α-rabbit Dynabeads (Invitrogen). Rabbit α-GR (24050-1-AP, Proteintech) antibody was used for immunoprecipitation. Enrichment was quantified using the Power SYBR Green Master Mix (Life Technologies) in a ViiA 7 or QuantStudio Real-Time PCR System (Thermo Fischer). Libraries were prepared and subjected to NGS on an Illumina HiSeq4000 following standard protocols as described (11).
Reads from two to four replicates were merged and aligned to the mouse mm10 reference genome using BWA-MEM version 0.7.136 and duplicates were removed using Picard Tools version 2.8.3 (http://picard.sourceforge.net/). Reads were filtered for uniquely mapped read pairs with samtools7 (18) and downsampled to 12 mio read pairs for Dex + LPS, and 7 mio read pairs for Dex treated MEFs. Genome browser tracks were visualized with UCSC (https://genome.ucsc.edu/) or IGB (http://bioviz.org/igb) browsers after merging all replicates of one sample and downsampling to 64 mio reads pairs, or as individual replicates after downsampling as mentioned above. Peaks were called using MACS2 version 2.1.1.201603099 with an FDR threshold of 0.05 in paired-end mode. The union of peaks called in all replicates per genotype and treatment was used to compare GR peaks (GR universe), and the peak overlap (min. overlap 1 bp) was determined after resizing peaks to 294 bp around the peak center. Genomic regions called in GR knockout MEFs, as well as blacklisted regions (http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/mm10-mouse/mm10.blacklist.bed.gz) were removed from the GR universe for all further analyses. Gene Ontology and distance to TSS analyses were performed with GREAT version 3.0.0 (19). Motif enrichment and read distribution analysis around GR peaks were conducted with HOMER (20). Known HOMER motifs enriched within complete peaks are displayed as position-weight matrices. Heatmaps were generated with HOMER’s ‘annotatePeak’ function (version 4.98) and visualized with R, after log-transformation using the ‘heatmap.2’ function. In order to quantify GREs, GREs listed in the HOMER motif database were annotated per peak (palindromic: motif145 and motif6, or half site GRE: motif7). The percentage of peaks containing each motif per peak subset are reported as bar plots.
NGS data and annotated peak files can be accessed via the NCBI’s Gene Expression Omnibus Super Series accession number GSE126655. Please see List S1 for ChIP peak lists.
ChIP-qPCR
For MEFs and fetal liver macrophages, ChIP was performed on disuccinimidyl glutarate (DSG) and formaldehyde crosslinked chromatin as previously described (10). Embryonic livers were lysed in 10 mM Hepes, 10 mM KCl, 5 mM MgCl2, 0.5 mM DTT with proteinase/phosphatase inhibitors using a TissueLyser (Qiagen) with steel beads. Hepatocytes were passed through a 70 μm cell strainer and crosslinked in 1% formaldehyde for 15 min. These antibodies were used for immunoprecipitation: rabbit α-GR (24050-1-AP, Proteintech), rabbit α-GRIP-1/SRC2 (ab10491, Abcam), rabbit IgG (2729, Cell Signaling) and rabbit α-p65 (ab7970, Abcam). ChIP DNA was quantified using the Power SYBR Green Master Mix (Life Technologies) in a ViiA 7 or QuantStudio Real-Time PCR System (Thermo Fischer). Primers are listed in Supplementary Table S2.
Co-IP
MEFs were treated with 1 μM Dex overnight and 100 ng/μl LPS for 3 h. Nuclear protein extracts were pre-cleared using α-rabbit Dynabeads (Invitrogen) for 1 h in IP buffer (20 mM Tris pH 8, 100 mM KCl, 5 mM MgCl2, 0.2 mM EDTA, 20% glycerol and protease inhibitors). Protein lysates were pre-cleared for 1 h with Dynabeads. IPs were incubated with rabbit α-GR antibody (24050-1-AP, Proteintech) or rabbit IgG (2729, Cell Signaling) for 2 h and BSA-blocked rabbit Dynabeads were added for overnight immunoprecipitation at 4°C. Beads were washed 3× with IP buffer and eluted in Laemmli buffer with DTT at 37°C. Western blotting was performed according to standard protocols using mouse α-GR (sc-393232, Santa Cruz) and rabbit α-p65 (6956, Cell Signaling) antibodies.
Gel shift assay
Gel shift assays were performed similarly to Schauwaers et al. (21). In short, full-length mouse GR wild type or ΔZn was overexpressed in CV-1 cells. Twenty four hours after transfection, cells were washed twice with ice-cold PBS and resuspended in 200 μl lysis buffer (20 mM HEPES–KOH (pH 7.8), 450 mM NaCl2, 0.4 mM EDTA, 25% glycerol, 0.5 mM DTT and protease/phosphatase inhibitors). Cells were lysed by three freeze-thaw cycles in liquid N2, then centrifuged at 9000 × g and the supernatant was stored at –80°C. About 20 μg of CV-1 protein lysate were used per reaction. The following 5′-biotinylated oligos were used (Sigma): Per1 AGAGAACACGATGTTCCCTA (forward), Per1 TAGGGAACATCGTGTTCTCT (reverse) and negative control GATCGATCGATCGATCGATC.
Binding reactions were performed in 20 mM HEPES–KOH (pH 7.9), 60 mM KCl, 5 mM MgCl2, 2 mM DTT, 10% glycerol, 0.1 μg/μl Poly(dI/dC), 0.1 mg/μl BSA, 0.1 μM dexamethasone and 1 mM ZnCl2 during 20 min at room temperature. Protein:DNA complexes were run on a 6% acrylamide TBE gel (Thermo) and detected using the Chemiluminescent Nucleic Acid Detection Module Kit (Thermo) according to manufacturer's instructions.
Histology
Haematoxylin & Eosin stainings of E18.5 lungs were performed on 10 μm thorax paraffin sections following standard protocols. We used a Keyence BZ-9000 microscope at a magnification of 40× and 20×. The embryonic sex was GRWT/WT XY and GRΔZn XY.
Immunohistochemistry
MEFs were seeded onto cover slips and treated with vehicle, 1 μM Dex overnight and/or 100 ng/μl LPS for 3 h unless otherwise specified. Cells were fixed in 4% PFA, permeabilized with 0.1% TritonX-100 and blocked with 0.1% TritonX-100 and 1% FBS. Rabbit α-GR (12041, Cell Signaling), α-rabbit IgG Cy5 and DAPI staining was performed following standard procedure. Confocal images were taken on a Leica SP5 microscope.
Luciferase assays
Luciferase assays were performed in CV-1 cells treated overnight with either vehicle, 1 μM dexamethasone (Dex) or 100 ng/μl lipopolysaccharide (LPS), as previously described (10,12). Relative luciferase activity was normalized to vehicle and empty vector. cis-regulatory elements were cloned into pGL4.23, transfection efficiency was determined with pRL-TK Renilla, and luminescence was measured using the Dual Stop & Glo kit (Promega) according to manufacturer's instructions.
MMTV, Il6 and Btg1 reporters are published (10), the Lpin1 reporter was cloned into pGL4.23 using the following primers: CGAAGCTTGATATAGGTGCCCCATTTAG (forward), and CGCTCGAGATAGAAATCACACAGAGGTC (reverse).
The human GR DBD mutants (wild type GR, C421G, F444G, F445G, I465G and C473G) were kindly provided by the Evans lab (22).
