Abstract
Background
Lung adenocarcinoma (LAC) is the most prominent histological subtype of non‐small cell lung cancer (NSCLC) with a high rate of mortality and metastasis. Accumulating evidence has shown that long non‐coding RNAs (lncRNAs) play malfunctioning roles in the development of human tumors. Hence, this study aimed to determine the biological function of LINC00511 in LAC and to provide a novel diagnostic and therapeutic target for it.
Methods
LINC00511 expression in LAC tissues and cell lines (H1299 and A549) were detected by real time‐polymerase chain reaction (RT‐qPCR). Cell counting kit‐8 (CCK‐8) assay was employed to analyze cell proliferative ability. Cell metastasis change was measured using transwell assay. Moreover, we revealed a novel target gene of LINC00511 and elucidated the underlying competitive endogenous RNA regulatory mechanism in LAC cells.
Results
Data from our study demonstrated that LINC00511 expression was increased in LAC tissues and cells in comparison to their corresponding controls. Moreover, overexpression of LINC00511 indicated the poor prognosis of LAC patients. Overexpression of LINC00511 promoted proliferation, invasion and migration capacities of LAC cells. Moreover, LINC00511 promoted LAC progression via serving as a sponge of miR‐625‐5p and regulating PKM2 expression.
Conclusions
The present study showed that LINC00511 was involved in LAC progression by targeting miR‐625‐5p/PKM2, indicating that LINC00511/miR‐625‐5p/PKM2 may function as promising therapeutic targets for LAC.
Keywords: LINC00511, lung adenocarcinoma, miR‐625‐5p, PKM2
Taken all into account, upregulation of LINC00511 in LAC tissues and cell lines was validated. Upregulation of LINC00511 was closely correlated with shorter overall survival in LAC patients. We explored the biological function of LINC00511 in promoting LAC cell proliferation, migration and invasion. Moreover, LINC00511 knockdown could partially by sponging miR‐625‐5p, and then up‐regulate PKM2 to exert its tumor‐suppressive effect. Our studies confirmed the roles of a novel LINC00511/miR‐625‐5p/PKM2 axis in LAC progression, which might provide a promising application in LAC treatment.

Introduction
Lung cancer is one of the main causes of cancer‐related mortalities worldwide. 1 The five‐year survival rate of lung cancer is currently only 18% and is in sharp contrast to the steady rise in that of other cancers. 2 As is widely known, non‐small cell lung cancer (NSCLC) accounts for about 85% of all lung cancers. Moreover, it is worth noting that lung adenocarcinoma (LAC) is the most prominent histological subtype of NSCLC with a high rate of mortality and metastasis. 3 Furthermore, atypical cell proliferation in the alveoli and bronchioles of the lung has been reported to lead to LAC. 2 The carcinogenicity of LAC is related to inactivation of tumor suppressors and overexpression of oncogenes. 4 To make matters worse, the majority of lung cancer patients are usually diagnosed at a late stage when a tumor has started to metastasize. 5 Although the prognosis of LAC has improved with the advent of targeted drugs, the mortalities of LAC patients are still high as a result of a lack of effective prognostic and diagnostic biomarkers. 6 Hence, it is necessary to fully illustrate good prognostic indicators to guide the treatment and clinical management of LAC patients.
Data from human genome sequences indicates that 90% of the intronic DNA sequences are actively transcribed, and only 2% of these sequences could encode proteins, while the rest are non‐coding RNAs (ncRNAs). 7 According to the length of the sequences, ncRNAs is classified into long non‐coding RNA (lncRNA) and microRNA (miR). LncRNAs are a class of transcripts longer than 200 nucleotides and play crucial roles in the occurrence and progression of multiple tumors. At present, due to its application potential in tumor diagnosis and prognosis as well as its remarkable tissue‐specific characteristics in tumors, lncRNA has become a new hot spot in tumorigenesis. 8 Recent studies have shown that lncRNA can serve as competing endogenous RNAs (ceRNAs) through sponging miRs, playing important roles in epigenetically regulating cancers. 9
The significance of miR and lncRNA has been well‐stated in a variety of tumors, including lung cancer. 6 LncRNA exerts key functions in numerous pathological and physiological processes, and is closely related to gene regulation in lung cancer. 7 , 8 Recent evidence has shown that miR and lncRNA are implicated in regulating the expression of coding genes in tumors via forming an ncRNA regulatory network. 9 Furthermore, increasing studies have indicated that many lncRNAs serve key roles in LAC progression. lncRNA ZFPM2‐AS1 was found to promote LAC cell viability via miR‐18b‐5p/VMA21 axis. 10 LncRNA FTX could facilitate LAC tumorigenesis by targeting miR‐300. 11 MiR‐625‐5p has been proved to be involved in the progression of cancers. For instance, in cervical carcinoma, miR‐625‐5p was found to sponge lncRNA MALAT1 and suppress NF‐kappaB signaling to inhibit cervical carcinoma cell growth 12 ; in gastric cancer, miR‐625‐5p/NFIX axis was involved in the functions of LINC00511 in accelerating gastric cancer progression 13 ; in addition, miR‐625‐5p/LASP1 axis participated in the regulatory functions of LINC01123 in promoting colorectal cancer progression. 14 PMK2 is an enzyme pyruvate kinase and a transcriptional coactivator of STAT1 responsible for the induction of the expression of PDL‐1, which is a key effector in tumor development in terms of immune evasion. In the current study, bioinformation analysis predicted that there were binding sites between LINC00511 and miR‐625‐5p. Additionally, emerging studies have further highlighted that PKM2 may function as an unfavorable prognostic biomarker in various malignancies. 15 , 16 In the present study, we explored the functional mechanism of the interactions between LINC00511 and miR‐625‐5p/PKM2 in LAC.
