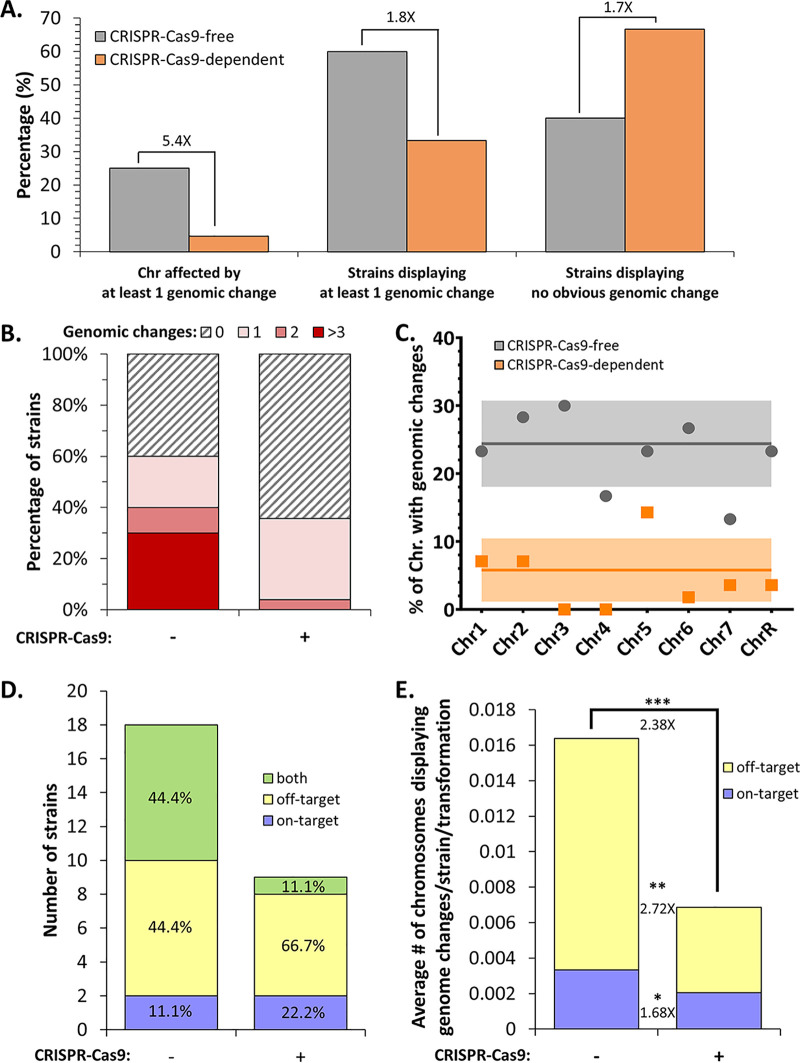
FIG 4.
Quantification of genome changes identified within C. albicans strains engineered using one of two transformation methods. (A) Percentages of chromosomes and strains impacted by transformation-associated genomic changes, as well as percentages of genomic-change-free strains. (B) Studying concurrent genomic changes within strains. (C) Percentage of each chromosome affected by at least one genomic change. (D) Representations of the numbers of strains displaying genomic change(s) on a targeted chromosome, a nontargeted chromosome, or both types. The fraction of each category of strains within genomic-change-displaying strains is indicated by a percentage. (E) Frequency of transformation-associated genomic changes tabulated as the average number of chromosomes displaying genome changes per strain per transformation. Differences in genome change frequencies between strains constructed using the two transformation strategies are represented as fold changes (*, P < 0.05; **, P < 0.01; ***, P < 0.001 by t test).