The IFN1 reporter construct was obtained from Switchgear Genomics (GR pathway) and transfected together with an expression vector for human GR (11), and the following IFN1 insert sequence: AGCAACAGTGGGATAAGATTGGAAAGGTATATTGGGGGCTTATTCAAGAGGAACTTAGAATATGGAACTGGGTATTTGTAATTCAAAGGACATTGTTAATGTAGAGATGTTCACATTAGTTTACCAGAGCATCACTATTCTAGGGAATATACGCTAACCAAATGCCCCCAATCTCTCTTCACATATAATTTTGTCATTATAATGGATATATATCATTTAATTATATGGCTGTACCAACATATATGTAATCATTCACCTGTCGTTGAACCTATAGGTTGTTTCTAATATTCTGCTTTGCAATAATATTATAACACTCTTCTTTTGCTAATTTTTTTGCATTGCTTCTATAGGCTAGAATCTTAGATATTAAACTGATAGGATAAGATATAAAATAATTTAAGATTGCTGATATATGTTTTAAAATTAATTATTTGCTCAAGCATTTGTGACAATTTACAGTTCTAATTAGGTTTTAAATTTAGTAGTTTGTAGGTATTTTAAGTTTTGCCCCTGAATTCTTTATAGGTGCTGATAAGCCTTTGGTAAGTTTTACTCCATGAAAGACTATTACTGAAAAAAACGTAATCTCAATAAAAGAACTTTAATAAGCTTGACTAAATATTTAGAAAGCACATTGTGTTCAGTGAAACTTTGTATATAATGAATAGAATAATAAAAGATTATGTTGGATGACTAGTCTGTAATTGCCTCAAGGAAAGCATACAATGAATAAGTTATTTTGGTACTTCCTCAAAATAGCCAACACAATAGGGAAATGGAGAAAATGTACTCTGAACACCATGAAAAGGGAACCTGAAAATCTAATGTGTAAACTTGGAGAAATGACATTAGAAAACGAAAGCAACAAAAGAGAACACTCTCCAAAATAATCTGAGATGCATGAAAGGCAAACATTCACTAGAGCTGGAATTTCCCTAAGTCTATGCAGGGATAAGTAGCATATTTGACCTTCACCATGATTATCAAGCACTTCTTTGGAACTGTGTTGGTGCTGCTGGCCTCTACCACTATCTTC.
Mutagenesis of the GRE (marked in bold and underlined) was performed using the Q5 site-directed mutagenesis kit from NEB, with the following primers: gcactcaTCAGTGAAACTTTGTATATAATG (forward) and aacacgctGCTTTCTAAATATTTAGTCAAGC (reverse).
Quantification and statistical analysis
For differences between two groups, unpaired two-tailed Student's t-test was performed. Results are given as mean ± SEM unless otherwise specified. A P-value < 0.05 was considered significant.
No statistical methods were used to predetermine sample size. The experiments were not randomized and investigators were not blinded to allocation during experiments and analyses.
RNA isolation, cDNA synthesis and RT-qPCR
Total RNA was extracted with RNeasy kits and cDNA was prepared with the QuantiTect Reverse Transcription Kit (Qiagen) following manufacturer's instructions. The Power SYBR Green Master Mix (Life Technologies) was used in a ViiA 7 or QuantStudio Real-Time PCR System (Thermo Fischer). Expression was normalized to U36b4 (or Rpl38 for the adrenal glands). Primers are listed in Supplementary Table S3.
RNA-Seq and data analysis
MEFs were treated with 1 μM Dex overnight and/or 100 ng/μl LPS for 6 h. Total RNA was isolated using the RNeasy kit (Qiagen) and quality-controlled on a 2100 Bioanalyzer (Agilent Technologies). Library preparation and rRNA depletion were performed using the Illumina TruSeq mRNA Library Prep Kit v2 chemistry in an automated system (Agilent Bravo liquid handling platform) from 1 μg total RNA. Libraries were sequenced on the Illumina HiSeq4000.
Sequencing quality was assessed with FastQC (http://www.bioinformatics.babraham.ac.uk/ projects/fastqc/). Reads were mapped to the mouse genome mm10 (Ensembl build 38.91) and reads per gene were counted using STAR version 2.4.2a2 (23). Gene count normalization and differential expression analysis was performed using DESeq2 (24). Genes were defined as expressed when their mean count across samples passed 200, and differentially expressed when the fold-change between treatment and reference group was greater than 1.5 at an FDR <0.05. For gene annotation, biomaRt (25) was used. Functional enrichment according to gene ontology was carried out using GOrilla (16). Heatmaps and volcano plots were generated in R (www.R-project.org). The heatmap was generated in R using DESeq2-normalized read counts.
NGS data can be accessed via the NCBI’s Gene Expression Omnibus Super Series accession number GSE126655. Please see List 2 for the list of differentially expressed genes.
siRNA knockdowns
Cells were seeded to 60% confluency and transfected with 10 nM siRNA oligonucleotides using Lipofectamine RNAiMAX (Thermo) according to the manufacturer's instructions. Cells were treated with vehicle, 100 ng/μl LPS for 3 h or 1 μM Dex overnight and 100 ng/μl LPS for 3 h. Cells were collected 72 h after transfection with siRNA. The following oligonucleotides were used: non-targeting scramble control (D-001206-13), siNr3c1 (M-045970-01), siArid1a (M-040694-01), siArid1b (M-053908-01), and siArid5b (M-054678-01) (Dharmacon siGENOME SMARTpool). qRT-PCR primers are listed in Supplementary Table S3.
Western blot
Cells were lysed with RIPA buffer (50 mM Tris–HCl pH 8, 150 mM NaCl, 1% NP40, 0.1% SDS and 0.5% sodium deoxycholate) supplemented with protease and phosphatase inhibitors. Western blotting on sonicated, snap-frozen lysates was performed according to standard protocols with these antibodies: mouse α-actin (sc-56459, Santa Cruz), mouse α-GR (sc-393232, Santa Cruz), rabbit α-GR S212 (mouse) / S203 (human) (abx011845, Abbexa), rabbit α-GR S220 (mouse)/S211 (human) (4161, Cell Signaling), rabbit α-GR S234 (mouse)/S226 (human) (97285, Cell Signaling), rabbit α-GRIP-1 (96687, Cell Signaling) and rabbit α-p65 (6956, Cell Signaling).
RESULTS
DNA binding by GR is required for survival
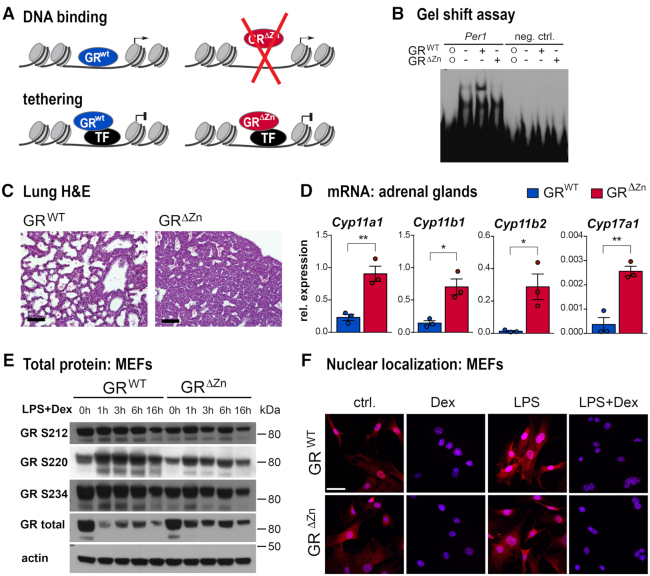
To investigate direct DNA binding dependent versus indirect modes of transcriptional regulation by GR, we generated a mouse model carrying a point mutation within the first zinc finger of the DNA binding domain (DBD) of GR (GRΔZn) (Figure 1A). As shown by Hollenberg and Evans (22) and by gel shift assay (Figure 1B), replacing a single amino acid, cysteine 437, with glycine abolishes direct DNA binding, but leaves all other domains intact. (Our experimental design, targeting strategy and validation are shown in Supplementary Figure S1A–D. We would like to point out that we do not know the oligomerization state of our GRΔZn mutant.)
Figure 1.