Methods
Tissue samples
Human tissue samples (LAC tissues and corresponding adjacent normal lung tissues) were collected from 45 LAC patients that were pathologically and surgically confirmed at the Tianjin Hospital. None of the enrolled LAC patients had undergone radiotherapy, chemotherapy, or other tumor‐related therapies prior to the operations. Written informed consents from all patients were obtained, and this study was approved by the Ethics committee of Tianjin Hospital.
Cell lines and cell culture
Human pulmonary epithelial cell line BEAS‐2B and LAC cell lines (H1299and A549) were purchased from ATCC (Manassas, VA, USA). The cells were cultured at 37°C in a humidified incubator with DMEM medium (11995073, Invitrogen, USA) containing 10% fetal bovine serum (FBS) with 5% CO2 at 37°C.
Cell transfection
Full‐length sequences of LINC00511 or PKM2 were synthesized and cloned into pcDNA 3.1 vector, termed pcDNA‐LINC00511 or pcDNA‐ PKM2. siRNAs targeting LINC00511 were purchased from GenePharma Biotechnology Co. Ltd. (Shanghai, China), termed si‐LINC00511. Specific miR‐625‐5p inhibitor or mimic was obtained from Genechem (Shanghai, China). Lentiviral vectors were transfected using polybrene (GenePharma Biotechnology Co. Ltd., Shanghai, China) according to the manufacturer's instructions. Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was applied for transfection of miR‐625‐5p mimics or inhibitor. The transfection efficiency was detected using quantitative real time‐polymerase chain reaction (RT‐qPCR).
RT‐qPCR assay
Total RNA was harvested and isolated from LAC tissues and cells with the application of Trizol reagent (Invitrogen, Carlsbad, CA, USA). The synthesis of cDNA was performed per the proposal of reverse transcription kit (Takara, Dalian, China). SYBR Premix Ex Taq (Takara) was employed for RT‐qPCR analysis, which was carried out on an Applied Biosystems 7500 sequence detection system. Glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) was utilized as an internal reference for LINC00511 and PKM2 while U6 was an internal control for miR‐625‐5p. The relative expression was calculated using the 2−ΔΔCt method. The primers sequences used were listed as following: LINC00511, forward: 5′‐CAGGCATGTGGGGCTTTACT‐3′; reverse: 5′‐TCACGTACCTCACCCAACAC‐3′. MiR‐625‐5p, forward: 5′‐AGGGGGAAAGTTCTATAGTCCGC‐3′; reverse: 5′‐CTCTACAGCTATATTGCCAGCCAC‐3′. PKM2, forward: 5′‐CTATCCTCTGGAGGCTGTGC‐3′; reverse: 5′‐GTGGGGTCGCTGGTAATG‐3′.
GAPDH, forward: 5′‐AATGGGCAGCCGTTAGGAAA‐3′; reverse: 5′‐GCGCCCAATACGACCAAATC‐3′. U6, forward: 5′‐CTCGCTTCGGCAGCACA‐3′; reverse: 5′‐AACGCTTCACGAATTTGCGT‐3′.
CCK‐8 assay
Cell viability was measured via cell counting kit‐8 (CCK‐8) assay. Forty‐eight hours post‐transfection, the transfected LAC cells were inoculated into a 96‐well plate. The cell viability was evaluated at four time points (24, 48, 72, 96 hours) with the addition of 10 μL CCK‐8 solution (Dojindo, Kumamoto, Japan). After CCK‐8 had been added into the cells, the cells underwent another cultural incubation for two hours at 37°C. The optical density (OD) values at 450 nm were detected using a microplate reader (Olympus, Japan).
Transwell assay
LAC cells with different transfections in serum‐free medium were seeded into the apical chamber (8 μm pore size, Corning) of a transwell plate (Corning Costar, Cambridge, Massachusetts, USA) precoated with or without Matrigel (A1569601, BD Biosciences) to evaluate cell invasion or migration capacities. The bottom chamber was filled with 10% FBS DMEM medium, and the cells were incubated for 48 hours. After that, the cells which remained in the apical chambers were wiped out, whereas those which had adhered to the lower chambers were fixed and stained with crystal violet. Finally, the captured images of invasive or migratory cells were counted under a microscope (Olympus, Japan) at 200 × magnification.
Western blot
Total proteins were extracted from LAC cells using radioimmunoprecipitation assay buffer (RIPA buffer, Thermo Fisher Scientific, Inc.). A Pierce BCA Protein Assay Kit (Thermo Scientific, USA) was used to assess the protein concentration in line with the manufacturer's instructions. Thereafter, 30 μg proteins from each sample were loaded on 10% SDS‐PAGE for separation and then transferred to PVDF membrane, followed by blocking with 5% skimmed milk for two hours in TBST at room temperature. After that, the membrane was incubated with primary antibodies against PKM2 (1:1000, Abcam, Cambridge, MA, USA) and GAPDH (1:1000, Abcam, Cambridge, MA, USA) at 4°C overnight. Subsequently, the membrane was incubated with HRP‐conjugated goat anti‐rabbit (1: 2000, Abcam, Cambridge, MA, USA) secondary antibody for two hours at room temperature. GAPDH was used as the internal control. Finally, the signals were detected using an enhanced chemiluminescence (ECL) kit (Millipore, Billerica, MA, USA).
Luciferase reporter assay
The wild‐type (WT) and the corresponding mutant (MUT) binding sites of LINC00511 and PKM2 were cloned into pGL3 promoter vectors (Promega) to construct WT‐LINC00511, WT‐PKM2, MUT‐LINC00511, and MUT‐PKM2, respectively. These recombinant plasmids were then cotransfected into LAC cells together with miR‐625‐5p mimics. Cells were harvested 48 hours after the transfection. Dual‐luciferase reporter assay system (Promega) was used to detect the luciferase activities in accordance with the manufacturer's protocols. Renilla activity was used to normalize firefly activity.
Statistical analysis
All the above experiments were performed in triplicate. All data are displayed as the mean ± SD. The SPSS 17.0 version (SPSS Inc. Chicago, IL, USA) was used to perform the statistical analysis. Data was tested using Student's t‐test or one‐way ANOVA analysis with Tukey's post hoc test. The overall survival of LAC patients was determined by the Kaplan‐Meier curve together with log‐rank test. P < 0.05 was considered to be statistically significant.
Results
High LINC00511 expression in LAC indicated poor prognosis
To examine the role of LINC00511 in LAC, RT‐qPCR was performed to detect LINC00511 expression in LAC tissues and adjacent normal tissues. As shown in Fig 1a, a conspicuous upregulation of LINC00511 was observed in LAC tissues compared with normal tissues (P < 0.01). Moreover, we also examined LINC00511 expression in LAC cells. The results of RT‐qPCR showed that LINC00511 was also remarkably elevated in LAC cells (H1299 and A549) compared with that in BEAS‐2B cells (Fig 1b) (P < 0.01; P < 0.001). Subsequently, the prognostic significance of LINC00511 in LAC patients was also evaluated, and the results indicated that LAC patients with higher LINC00511 levels had shorter survival than those with lower LINC00511 levels (Fig 1c) (P < 0.05). According to these findings, we were able to confirm that strong expression of LINC00511 forecasted a poor prognosis in LAC.
Figure 1.

High LINC00511 expression in LAC indicated poor prognosis. (a) RT‐qPCR analysis showed that LINC00511 was increased in LAC tissues. (b) Upregulated LINC00511 in LAC cells was also confirmed by RT‐qPCR analysis. (c) High LINC00511 expression in LAC patients indicated poor prognosis ( ) LINC00511 (−), (
) LINC00511 (−), ( ) LINC00511 (+). **P < 0.01, ***P < 0.001.
) LINC00511 (+). **P < 0.01, ***P < 0.001.
LINC00511 accelerated LAC cell proliferation, invasion and migration
Following confirmation of the upregulation of LINC00511 in LAC tissues and cell lines, the function of LINC00511 was further explored by transfecting pcDNALINC00511, pcDNA3.1, si‐LINC00511 or si‐NC into LAC cells to augment or silence LINC00511 expression. The transfection efficiencies were confirmed by RT‐qPCR. We found that pcDNALINC00511 in H1299 successfully upregulated LINC00511 expression, while si‐LINC00511 significantly reduced LINC00511 levels in A549 cells (Fig 2a) (P < 0.01; P < 0.001). In addition, the effects of LINC00511 on LAC cell growth, invasion and migration was then investigated. Results of CCK‐8 assays showed that LINC00511 upregulation prominently facilitated H1299 cell proliferation (Fig 2b) (P < 0.01; P < 0.001). Furthermore, overexpression of LINC00511 also enhanced the cell invasion and migration abilities in H1299 cells; on the contrary, the invasion and migration of A549 cells were inhibited by LINC00511 knockdown (Fig 2c and d) (P < 0.01). Based on these findings, we were able to confirm that LINC00511 exerted an oncogenic role in LAC development.
Figure 2.