DNA binding of GR is required for survival. (A) Schematic illustrating the design of the GRΔZn mutant, separating direct DNA binding from tethering to other proteins. (B) EMSA with wild type or GRΔZn protein and labeled oligos containing the Per1 GRE sequence (neg. ctrl.: random DNA sequence negative control; o: oligo only, no protein). (C) Lung H&E histology of wild type and homozygous mutant E18.5 embryos. Representative images from n = 3, scale bar = 100 μm. (D) Steroidogenic enzyme mRNA expression by qRT-PCR in the adrenal glands of E18.5 wild type and homozygous mutants (normalized to Rpl38). n = 3, values are mean ± SEM, *P< 0.05, **P< 0.01. (E) Western blot in MEFs detecting phosphorylated mouse GR S212, S220 and S234, total GR and actin as loading control. Cells were either untreated (0 h) or treated with LPS (lipopolysaccharide) and Dexamethasone (Dex) for 1–16 h. Representative blot from n = 3. (F) Immunofluorescent detection of GR (red) in MEFs treated with Dex and/or LPS (blue = DAPI). Representative images from n = 3, scale bar = 50 μm.
Strikingly, our GRΔZn mutants displayed perinatal lethality similar to GR null mice (26). While heterozygous mutants were born with the expected Mendelian ratio, no viable homozygous GRΔZn pups were obtained. Because of the perinatal lethality, we set up timed matings to generate MEFs (mouse embryonic fibroblasts) and to isolate primary embryonic cells and tissues for functional analyses. Phenocopying complete loss of GR function, GRΔZn mice died of atelectasis and respiratory failure, and presented with adrenal hyperplasia accompanied by increased expression of steroidogenic cytochrome P450 enzymes (Figure 1C, D and Supplementary Figure S1E–G).
This broad loss of glucocorticoid responses was not explained by reduced expression of GR mRNA or protein itself (Figure 1E). Importantly, two indicators of protein functionality (subsequent to full length translation, folding, transport etc.), namely phosphorylation and nuclear localization of the receptor in response to ligand, were not affected by the GRΔZn mutation (27–29). MEFs generated from wild type and GRΔZn mutant embryos showed no visible differences in GR S212, S220 or S234 phosphorylation status or in cytoplasmic—nuclear translocation when treated with the synthetic glucocorticoid Dexamethasone (Figure 1F and Supplementary Figure S1H, I). These controls suggest that the GRΔZn mutation does not cause major alterations of protein conformation or interactions with heat shock proteins, kinases or ligands.
Taken together, we found that mutating the first zinc finger of GR in vivo mirrored the phenotype of GR null animals.
Tethered binding sites are found near inflammatory genes in GRΔZn MEFs
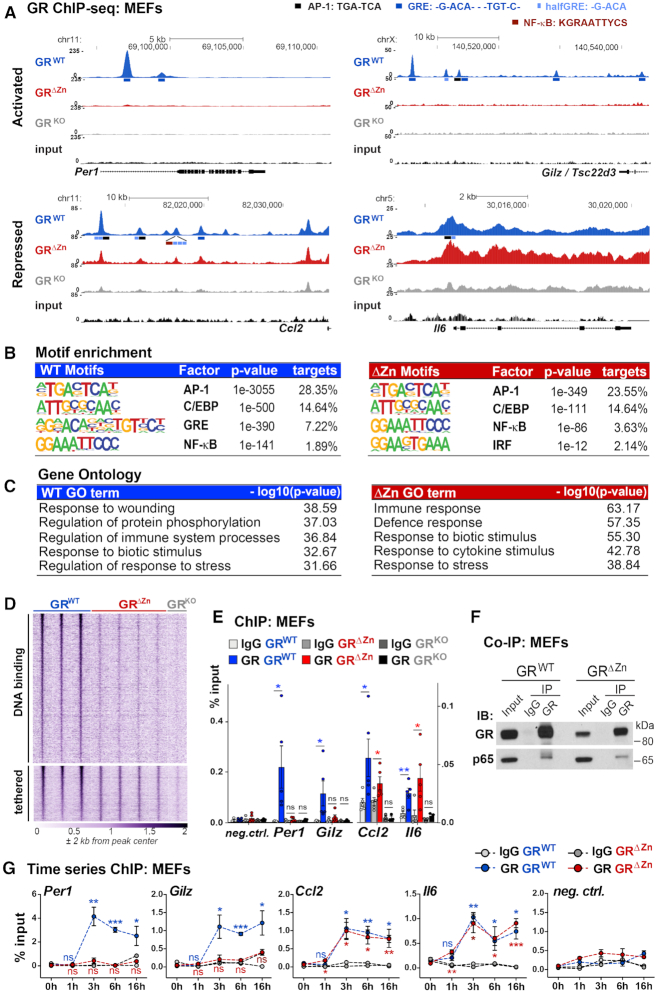
Since mutation of the first zinc finger of GR resulted in perinatal lethality, we used primary MEFs for further functional analyses. As shown in Figure 2A, we first performed ChIP-Seq for GR in wild type and mutant MEFs treated with both LPS to activate inflammatory Tlr4 signaling, and with the GR ligand dexamethasone (Dex) to induce GR nuclear translocation and genomic occupancy. We also used GR null MEFs as background to validate the specificity of our GR ChIP-Seq data, by removing signals still present in the knockout from further examination (Supplementary Figure S2A–C).
Figure 2.
Tethered binding sites are found near inflammatory genes in GRΔZn MEFs. (A) Representative GR ChIP-Seq tracks from wild type, homozygous mutant and knockout MEFs treated with LPS for 3 h and Dex overnight, with the positions of predicted motifs (not to scale!), (wild type n = 3, mutant n = 4 and knockout n = 2). (B) Enriched consensus motifs in GR ChIP-sequences. Known motifs were identified among wild type or mutant peaks. (C) Functional annotation of GR ChIP peaks assigned to the nearest gene, for either wild type or ΔZn cistromes. (D) Heatmap of GR ChIP-Seq coverage, for both wild type specific – and common (wild type and mutant) peaks (n = 2–4). (E) GR ChIP-qPCR for selected loci in MEFs treated with LPS for 3 h and Dex overnight (n = 5), shown as % input together with a negative site. Values are mean ± SEM, ns = not significant, *P< 0.05, **P< 0.01. (F) Western blot of endogenous co-IPs in MEFs treated with LPS for 3 h and with Dex overnight, representative example from n = 3. (G) GR ChIP-qPCR for selected loci in MEFs treated with LPS and Dex for 0–16 h. t-test for each time point versus untreated (0 h). Values are mean ± SEM, ns = not significant, *P< 0.05, **P< 0.01, ***P< 0.001.
In wild type MEFs, we observed GR binding to its known targets such as the Per1, Gilz (also known as Tsc22d3), Ccl2, Il6, Ccl20, Vcam1, Mmp8, Cxcl2, Fkbp5 and Dusp1 loci as expected (Figure 2A and Supplementary Figure S2A). In line with this profile, bioinformatic motif analyses revealed an enrichment for GRE, AP-1, NF-κB, IRF and C/EBP consensus motifs in the GR ChIP-sequences (Figure 2B). Functional annotation to nearby target genes displayed enrichment of GO terms referring to inflammatory responses (Figure 2C).
In contrast, in GRΔZn homozygous mutant MEFs, GR binding was almost undetectable at promoters and enhancers containing classical palindromic GREs, such as Per1 and Gilz, consistent with its inability to directly bind DNA. However, we still found a significant number of maintained, tethered binding sites, for example at the Ccl2 and Il6 loci (Figure 2A, D, E). Similarly, NF-κB, AP-1, IRF and C/EBP motifs were enriched among these GR ChIP-sequences obtained from GRΔZn MEFs, while GRE full palindromes or half sites were absent (Figure 2B). Interestingly, NF-κB motifs were the most highly enriched in the GRΔZn cistrome, corresponding to its preserved tethering potential. These tethered binding sites could be assigned to nearby target genes involved in inflammation, immune responses and cytokine/chemokine production (Figure 2C), agreeing with previous reports (4,9,30–32).
To further verify that GRΔZn retains the ability to be tethered but no longer binds directly to DNA, we performed protein–protein interaction experiments in both wild type and GRΔZn MEFs. We found that the endogenous NF-κB subunit p65 co-immunoprecipitated both wild type GR and GRΔZn (Figure 2F), which points at p65 as one potential tethering partner in this condition.
Importantly, this specific signature was not detected in control MEFs lacking the GR protein (Figure 2A, D and Supplementary Figure S2A–C). In addition, ChIP-Seq revealed only wild type GR, but not GRΔZn, binding directly to GREs near known repressed genes including Mmp8 or Cxcl2 (Supplementary Figure S2A). This agrees with GR binding directly to DNA to repress inflammatory gene expression.