LINC00511 accelerated LAC cell proliferation, invasion and migration. (a) LINC00511 expression was successfully overexpressed or inhibited by pcDNALINC00511 or si‐LINC00511. (b) CCK‐8 assays showed that LINC00511 upregulation inhibited LAC cell proliferation ( ) pcDNA3.1, (
) pcDNA3.1, ( ) pcDNALINC00511, (
) pcDNALINC00511, ( ) si‐NC, (
) si‐NC, ( ) si‐LINC00511. (c, d) LINC00511 upregulation inhibited LAC invasion and migration as demonstrated by transwell assays.*P < 0.05, **P < 0.01, ***P < 0.001.
) si‐LINC00511. (c, d) LINC00511 upregulation inhibited LAC invasion and migration as demonstrated by transwell assays.*P < 0.05, **P < 0.01, ***P < 0.001.
LINC00511 acted as a sponge of miR‐625‐5p in LAC cells
A number of studies introduced a competing endogenous RNA (ceRNA) regulatory system where lncRNAs were treated as rivals to mRNAs to bind with miRNAs so that those mRNAs lost their ability to translate into protein. 13 Subsequently, the starBase database (http://starbase.sysu.edu.cn/index.php) was utilized to explore the potential mechanisms of LINC00511 in LAC progression. Results indicated that miR‐625‐5p complementarily paired with LINC00511 and the binding sites are presented in Fig 3a. Thereafter, the luciferase reporter assay was performed by cotransfecting miR‐625‐5p mimic and WT‐LINC00511 or MUT‐LINC00511 vector into LAC cells to verify whether LINC00511 was a sponge of miR‐625‐5p. As presented in Fig 3b, the luciferase activity of WT‐LINC00511 was strikingly decreased by miR‐625‐5p mimic while MUT‐LINC00511 constrained the reduction function caused by miR‐625‐5p mimic (P < 0.001). In addition, the interaction between LINC00511 and miR‐625‐5p was further verified by RT‐qPCR analysis. As shown in Fig 3c, miR‐625‐5p expression was dramatically downregulated by LINC00511 overexpression in H1299 cells, and conversely, LINC00511 knockdown in A549 cells significantly increased miR‐625‐5p expression (P < 0.01; P < 0.001). Similarly, the regulatory effect of miR‐625‐5p on LINC00511 expression was also detected. As expected, miR‐625‐5p upregulation conspicuously increased LINC00511 expression, whereas miR‐625‐5p silence had the opposite effect (Fig 3d) (P < 0.05; P < 0.001). Obviously decreased miR‐625‐5p levels in LAC tissues were also confirmed (Fig 3e) (P < 0.01). For further proof, Pearson correlation analysis was applied to reveal that there was a negative correlation between miR‐625‐5p and LINC00511 (Fig 3f) (P < 0.0001). These results suggest that LINC00511 and miR‐625‐5p may function as a competing endogenous RNA (ceRNA) regulatory system whereby LINC00511 are treated as rivals to mRNAs to bind with miR‐625‐5p so that those mRNAs lose their ability to translate into protein.
Figure 3.
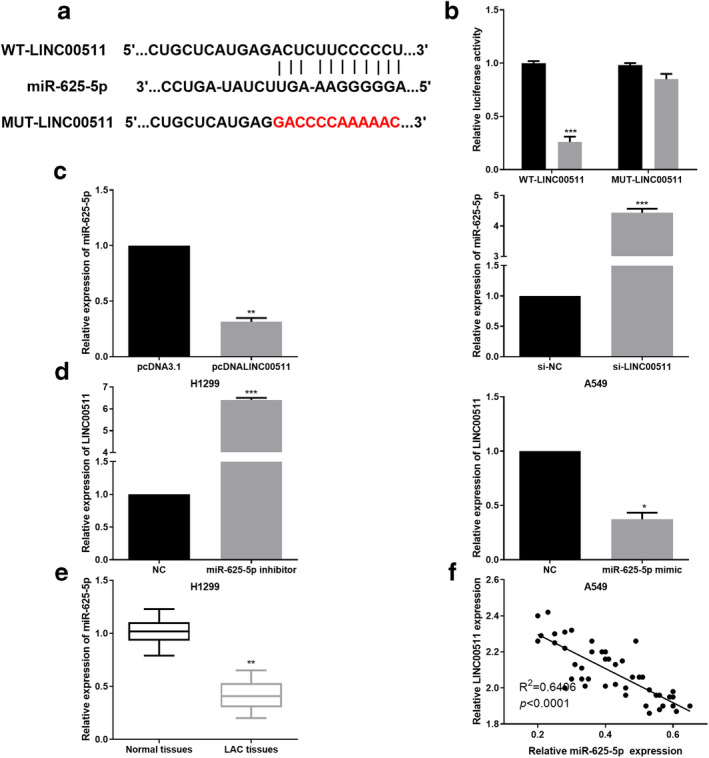
LINC00511 acted as a sponge of miR‐625‐5p in LAC cells. (a) The binding sites between LINC00511 and miR‐625‐5p were predicted. (b) The luciferase activity of WT‐LINC00511 was strikingly decreased by miR‐625‐5p mimic while luciferase activity of MUT‐LINC00511 was not significantly affected ( ) NC, (
) NC, ( ) miR‐625‐5p mimic. (c) Regulatory functions of LINC00511 in miR‐625‐5p expression was determined by RT‐qPCR analysis. (d) miR‐625‐5p regulated LINC00511 expression in LAC cells as demonstrated by RT‐qPCR analysis. (e) miR‐625‐5p was significantly downregulated in LAC tissues. (f) A reverse correlation between miR‐625‐5p and LINC00511 in LAC tissues was validated. *P < 0.05,**P < 0.01,***P < 0.001.