We also measured GR wild type and GRΔZn occupancy by ChIP-qPCR during a time course experiment in MEFs co-treated with LPS and Dex for zero to sixteen hours (overnight), to characterize binding profile dynamics (Figure 2G). These data confirmed our observations from the ChIP-Seq studies, with GRΔZn still binding to certain sites near negative target genes, but being undetectable near GRE-containing cis-regulatory elements at all time points.
Finally, we performed an analogous GR ChIP-Seq experiment in MEFs treated only with Dex, without the inflammatory LPS stimulus. Under these conditions, we detected wild type GR binding near developmental genes and classical targets like Per1, but there was barely any measurable signal from GRΔZn samples. This result was confirmed by ChIP-qPCR, which showed wild type GR binding only to Per1, Gilz and Fkbp5, but not to Ccl2 or Il6 loci, none of which were occupied by the mutant (Supplementary Figure S2D–F. Inflammatory genes like Ccl2 or Il6 do not need to be transcriptionally repressed in unstimulated cells, and consequently were not bound by GR, which again indicates that tethering of GR might require the presence of NF-κB or other protein partners.
In conclusion, under inflammatory conditions, GRΔZn remained tethered to a subset of binding sites occupied by NF-κB, AP-1 and IRFs, whereas direct binding to DNA sequences associated with either activated or repressed targets was lost. This provides a new tool to analyze the functional relationship between tethering and direct DNA binding of GR.
Target gene regulation by GR requires DNA binding
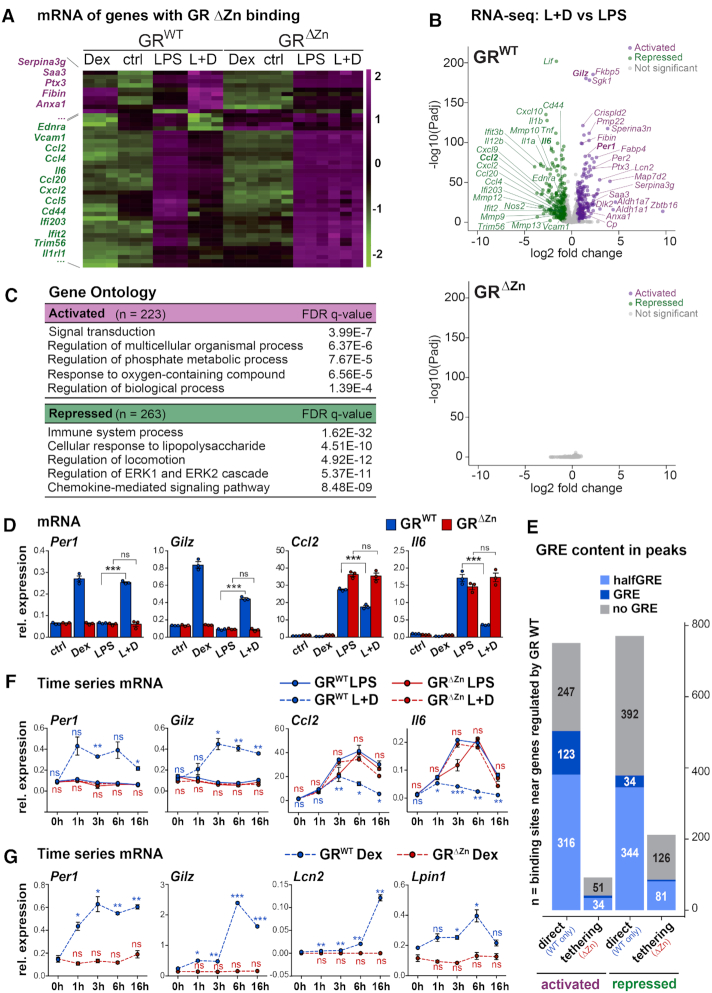
To understand the functionality of tethered versus DNA-dependent binding sites, we performed total RNA-Seq for wild type and GRΔZn MEFs under different conditions. Unexpectedly, in GRΔZn cells, there were no significant changes in gene expression in response to GR ligand, which would explain the lethality of these mice. None of the genes harboring a nearby tethered GRΔZn site showed any difference in expression levels upon stimulation with Dex, neither in quiescent nor in activated (LPS treated) MEFs (Figure 3A and B). In contrast, in LPS-activated wild type MEFs, Dex-activated GR targets corresponded to genes involved in signal transduction and phosphorylation, for example, while repressed targets included genes important for inflammation, chemotaxis and immune responses (Figure 3C, D and Supplementary Figure S3A–C). Specifically, wild type MEFs responded to Dex by induction of Gilz, Fkbp5, Per1 and Sgk1 and by suppression of inflammatory mediators like Ccl and Cxcl family members, interleukins and matrix metalloproteinases. The GRΔZn profiles, however, did not differ from vehicle treated samples.
Figure 3.
Target gene regulation by GR requires DNA binding. (A) Clustering for transcripts near tethered sites, in MEFs treated with vehicle, Dex and/or LPS, n = 3, values are RNA-Seq z-scores. (B) Volcano plot showing transcripts with significant fold changes in wild type MEFs (top) and GRΔZn (bottom), LPS + Dex compared to LPS, n = 3. (C) Functional annotation of differentially regulated genes from B. (D) qRT-PCR for GR target genes upon treatment with vehicle, Dex overnight and/or LPS for 6 h. (E) Numbers of predicted GRE motifs in GR wild type or tethered (common, wild type plus GRΔZn) ChIP-Seq peaks associated with nearby activated or repressed mRNAs. (F) qRT-PCR for GR target genes upon treatment with Dex and/or LPS for 0–16 h. t-test for Dex+LPS versus LPS. (G) qRT-PCR for GR target genes upon treatment with Dex alone or vehicle for 0–16 h. t-test for Dex time points versus 0 h. (D, F, G) Values are normalized to U36b4 and represent mean ± SEM, for D and F, n = 3 and G, n = 2, ns = not significant, *P< 0.05, **P< 0.01, ***P< 0.001.
It should be noted, though, that while the mutants responded to the LPS stimulus and clustered together with the vehicle treated samples in the PCA analysis, there was a baseline difference in mRNA expression profiles between GRΔZn and wild type MEFs (irrespective of the ligand). Consistent with GR’s known role in development and immunomodulation, mutant cells displayed de-repression of certain inflammatory genes and down-regulation of developmental genes. Again, the GR mRNA levels themselves were not significantly different between wild type and GRΔZn (Supplementary Figure S3D, E).
In part, this failure to respond could be explained by the large fraction of GR binding events near positive or negative target genes, that were lost in GRΔZn mutants (Figure 3E, Supplementary Figure S3F). However, an appreciable number of tethered (GRΔZn maintained) binding events were preserved, primarily near genes repressed by GR, in line with previous models associating protein–protein interactions with trans-repression (33). Yet our data suggest that, by itself, GR tethering was not sufficient to affect gene regulation, since there were no detectable changes in mRNA expression in response to GR ligand in GRΔZn cells.
To rule out the possibility that the GRΔZn mutant cells simply showed a delayed or diminished response, we performed a time series by stimulating wild type or GRΔZn MEFs for different lengths between 1 and 16 h. Per1, Gilz, Ccl2 and Il6 were activated and repressed, respectively, in response to Dex in wild type, but not in mutant cells, regardless of the time point, under any of the conditions we tested (Figure 3F and G).
In summary, we found that direct DNA binding by GR was globally required for its transcriptional activity in MEFs.
GR tethering can be detected under various conditions
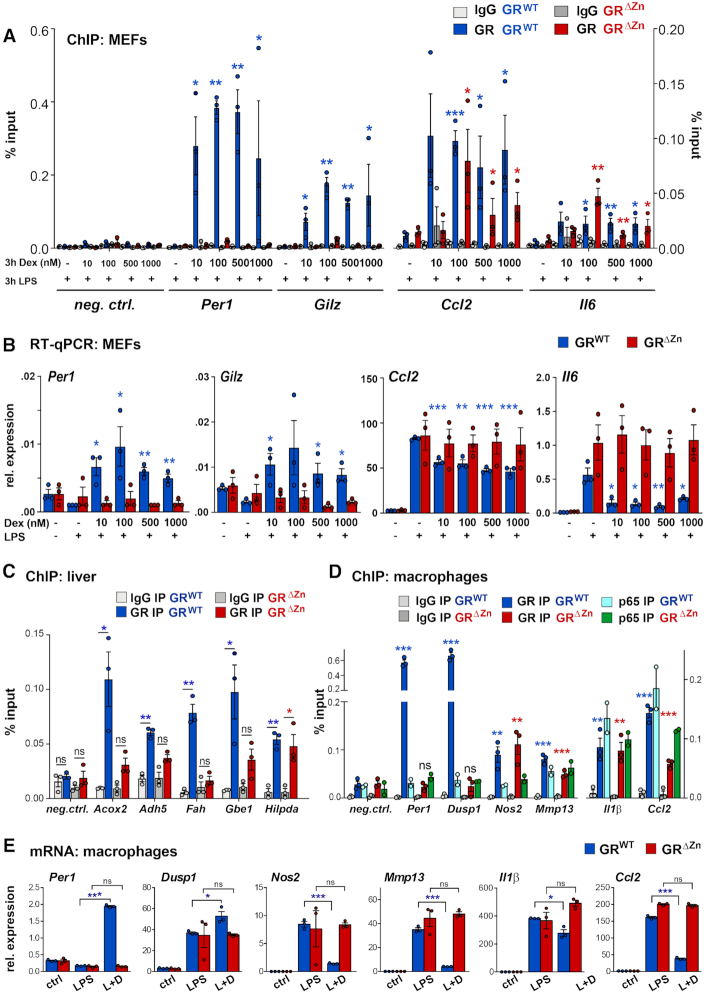
To validate our observations of partially maintained chromatin occupancy but complete lack of transcriptional activity in our zinc finger mutant, we carried out qPCR experiments with different concentrations of the GR ligand Dexamethasone (Figure 4A, B). In MEFs treated with LPS and varying doses of Dex, we again detected wild type GR binding to and up- or down-regulating the expression of the Per1, Gilz, Ccl2 and Il6 loci irrespective of the concentration. While occupancy near Ccl2 and Il6 was retained by GRΔZn, we could not observe transcriptional repression of these inflammatory genes in any of our settings without DNA binding.
Figure 4.
GR tethering occurs at different ligand concentrations and in various cell types. (A) GR ChIP-qPCR for selected loci in MEFs treated with different concentrations between 10 nM and 1 μM Dex and LPS for 3 h (n = 3), shown as % input with t-test for GR IPs over IgG. (B) qRT-PCR for GR target genes in MEFs treated as in (A) normalized to U36b4, n = 3. t-test for different Dex amounts over LPS only. (A, B) Values are mean ± SEM, ns = not significant, *P< 0.05, **P< 0.01, ***P< 0.001. (C) GR ChIP-qPCR in wild type and homozygous E18.5 livers, n = 3. (D) GR and p65 ChIP-qPCR in fetal macrophages, n = 3 for GR and n = 2 for p65, treated with LPS for 3 h and with Dex overnight. Values in (C) and (D) are shown as %input, mean ± SEM, ns = not significant, *P< 0.05, **P< 0.01, ***P< 0.001. (E) qRT-PCR for GR target genes (normalized to U36b4) in macrophages treated with vehicle, LPS for 6 h or LPS for 6 h and Dex overnight (L + D). Values represent mean ± SEM, n = 3. ns = not significant, *P< 0.05, ***P< 0.001.
Testing another scenario, we further confirmed these results by qRT-PCR in MEFs treated with TPA (12-O-tetradecanoylphorbol-13-acetate), which activates protein kinase C (instead of activation of Tlr4 by LPS). Results again demonstrated the expected changes in gene expression of known GR targets such as Per1, Gilz, Ccl2 or Il6 in wild type MEFs, with no changes in GRΔZn cells, independently of the stimulus (Supplementary Figure S4A).
Having characterized our new model in vitro, we next studied the importance of GR DNA binding for metabolic and inflammatory gene regulation in vivo. We harvested embryos and performed ChIP-qPCR in wild type and GRΔZn livers in the presence of endogenous ligand. Corresponding to its known functions in hepatic lipid, glycogen, amino-, fatty- and bile acid metabolism, we detected specific GR binding to the Acox2, Adh5, Fah, Gbe1 and Hilpda cis-regulatory loci (34) (Figure 4C). Occupancy of most of these GRE-containing sites was lost in fetal GRΔZn livers, with the exception of the Hilpda locus, which appeared to be tethered (i.e. does not harbor a GRE motif).
When we analyzed GR occupancy in fetal liver-derived macrophages, we similarly found wild type GR binding to its known target sites near Per1, Dusp1, Nos2, Mmp13, Il1β and Ccl2 by ChIP-qPCR (10) (Figure 4D). Per1 and Dusp1 are GRE-encompassing targets induced by GR, while Nos2, Mmp13, Il1β and Ccl2 represent important inflammatory genes repressed by Dex (Figure 4E). Confirming our results from LPS-treated MEFs, GR binding at Per1 and Dusp1 was greatly reduced in GRΔZn macrophages, while Nos2, Mmp13, Il1β and Ccl2 maintained GR occupancy independently of its zinc finger. Binding near these negative target genes mainly coincided with NF-κB occupancy, as measured by ChIP-qPCR for p65 (Figure 4D). However, consistent with our MEF results, we could not detect any changes in the mRNA levels of these genes in response to Dex, neither positively nor negatively (Figure 4E and Supplementary Figure S4B).
Thus, while tethering of GR to negative sites occurred, subsequent transcriptional repression required direct DNA binding, conceivably to nearby inflammatory cis-regulatory elements or to proximal sequence motifs (e.g. GREs or novel sites within the same or a second peak close by).
GR DNA binding is required for the recruitment of co-regulators
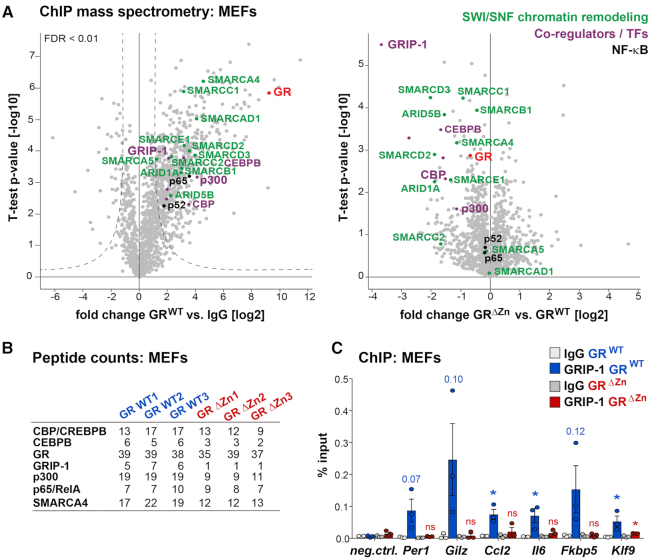
We next sought to determine how GR DNA binding mediates both transcriptional activation and repression. Therefore, we performed GR ChIP-MS to chart the differential assembly of transcriptional co-regulator complexes by proteomics. In wild type MEFs treated with LPS and Dex, we measured robust interactions between GR and its known co-regulators GRIP-1, C/EBPβ, p300, CBP, p65, SWI/SNF nucleosome remodelers etc. While the protein–protein interactions between GRΔZn and NF-κB as well as many others were maintained, recruitment of GRIP-1, some components of the SWI/SNF complex, and the histone acetyl transferases (HATs) CBP and p300 were significantly reduced (Figure 5A and B and Supplementary Figure S5A, B). Of note, the mRNA expression of these nuclear proteins was not lowered in mutant MEFs, and the majority of interactions between GRΔZn and its chromatin interactors was preserved (Supplementary Figure S5A, C). These interactions need not necessarily be direct, however, as presence in the proteomics purification could also simply have indicated nucleosomal proximity.