) miR‐625‐5p mimic. (c) Regulatory functions of LINC00511 in miR‐625‐5p expression was determined by RT‐qPCR analysis. (d) miR‐625‐5p regulated LINC00511 expression in LAC cells as demonstrated by RT‐qPCR analysis. (e) miR‐625‐5p was significantly downregulated in LAC tissues. (f) A reverse correlation between miR‐625‐5p and LINC00511 in LAC tissues was validated. *P < 0.05,**P < 0.01,***P < 0.001.
miR‐625‐5p inhibited proliferation, invasion and migration in LAC cells
We further performed gain or loss assays of miR‐625‐5p to determine its role in LAC cells. The results of RT‐qPCR showed that miR‐625‐5p was downregulated in LAC cells (H1299 and A549) compared with that in BEAS‐2B cells (Fig 4a) (P < 0.05; P < 0.01). miR‐625‐5p mimic and inhibitor was then transfected into A549 and H1299 cells, respectively. Successful miR‐625‐5p overexpression or knockdown in LAC cells were confirmed by RT‐qPCR (Fig 4b) (P < 0.01; P < 0.001). Subsequently, functional assays, including CCK‐8 and transwell assays, were performed to evaluate the effect of miR‐625‐5p on LAC cell viability, invasion and migration. As expected, miR‐625‐5p overexpression inhibited A549 cell proliferation while miR‐625‐5p inhibition promoted the proliferation ability in H1299 cells (Fig 4c) (P < 0.01; P < 0.001). Moreover, the invasion and migration abilities of A549 cells were suppressed by miR‐625‐5p overexpression whereas the invasion and migration capacities of H1299 cells were facilitated by miR‐625‐5p silence as shown in Fig 4d (P < 0.05; P < 0.01). The findings indicated that miR‐625‐5p had a suppressive effect on the capacity of LAC proliferation and metastasis.
Figure 4.

miR‐625‐5p inhibited proliferation, invasion and migration in LAC cells. (a) RT‐qPCR analysis showed that miR‐625‐5p was downregulated in LAC cells. (b) miR‐625‐5p was successfully overexpressed or inhibited by miR‐625‐5p mimic or inhibitor. (c) miR‐625‐5p overexpression inhibited LAC cell proliferation ( ) NC, (
) NC, ( ) miR‐625‐5p mimic, (
) miR‐625‐5p mimic, ( ) NC, (
) NC, ( ) miR‐625‐5p inhibitor. (d) miR‐625‐5p overexpression suppressed LAC cell invasion and migration. *P < 0.05,**P < 0.01,***P < 0.001.
) miR‐625‐5p inhibitor. (d) miR‐625‐5p overexpression suppressed LAC cell invasion and migration. *P < 0.05,**P < 0.01,***P < 0.001.
PKM2 served as a target of miR‐625‐5p in LAC cells
The binding sequences of miR‐625‐5p were predicted on PKM2 3′UTR using TargetScan (Fig 5a). Similarly, luciferase reporter assays identified that PKM2 was a direct target of miR‐625‐5p (Fig 5b) (P < 0.01). The regulatory relationship between PKM2 and miR‐625‐5p was studied and RT‐qPCR analysis showed that PKM2 expression was significantly inhibited by miR‐625‐5p mimic and elevated by miR‐625‐5p inhibitor in LAC cells (Fig 5c and d) (P < 0.05; P < 0.01). Similarly, we also detected PKM2 expression in LAC cells transfected with pcDNALINC00511 or si‐LINC00511. As shown in Fig 5e and f, LINC00511 overexpression led to elevated PKM2 expression and LINC00511 silence inhibited PKM2 expression (P < 0.05; P < 0.01). Additionally, PKM2 expression in LAC tissues was also measured and the results demonstrated a significant increase compared to surrounding healthy tissues (Fig 5g) (P < 0.01). In LAC tissues, PKM2 expression was positively associated with LINC00511 expression (Fig 5h) (P < 0.0001). All these data suggest that miR‐625‐5p can modulate PKM2 expression by directly targeting its 3′UTR.
Figure 5.