Figure 5.
GR DNA binding is required for the recruitment of co-regulators. (A) ChIP-MS proteomics peptide counts enriched in GR wild type (left) purifications, and statistical comparison between peptides present in GR and GRΔZn samples (right), with functional annotation. MEFs were treated with LPS for 3 h and Dex overnight. (B) Peptide counts for selected proteins from a. (C) GRIP-1 ChIP-qPCR in MEFs treated with LPS for 3 h and Dex overnight. Values are mean ± SEM % input, from n = 3. t-test for GRIP-1 over IgG, *P< 0.05, ns = not significant.
Nevertheless, failure to recruit the p160/SRC family member GRIP-1 could well explain our phenotype, since it plays a central role in the suppression of inflammatory responses by GCs (10,35). Importantly, the GRIP-1 protein expression levels themselves were similar between wild type and mutant MEFs treated with LPS+Dex (Supplementary Figure S5D). We successfully validated the loss of GRIP-1 recruitment by GRΔZn by ChIP-qPCR in MEFs treated with LPS+Dex (Figure 5C).
Similarly, it is well established that nuclear receptors assemble HAT complexes to activate target genes, so loss of p300/CBP recruitment could underlie the transcriptional inertia of GRΔZn (10). Interestingly, it is conceivable that SWI/SNF might likewise be implicated in GR-mediated repression (36). Arid1a and Arid1b are DNA-binding BAFs (BRG1-associated factors), a class of SWI/SNF subunits implicated in ligand-dependent transcriptional activation by nuclear receptors (37). However, we could not identify a significant role for the ARID proteins Arid1a, Arid1b or Arid5a in the transcriptional repression of inflammatory genes by GR, when we performed siRNA knockdowns of these candidates in MEFs treated with LPS plus Dex (Supplementary Figure S5E).
Taken together, our data suggest that DNA binding by GR is required for the recruitment of essential co-regulators such as GRIP-1 to both positive and negative binding sites.
Regulation of functional enhancers requires sequence recognition by GR
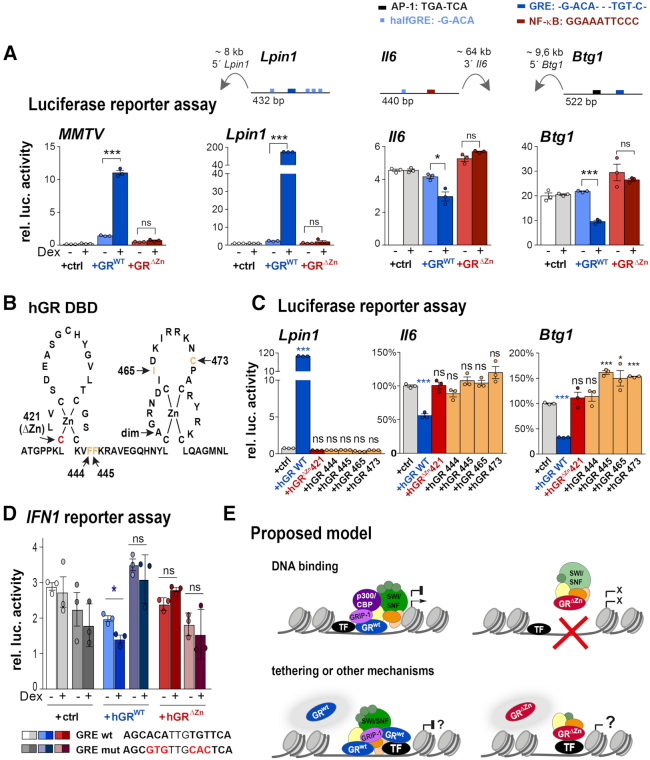
Following up on our NGS, qPCR and proteomics results, we next verified that the GRΔZn mutation was indeed blocked in both transcriptional activation and repression. We therefore used in vitro assays with isolated GRE-containing positive and negative luciferase reporter sequences. As expected, wild type GR activated classical GR responsive cis-regulatory elements such as MMTV and Lpin1 and repressed macrophage enhancers such as Il6 and Btg1 in transient transfections (10) (Figure 6A). Again, the GRΔZn expression constructs showed neither positive nor negative regulatory activity in these assays.
Figure 6.
Positive and negative regulation of GR reporters involves DNA binding. (A) Normalized luciferase activity in CV-1 cells overexpressing empty vector (+ctrl), GR wild type or ΔZn mutant and MMTV, Lpin1, Il6 or Btg1 reporters. Cells were treated with LPS and either vehicle or Dex (− or +) overnight (n = 3 biological replicates, values are mean ± SEM, ns = not significant, *P< 0.05, ***P< 0.001). The cartoons depict enhancer positions and composition. (B) Human GR DNA-binding domain (DBD) showing the mutations used in C. (C) Normalized luciferase activity in CV-1 cells overexpressing empty vector (+ctrl), human GR wild type, ΔZn (hGR C421G) and F444G, F445G, I465G and C473G mutants. Cells were treated with vehicle or Dex overnight (n = 3 biological replicates). Values represent mean ± SEM. ns = not significant, *P< 0.05, ***P< 0.001. (D) Normalized luciferase activity in CV-1 cells overexpressing empty vector (+ctrl), human GR wild type or ΔZn (hGR C421G) mutant. Cells were treated with LPS and either vehicle or Dex (− or +) overnight (n = 3 biological replicates, values are mean ± SEM, ns = not significant, *P< 0.05). (E) Proposed mechanism: GR binding to DNA leads to the recruitment of GRIP-1, HATs, the SWI/SNF complex and other co-regulators to activate or repress transcription. The transcriptional inertia of GRΔZn mutants could be explained by the failure to recruit co-regulators, or by other mechanisms such as squelching or non-genomic actions. While tethering still occurs GRΔZn mice, it is not sufficient to regulate gene expression either positively or negatively.
To demonstrate conservation and clinical relevance, we then performed the same assay using human GR expression constructs (instead of mouse). Indeed, both the activating and repressive effects observed with wild type human GR were absent with the analogous GRΔZn GRC421G mutation (Figure 6B, C). Furthermore, four other single point mutants that were shown to disable GR DNA binding without disrupting the structure of the C4-type zinc finger (22), also abolished Dex-mediated repression of either the Btg1 or Il6 GRE- and AP-1 or NF-κB-motif containing enhancers (10) (Figure 6C).
Finally, when interrogating the human type 1 interferon (IFN1) cis-regulatory element, which harbors a GRE, we measured repression by wild type, but not by our Zinc finger mutant GR, in a luciferase reporter assay. This repressive effect was lost upon mutation of the GRE sequence (Figure 6D).
Thus, our results suggest that direct DNA binding by GR is required for the assembly of a functional coactivator/corepressor complex to regulate both transcriptional activation and repression (Figure 6E).
DISCUSSION
In conclusion, we found that direct DNA binding by GR was absolutely required for both transcriptional activation and, importantly, repression of inflammatory genes under the conditions we examined. GRΔZn mice and cells showed no response to glucocorticoids in any of the situations tested, despite the fact that ligand binding, nuclear localization, protein expression, protein–protein interactions and tethering to chromatin-bound factors were preserved.
These observations were very unexpected (8). Other nuclear receptors, such as Rev-erbα or PPARα, for example, have been shown to exert DNA binding-independent regulation of hepatic metabolism in vivo. For instance, mice with an in-frame deletion of the Rev-erbα DNA binding domain still regulate a subset of metabolic target genes, including repressed ones, via tethered recruitment of HDAC3 (38). Similarly, a point mutation in the second zinc finger of PPARα abolishes PPARE driven activation of metabolic targets, while maintaining tethered trans-repression of pro-inflammatory genes in the liver (39). In contrast, our GRΔZn mutant does not discriminate between activation and repression: all transcriptional effects are lost when GR is no longer able to directly bind DNA.
Of note, the original papers reporting repression of POMC, osteocalcin and prolactin transcription by GR, also refer to direct binding at specific sequences. These motifs did not match the GRE consensus perfectly, which led to the concept of negative nGREs, and they now support our model. The importance of DNA binding has further been advocated by other studies showing direct binding of GR to genomic NF-κB and AP-1 response elements during the repression of inflammatory gene expression (14,15). Also, GR DNA binding sequences with distinct motifs were found near several repressed mouse and human GR target genes (40).
In addition to other anti-inflammatory mechanisms such as the induction of Dusp1, Gilz, KLFs etc., GR has been shown to activate IκBα expression, which subsequently acts as a negative regulator of NF-κB enhancer/promoter binding in macrophages (41,42). While we did not observe changes in p65 occupancy between our GRΔZn mutant and wild type macrophages (Figure 4d), this mechanism is consistent with our model, as it also requires direct DNA binding of GR to induce IκBα transcription. A recent genomic study in BEAS-2B cells further argued against tethering in repression, as ‘dex-induced genome-wide binding of the GR to canonical binding sites’ was shown to be required for repressive alterations of local chromatin structures (43).
One possibility to consider might be that coactivator squelching (competition for limited cofactors) or binding together with other factors at composite elements, may be abrogated in our mutant, especially since it fails to recruit GRIP-1 (44). Specifically, the inability of GRΔZn to bind DNA may reduce its residence time on chromatin or in the nucleus, which could affect squelching, sequestration or repression (45). Similarly, our GRΔZn mutation might affect recently described phenomena such as p300 cofactor reshuffling or coregulator reprogramming, which would repeal its gene regulatory functions (46). Correspondingly, our zinc finger mutation might reduce or abrogate GR’s ability to regulate mRNA stability post-transcriptionally by RNA binding, as has been reported for chemokines like Ccl2 (47).
While our study suggests that tethering of GR to AP-1 or NF-κB is not sufficient for its anti-inflammatory actions, this DNA-independent interface might specify a conformational change or strengthen interactions that are necessary for repression. The total lack of GC responses in GRΔZn mutants (despite tethering to a distinct fraction of sites) might be explained by the additional requirement for GREs, half sites or other DNA motifs within the same or a nearby ChIP-Seq peak (12). Since our GRΔZn mutation does not discriminate between GRE-dependent and -independent DNA interactions, these putative scenarios will have to be further investigated in detail, as they may represent multiple different subset-specific mechanisms.
Finally, our discovery has important implications for the development of novel glucocorticoid receptor agonists or modulators with reduced side effect profiles, and for our understanding of transcriptional regulation by GR (6).
DATA AVAILABILITY
The NGS data sets have been deposited in the GEO (NCBI) under super series ID code: GSE126655. Single data sets ID codes ChIP-Seq: GSE126654 & RNA-Seq: GSE124919 (Figures 2 and 3).
The proteomics data have been deposited in the ProteomeXchange (PRIDE) under ID code PXD013772 (Figure 5).
For data analysis, we used FastQC: http://www.bioinformatics.babraham.ac.uk/ projects/fastqc/ and removed read duplicates using Picard Tools: http://picard.sourceforge.net/. Heatmaps and boxplots were generated in R: www.R-project.org/.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Evans and J. Tuckermann for providing reagents; O. García-Gonzáles, T. Horn, M. J. Hubert, M. C. Hemmer, K. Beresowski, C. Sportelli, S. Regn, I. Guderian, K. Biniossek and M. Tschoep for their contributions; E. Graf, T. Schwarzmayr and T. Strom for their NGS support; and Life Science Editors for editing assistance.
Contributor Information
Laura Escoter-Torres, Molecular Endocrinology, Institutes for Diabetes and Obesity & Diabetes and Cancer IDO & IDC, Helmholtz Zentrum Muenchen (HMGU) and German Center for Diabetes Research (DZD), Munich 85764, Germany.
Franziska Greulich, Molecular Endocrinology, Institutes for Diabetes and Obesity & Diabetes and Cancer IDO & IDC, Helmholtz Zentrum Muenchen (HMGU) and German Center for Diabetes Research (DZD), Munich 85764, Germany; Metabolic Programming, TUM School of Life Sciences Weihenstephan and ZIEL Institute for Food & Health, Munich 85354, Germany.
Fabiana Quagliarini, Molecular Endocrinology, Institutes for Diabetes and Obesity & Diabetes and Cancer IDO & IDC, Helmholtz Zentrum Muenchen (HMGU) and German Center for Diabetes Research (DZD), Munich 85764, Germany.
Michael Wierer, Department of Proteomics and Signal Transduction, Max Planck Institute for Biochemistry, Munich 82152, Germany.
Nina Henriette Uhlenhaut, Molecular Endocrinology, Institutes for Diabetes and Obesity & Diabetes and Cancer IDO & IDC, Helmholtz Zentrum Muenchen (HMGU) and German Center for Diabetes Research (DZD), Munich 85764, Germany; Metabolic Programming, TUM School of Life Sciences Weihenstephan and ZIEL Institute for Food & Health, Munich 85354, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation (Emmy Noether Programme) [UH 275/1-1]; European Research Council [ERC-2014-StG 638573 SILENCE to N.H.U.]. Funding for open access charge: ERC.
Conflict of interest statement. None declared.
REFERENCES
- 1. Evans R.M., Mangelsdorf D.J.. Nuclear receptors, RXR, and the big bang. Cell. 2014; 157:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mangelsdorf D.J., Thummel C., Beato M., Herrlich P., Schutz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P. et al.. The nuclear receptor superfamily: the second decade. Cell. 1995; 83:835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Evans R.M. The steroid and thyroid hormone receptor superfamily. Science. 1988; 240:889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cain D.W., Cidlowski J.A.. Immune regulation by glucocorticoids. Nat. Rev. Immunol. 2017; 17:233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Greulich F., Hemmer M.C., Rollins D.A., Rogatsky I., Uhlenhaut N.H.. There goes the neighborhood: Assembly of transcriptional complexes during the regulation of metabolism and inflammation by the glucocorticoid receptor. Steroids. 2016; 114:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sundahl N., Bridelance J., Libert C., De Bosscher K., Beck I.M.. Selective glucocorticoid receptor modulation: new directions with non-steroidal scaffolds. Pharmacol. Ther. 2015; 152:28–41. [DOI] [PubMed] [Google Scholar]
- 7. Schule R., Rangarajan P., Kliewer S., Ransone L.J., Bolado J., Yang N., Verma I.M., Evans R.M.. Cell. 1990; 62:United States: 1217–1226. [DOI] [PubMed] [Google Scholar]
- 8. Reichardt H.M., Kaestner K.H., Tuckermann J., Kretz O., Wessely O., Bock R., Gass P., Schmid W., Herrlich P., Angel P. et al.. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998; 93:531–541. [DOI] [PubMed] [Google Scholar]