PKM2 served as a target of miR‐625‐5p in LAC cells. (a) The binding sequences of miR‐625‐5p were predicted on PKM2 3′UTR using TargetScan. (b) Luciferase reporter assays indicated that miR‐625‐5p mimic significantly decreased luciferase activity of WT‐PKM2 while had no obvious influence on luciferase activity of MUT‐PKM2 ( ) NC, (
) NC, ( ) miR‐625‐5p mimic. (c, d) miR‐625‐5p regulated PKM2 expression in LAC cells. (e,f) The function of LINC00511 in regulating PKM2 expression was detected by RT‐qPCR. (g) PKM2 expression was notably increased in LAC tissues. (h) A positive correlation between PKM2 expression and LINC00511 expression in LAC tissues was observed. *P < 0.05, **P < 0.01.
) miR‐625‐5p mimic. (c, d) miR‐625‐5p regulated PKM2 expression in LAC cells. (e,f) The function of LINC00511 in regulating PKM2 expression was detected by RT‐qPCR. (g) PKM2 expression was notably increased in LAC tissues. (h) A positive correlation between PKM2 expression and LINC00511 expression in LAC tissues was observed. *P < 0.05, **P < 0.01.
LINC00511 promoted LAC tumorigenesis by sponging miR‐625‐5p to upregulate PKM2
To identify the interaction between LINC00511 and miR‐625‐5p/PKM2 axis in LAC cells, we transfected si‐LINC00511, miR‐625‐5p inhibitor and pcDNA‐PKM2 into A549 cells as indicated. The results of western blot and RT‐qPCR showed that si‐LINC00511 significantly decreased PKM2 expression, while in cells with cotransfection of si‐LINC00511 and miR‐625‐5p inhibitor/pcDNA‐PKM2, the reduced PKM2 expression was recovered (Fig 6a) (P < 0.001). Moreover, we also detected the PDL‐1 expression level, a downstream factor of PMK2, to further confirm our conclusions. Results showed that si‐LINC00511 significantly decreased PDL‐1 expression, while in cells with cotransfection of si‐LINC00511 and miR‐625‐5p inhibitor/pcDNA‐PKM2, the reduced PDL‐1 expression was recovered (Fig 6b) (P < 0.01). CCK‐8 assays demonstrated that miR‐625‐5p silence or PKM2 upregulation in si‐LINC00511‐transfected cells could restore cell proliferation in comparison with the controls (Fig 6c) (P < 0.01). Cotransfection with si‐LINC00511 and miR‐625‐5p inhibitor or pcDNA‐PKM2 led to an obvious increase in the number of cell invasion and migration compared to the corresponding si‐LINC00511‐transfected cells (Fig 6d) (P < 0.01). These findings implied that LINC00511 promoted LAC tumorigenesis by sponging miR‐625‐5p to upregulate PKM2.
Figure 6.

LINC00511 promoted LAC tumorigenesis by sponging miR‐625‐5p to upregulate PKM2. (a) Decreased PKM2 expression in LAC cells caused by si‐LINC00511 were restored by miR‐625‐5p inhibitor or pcDNA‐PKM2 ( ) si‐NC, (
) si‐NC, ( ) si‐LINC00511, (
) si‐LINC00511, ( ) si‐LINC00511+miR‐625‐5p inhibitor, (
) si‐LINC00511+miR‐625‐5p inhibitor, ( ) si‐LINC00511+pcDNA‐PKM2. (b) Decreased PDL‐1 expression in LAC cells caused by si‐LINC00511 were restored by miR‐625‐5p inhibitor or pcDNA‐PKM2. (c, d) The impaired proliferation, invasion and migration abilities of LAC cells caused by si‐LINC00511 were recovered by miR‐625‐5p inhibitor or pcDNA‐PKM2 (
) si‐LINC00511+pcDNA‐PKM2. (b) Decreased PDL‐1 expression in LAC cells caused by si‐LINC00511 were restored by miR‐625‐5p inhibitor or pcDNA‐PKM2. (c, d) The impaired proliferation, invasion and migration abilities of LAC cells caused by si‐LINC00511 were recovered by miR‐625‐5p inhibitor or pcDNA‐PKM2 ( ) si‐NC, (
) si‐NC, ( ) si‐LINC00511, (
) si‐LINC00511, ( ) si‐LINC00511+miR‐625‐5p inhibitor, (
) si‐LINC00511+miR‐625‐5p inhibitor, ( ) si‐LINC00511+pcDNA‐PKM2, (
) si‐LINC00511+pcDNA‐PKM2, ( ) si‐NC, (
) si‐NC, ( ) si‐LINC00511, (
) si‐LINC00511, ( ) si‐LINC00511+miR‐625‐5p inhibitor, (
) si‐LINC00511+miR‐625‐5p inhibitor, ( ) si‐LINC00511+pcDNA‐PKM2, (
) si‐LINC00511+pcDNA‐PKM2, ( ) si‐NC, (
) si‐NC, ( ) si‐LINC00511, (
) si‐LINC00511, ( ) si‐LINC00511+miR‐625‐5p inhibitor, (
) si‐LINC00511+miR‐625‐5p inhibitor, ( ) si‐LINC00511+pcDNA‐PKM2. **P < 0.01,***P < 0.001.