- 9. Ogawa S., Lozach J., Benner C., Pascual G., Tangirala R.K., Westin S., Hoffmann A., Subramaniam S., David M., Rosenfeld M.G. et al.. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005; 122:707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uhlenhaut N.H., Barish G.D., Yu R.T., Downes M., Karunasiri M., Liddle C., Schwalie P., Hubner N., Evans R.M.. Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol. Cell. 2013; 49:158–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hemmer M.C., Wierer M., Schachtrup K., Downes M., Hubner N., Evans R.M., Uhlenhaut N.H.. E47 modulates hepatic glucocorticoid action. Nat. Commun. 2019; 10:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lim H.W., Uhlenhaut N.H., Rauch A., Weiner J., Hubner S., Hubner N., Won K.J., Lazar M.A., Tuckermann J., Steger D.J.. Genomic redistribution of GR monomers and dimers mediates transcriptional response to exogenous glucocorticoid in vivo. Genome Res. 2015; 25:836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Presman D.M., Ogara M.F., Stortz M., Alvarez L.D., Pooley J.R., Schiltz R.L., Grontved L., Johnson T.A., Mittelstadt P.R., Ashwell J.D. et al.. Live cell imaging unveils multiple domain requirements for in vivo dimerization of the glucocorticoid receptor. PLoS Biol. 2014; 12:e1001813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weikum E.R., de Vera I.M.S., Nwachukwu J.C., Hudson W.H., Nettles K.W., Kojetin D.J., Ortlund E.A.. Tethering not required: the glucocorticoid receptor binds directly to activator protein-1 recognition motifs to repress inflammatory genes. Nucleic Acids Res. 2017; 45:8596–8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hudson W.H., Vera I.M.S., Nwachukwu J.C., Weikum E.R., Herbst A.G., Yang Q., Bain D.L., Nettles K.W., Kojetin D.J., Ortlund E.A.. Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappaB response elements. Nat. Commun. 2018; 9:1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z.. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009; 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M. et al.. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019; 47:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schauwaers K., De Gendt K., Saunders P.T., Atanassova N., Haelens A., Callewaert L., Moehren U., Swinnen J.V., Verhoeven G., Verrijdt G. et al.. Loss of androgen receptor binding to selective androgen response elements causes a reproductive phenotype in a knockin mouse model. Proc. Natl Acad. Sci. U.S.A. 2007; 104:4961–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hollenberg S.M., Evans R.M.. Multiple and cooperative trans-activation domains of the human glucocorticoid receptor. Cell. 1988; 55:899–906. [DOI] [PubMed] [Google Scholar]
- 23. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Durinck S., Spellman P.T., Birney E., Huber W.. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009; 4:1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cole T.J., Blendy J.A., Monaghan A.P., Krieglstein K., Schmid W., Aguzzi A., Fantuzzi G., Hummler E., Unsicker K., Schutz G.. Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev. 1995; 9:1608–1621. [DOI] [PubMed] [Google Scholar]
- 27. Chen W., Dang T., Blind R.D., Wang Z., Cavasotto C.N., Hittelman A.B., Rogatsky I., Logan S.K., Garabedian M.J.. Glucocorticoid receptor phosphorylation differentially affects target gene expression. Mol. Endocrinol. 2008; 22:1754–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blind R.D., Garabedian M.J.. Differential recruitment of glucocorticoid receptor phospho-isoforms to glucocorticoid-induced genes. J. Steroid Biochem. Mol. Biol. 2008; 109:150–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Webster J.C., Jewell C.M., Bodwell J.E., Munck A., Sar M., Cidlowski J.A.. Mouse glucocorticoid receptor phosphorylation status influences multiple functions of the receptor protein. J. Biol. Chem. 1997; 272:9287–9293. [DOI] [PubMed] [Google Scholar]
- 30. Hua G., Ganti K.P., Chambon P.. Glucocorticoid-induced tethered transrepression requires SUMOylation of GR and formation of a SUMO-SMRT/NCoR1-HDAC3 repressing complex. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E635–E643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sacta M.A., Tharmalingam B., Coppo M., Rollins D.A., Deochand D.K., Benjamin B., Yu L., Zhang B., Hu X., Li R. et al.. Gene-specific mechanisms direct glucocorticoid-receptor-driven repression of inflammatory response genes in macrophages. Elife. 2018; 7:e34864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rao N.A., McCalman M.T., Moulos P., Francoijs K.J., Chatziioannou A., Kolisis F.N., Alexis M.N., Mitsiou D.J., Stunnenberg H.G.. Coactivation of GR and NFKB alters the repertoire of their binding sites and target genes. Genome Res. 2011; 21:1404–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Desmet S.J., De Bosscher K.. Glucocorticoid receptors: finding the middle ground. J. Clin. Invest. 2017; 127:1136–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rando G., Tan C.K., Khaled N., Montagner A., Leuenberger N., Bertrand-Michel J., Paramalingam E., Guillou H., Wahli W.. Glucocorticoid receptor-PPARalpha axis in fetal mouse liver prepares neonates for milk lipid catabolism. Elife. 2016; 5:e11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chinenov Y., Gupte R., Dobrovolna J., Flammer J.R., Liu B., Michelassi F.E., Rogatsky I.. Role of transcriptional coregulator GRIP1 in the anti-inflammatory actions of glucocorticoids. Proc. Natl Acad. Sci. U.S.A. 2012; 109:11776–11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. John S., Sabo P.J., Johnson T.A., Sung M.H., Biddie S.C., Lightman S.L., Voss T.C., Davis S.R., Meltzer P.S., Stamatoyannopoulos J.A. et al.. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol. Cell. 2008; 29:611–624. [DOI] [PubMed] [Google Scholar]
- 37. Nagarajan S., Rao S.V., Sutton J., Cheeseman D., Dunn S., Papachristou E.K., Prada J.G., Couturier D.L., Kumar S., Kishore K. et al.. ARID1A influences HDAC1/BRD4 activity, intrinsic proliferative capacity and breast cancer treatment response. Nat. Genet. 2020; 52:187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Y., Fang B., Emmett M.J., Damle M., Sun Z., Feng D., Armour S.M., Remsberg J.R., Jager J., Soccio R.E. et al.. GENE REGULATION. Discrete functions of nuclear receptor Rev-erbalpha couple metabolism to the clock. Science. 2015; 348:1488–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pawlak M., Bauge E., Bourguet W., De Bosscher K., Lalloyer F., Tailleux A., Lebherz C., Lefebvre P., Staels B.. The transrepressive activity of peroxisome proliferator-activated receptor alpha is necessary and sufficient to prevent liver fibrosis in mice. Hepatology. 2014; 60:1593–1606. [DOI] [PubMed] [Google Scholar]
- 40. Surjit M., Ganti K.P., Mukherji A., Ye T., Hua G., Metzger D., Li M., Chambon P.. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011; 145:224–241. [DOI] [PubMed] [Google Scholar]
- 41. Sasse S.K., Gruca M., Allen M.A., Kadiyala V., Song T., Gally F., Gupta A., Pufall M.A., Dowell R.D., Gerber A.N.. Nascent transcript analysis of glucocorticoid crosstalk with TNF defines primary and cooperative inflammatory repression. Genome Res. 2019; 29:1753–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Escoter-Torres L., Caratti G., Mechtidou A., Tuckermann J., Uhlenhaut N.H., Vettorazzi S.. Fighting the fire: mechanisms of inflammatory gene regulation by the glucocorticoid receptor. Front. Immunol. 2019; 10:1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oh K.S., Patel H., Gottschalk R.A., Lee W.S., Baek S., Fraser I.D.C., Hager G.L., Sung M.H.. Anti-Inflammatory chromatinscape suggests alternative mechanisms of glucocorticoid receptor action. Immunity. 2017; 47:298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schmidt S.F., Larsen B.D., Loft A., Mandrup S.. Cofactor squelching: artifact or fact. Bioessays. 2016; 38:618–626. [DOI] [PubMed] [Google Scholar]
- 45. Clauss K., Popp A.P., Schulze L., Hettich J., Reisser M., Escoter Torres L., Uhlenhaut N.H., Gebhardt J.C.M.. DNA residence time is a regulatory factor of transcription repression. Nucleic Acids Res. 2017; 45:11121–11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dendoncker K., Timmermans S., Vandewalle J., Eggermont M., Lempiainen J., Paakinaho V., Van Hamme E., Dewaele S., Vandevyver S., Ballegeer M. et al.. TNF-alpha inhibits glucocorticoid receptor-induced gene expression by reshaping the GR nuclear cofactor profile. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:12942–12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Panganiban R.P., Vonakis B.M., Ishmael F.T., Stellato C.. Coordinated post-transcriptional regulation of the chemokine system: messages from CCL2. J. Interferon Cytokine Res. 2014; 34:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NGS data sets have been deposited in the GEO (NCBI) under super series ID code: GSE126655. Single data sets ID codes ChIP-Seq: GSE126654 & RNA-Seq: GSE124919 (Figures 2 and 3).
The proteomics data have been deposited in the ProteomeXchange (PRIDE) under ID code PXD013772 (Figure 5).
For data analysis, we used FastQC: http://www.bioinformatics.babraham.ac.uk/ projects/fastqc/ and removed read duplicates using Picard Tools: http://picard.sourceforge.net/. Heatmaps and boxplots were generated in R: www.R-project.org/.