) si‐LINC00511+pcDNA‐PKM2. **P < 0.01,***P < 0.001.
Discussion
Lung cancer is the most common malignant tumor worldwide, among which LAC has the highest degree of diversities. Metastasis‐related deaths account for approximately 90% of total mortality. 17 In recent years, accumulating studies have proposed that aberrant lncRNA expression participates in a variety of cancer‐related processes. 18 Additionally, it has been identified that certain lncRNAs are specific to developmental stages, disease types and tissues. 19 , 20 Therefore, further research of the relationship between tumorigenesis and lncRNAs is very important to better understand cancer. Herein, we discovered that LINC00511 was highly expressed in LAC tissues compared with that in corresponding noncancerous tissues, and higher LINC00511 expression was closely connected with worse survival in LAC patients. We also confirmed that silencing LINC00511 suppressed LAC cell proliferation, invasion and migration. Mechanistically, we revealed that LINC00511 directly interacted with miR‐625‐5p at recognized seeds. LINC00511 exerted its oncogenic function on LAC via its role to act as a ceRNA for miR‐625‐5p, and subsequently activate the PKM2 signaling pathway. These findings indicate that LINC00511 may exert an oncogenic function and play a crucial role in LAC development.
LINC00511 is a newly discovered lncRNA reported to exert important functions in tumorigenesis. In cervical cancer, inhibition of LINC00511 was found to promote cell apoptosis and autophagy via RXRA‐regulated PLD1. 21 In hepatocellular carcinoma, overexpression of LINC00511 was correlated with poor prognosis, and promoted cell metastasis and proliferation by regulating of miR‐424. 22 LINC00511 functioned as a competing endogenous RNA to regulate VEGFA via sponging miR‐29b‐3p in pancreatic ductal adenocarcinoma. 23 All these results support the potential oncogenic roles of LINC00511 in tumors. However, there are no relevant studies which focus on the effects of LINC00511 on LAC. In our study, upregulation of LINC00511 was identified and validated in LAC cells and tissues. Further, Kaplan Meier analysis determined that LINC00511 expression is an indicator for overall survival, and is markedly correlated with shorter survival and poor prognosis. Moreover, we also found that overexpression of LINC00511 promoted LAC cell proliferation, invasion and migration.
Identifying the molecular events associated with the carcinogenic effects of LINC00511 on LAC will help to explore effective treatment strategies. Competitive endogenous RNAs (ceRNAs) are considered to be a kind of post‐transcriptional regulator in altering gene expression by miRs. 24 Recently, lncRNAs have been identified to function as ceRNAs in mediating the gene expressions which are involved in tumor progression. 25 Data in our study indicated that LINC00511 acted as a ceRNA of miR‐625‐5p via regulation of PKM2 in LAC. Underexpression of miR‐625‐5p has been identified in multiple human cancer types, including cervical cancer 26 and gastric cancer. 13 Here, our solid data demonstrated that miR‐625‐5p was downregulated in LAC tissues and cell lines. MiR‐625‐5p overexpression inhibited LAC cell malignancy by inhibiting proliferation, invasion and migration. Another important finding in our study was that the oncogenic functions of LINC00511 in LAC cells may depend on sponging miR‐625‐5p. Importantly, Pearson correlation analysis was applied to reveal that there was a negative correlation between miR‐625‐5p and LINC00511 in LAC tissues. In brief, we found that LINC00511 expression was inhibited by miR‐625‐5p overexpression and promoted by silencing miR‐625‐5p, suggesting that miR‐625‐5p is involved in the functions of LINC00511 in LAC. miR may exert its function via direct interaction with the 3′‐UTRs of the target mRNAs. Therefore, the mechanism underlying miR‐625‐5p inhibited LAC progression was further investigated in the current study. PKM2 was confirmed as a direct target of miR‐625‐5p and was involved in the regulatory functions of miR‐625‐5p/LINC00511 axis in LAC. Consequently, we found that silencing LINC00511 and miR‐625‐5p restoration could inhibit LAC cell proliferation, invasion and migration via downregulation of PKM2.
In conclusion, upregulation of LINC00511 in LAC tissues and cell lines was validated. Upregulation of LINC00511 was closely correlated with shorter overall survival in LAC patients. We explored the biological function of LINC00511 in promoting LAC cell proliferation, migration and invasion. Moreover, LINC00511 knockdown could significantly suppress LAC malignancy, partially by sponging miR‐625‐5p, and then upregulate PKM2 to exert its tumor‐suppressive effect. Our studies confirmed the roles of a novel LINC00511/miR‐625‐5p/PKM2 axis in LAC progression, which might provide a promising application in LAC treatment.
Disclosure
The authors declare that there are no conflicts of interest.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019; 69: 7–34. [DOI] [PubMed] [Google Scholar]
- 2. Miller KD, Nogueira L, Mariotto AB et al Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 2019; 69: 363–85. [DOI] [PubMed] [Google Scholar]
- 3. Pisapia P, Pepe F, Troncone G, Malapelle U. Predictive biomarkers for molecular pathology in lung cancer. Biomark Med 2020; 14: 253–7. [DOI] [PubMed] [Google Scholar]
- 4. Zhao T, Qian K, Zhang Y. High expression of FGF5 is an independent prognostic factor for poor overall survival and relapse‐free survival in lung adenocarcinoma. J Comput Biol 2019; 27: 948–57. [DOI] [PubMed] [Google Scholar]
- 5. Zablockis R, Zurauskas E, Danila E, Gruslys V. Prognostic value of thyroid transcription factor‐1 expression in patients with advanced lung adenocarcinoma. In Vivo 2018; 32: 1571–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kastner J, Hossain R, White CS. Epidemiology of lung cancer. Semin Roentgenol 2020; 55: 23–40. [DOI] [PubMed] [Google Scholar]
- 7. Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non‐coding regions of the genome. Nat Rev Genet 2010; 11: 559–71. [DOI] [PubMed] [Google Scholar]
- 8. Clark MB, Johnston RL, Inostroza‐Ponta M et al Genome‐wide analysis of long noncoding RNA stability. Genome Res 2012; 22: 885–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta stone of a hidden RNA language? Cell 2011; 146: 353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xue M, Tao W, Yu S et al lncRNA ZFPM2‐AS1 promotes proliferation via miR‐18b‐5p/VMA21 axis in lung adenocarcinoma. J Cell Biochem 2020; 121: 313–21. [DOI] [PubMed] [Google Scholar]
- 11. Jiang W, Zhang B, Sun J, Liu Y, Bi Y, Wei H. LncRNA FTX promotes the tumorigenesis of lung adenocarcinoma by targeting miR‐300. Panminerva Med 2020. 10.23736/S0031-0808.19.03823-0 [DOI] [PubMed] [Google Scholar]
- 12. Li Y, Ding Y, Ding N et al MicroRNA‐625‐5p sponges lncRNA MALAT1 to inhibit cervical carcinoma cell growth by suppressing NF‐kappaB signaling. Cell Biochem Biophys 2020; 78: 217–25. [DOI] [PubMed] [Google Scholar]
- 13. Chen Z, Wu H, Zhang Z, Li G, Liu B. LINC00511 accelerated the process of gastric cancer by targeting miR‐625‐5p/NFIX axis. Cancer Cell Int 2019; 19: 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shang T, Zhou X, Chen W. LINC01123 promotes progression of colorectal cancer via miR‐625‐5p/LASP1 axis. Cancer Biother Radiopharm 2020. 10.1089/cbr.2020.3740 [DOI] [PubMed] [Google Scholar]
- 15. James AD, Richardson DA, Oh IW et al Cutting off the fuel supply to calcium pumps in pancreatic cancer cells: Role of pyruvate kinase‐M2 (PKM2). Br J Cancer 2020; 122: 266–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sfakianaki M, Papadaki C, Tzardi M et al PKM2 expression as biomarker for resistance to oxaliplatin‐based chemotherapy in colorectal cancer. Ann Oncol 2019; 30 (Suppl 4): iv21–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mehlen P, Puisieux A. Metastasis: A question of life or death. Nat Rev Cancer 2006; 6: 449–58. [DOI] [PubMed] [Google Scholar]
- 18. Forrest ME, Khalil AM. Review: Regulation of the cancer epigenome by long non‐coding RNAs. Cancer Lett 2017; 407: 106–12. [DOI] [PubMed] [Google Scholar]
- 19. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci 2016; 73: 2491–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alvarez‐Dominguez JR, Lodish HF. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood 2017; 130: 1965–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shi Y, Liu M, Huang Y, Zhang J, Yin L. Promotion of cell autophagy and apoptosis in cervical cancer by inhibition of long noncoding RNA LINC00511 via transcription factor RXRA‐regulated PLD1. J Cell Physiol 2020. 10.1002/jcp.29529 [DOI] [PubMed] [Google Scholar]
- 22. Wang WL, Yu DJ, Zhong M. LncRNA HAGLROS accelerates the progression of lung carcinoma via sponging microRNA‐152. Eur Rev Med Pharmacol Sci 2019; 23: 6531–8. [DOI] [PubMed] [Google Scholar]
- 23. Zhao X, Liu Y, Li Z et al Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa‐miR‐29b‐3p in pancreatic ductal adenocarcinoma. J Cell Mol Med 2018; 22: 655–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zheng L, Xiang C, Li X et al STARD13‐correlated ceRNA network‐directed inhibition on YAP/TAZ activity suppresses stemness of breast cancer via co‐regulating Hippo and Rho‐GTPase/F‐actin signaling. J Hematol Oncol 2018; 11: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014; 505: 344–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu B, Jin X, Yang T et al Upregulated lncRNA THRIL/TNF‐alpha signals promote cell growth and predict poor clinical outcomes of osteosarcoma. Onco Targets Ther 2020; 13: 119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]


