Abstract
Background
The status of targeted genes and the association between targeted genes and clinicopathological features in Chinese lung cancer patients remains to be elucidated.
Methods
The status of 10 targeted genes was evaluated by next‐generation sequencing (NGS) in 884 non‐small cell lung cancer (NSCLC) patients. The relationship between gene alterations and clinicopathological characters was analyzed.
Results
Overall, 684 (77.4%) patients harbored gene alterations, and EGFR (510, 57.7%) was found to be the most common type of mutation followed by KRAS (91, 10.3%), HER2 (38, 4.3%), PIK3CA (32, 3.6%), ALK (21, 2.4%), BRAF (10, 1.1%), ROS1 (5, 0.6%), RET (5, 0.6%), MET (4, 0.5%) and NRAS (1, 0.1%). Gene alterations were more frequent in females, non‐smokers and adenocarcinoma (P < 0.001). EGFR mutations were associated with women, non‐smokers, normal level of serum tumor markers, and adenocarcinoma (P < 0.001). Patients without lymph node metastasis (P = 0.012), or early stage disease (P < 0.001) exhibited a higher EGFR mutation rate. KRAS mutations tended to arise in men (P < 0.001), smokers (P < 0.001) and patients with higher levels of serum tumor markers (P = 0.048). A mucus‐producing component was associated with KRAS (P < 0.001), ROS1 (P = 0.033) and ALK (P < 0.001) alterations. ALK and ROS1 rearrangements were more frequent in micropapillary structures (P = 0.004, P = 0.012). BRAF mutation was associated with advanced disease patients and micropapillary structure (P < 0.001). PIK3CA mutation was more likely to be found in elderly patients (P = 0.014). Some patients had synchronous gene alterations, including EGFR/PIK3CA, EGFR/HER2, HER2/KRAS, EGFR/KRAS, EGFR/ROS1, EGFR/NRAS, KRAS/PIK3CA, KRAS/PIK3CA/HER2.
Conclusions
Most patients had at least one genetic alteration, and individual patients harbored synchronous mutation. Each gene alteration had unique clinicopathological characteristics.
Key points
Significant findings of the study
This study revealed the frequency and distribution of 10 targeted gene abnormalities and their association with clinicopathological parameters of Chinese non‐small cell lung cancer (NSCLC) patients in eastern China.
What this study adds
Some rare synchronous mutations were detected in our study by next‐generation sequencing (NGS).
Keywords: Clinicopathological characteristics, gene mutation, next‐generation sequencing, non‐small cell lung cancer
In this research, we investigated the frequency of common driving genes in 884 patients with non‐small cell lung cancer and analyzed their association with clinicopathologic parameters in in eastern China. Most patients had at least one genetic alteration, and individual patients harbored synchronous mutation. Each gene alteration has unique clinicopathologic characteristics.
Introduction
Lung cancer is a common malignant tumor and causes the largest number of cancer‐related deaths worldwide. 1 , 2 Non‐small cell lung cancer (NSCLC) is the most common histologic type of lung cancer, accounting for approximately 80% of all cases of lung cancer. NSCLC mainly includes adenocarcinoma, adenosquamous carcinoma, squamous cell carcinoma and large cell carcinoma. 3 In recent years, oncogene aberration has been widely studied in various cancers, especially in lung carcinoma. 4 , 5 New and emerging molecular targeted therapies have achieved substantial progress in NSCLC due to understanding the molecular origins, signaling transduction pathway and metabolic process in lung cancer. Research into the role of tyrosine kinase inhibitors in EGFR gene mutations has greatly improved the prognosis of lung cancer patients with EGFR activating mutation6, 7 and with the development of molecular targeted drug research, except for EGFR mutation, many other types of targeted gene aberrations have been identified, including HER2, 8 BRAF, 9 KRAS mutations, 10 and ALK rearrangement. 11 Targeted drugs can provide optimal efficacy and prolong survival time in activating mutation NSCLC patients. 12
There are many methods for gene detection for targeted drugs. Compared with traditional methods, next‐generation sequencing (NGS) reduces the cost of testing and can comprehensively screen different gene alterations to provide a reliable and effective therapeutic schedule for individualized treatment. Here, we examined the status of EGFR, HER2, BRAF, KRAS, ALK, MET, NRAS, PIK3CA, RET, and ROS1 in 884 patients with NSCLC who had undergone surgical resection, bronchoscopy biopsy or percutaneous transthoracic biopsy by NGS in our institution. We attempted to investigate the frequency and distribution of 10 targeted gene abnormalities and analyze their association with clinicopathological parameters.
Methods
Patients
This study included all NSCLC patients who underwent surgical resection (714/884) and bronchoscopy biopsy and percutaneous transthoracic biopsy (170/884) from June 2017 to September 2018 in the Affiliated Hospital of Qingdao University. A histological diagnosis had been sufficiently confirmed by two experienced pulmonary pathologists. A total of 884 patients (486 female and 398 males) were included in the present study. Bronchoscopy and puncture biopsy specimens from patients diagnosed with NSCLC‐NOS were not included in this study. This study was approved by the Ethics Committee of the Affiliated Hospital of Qingdao University (No. QYFY WZLL 25784). Written informed consent was obtained from all patients.
Clinicopathological characteristics
Histologic subtypes were classified according to the 2015 WHO histologic classification of lung cancer. 3 All patients were staged according to the eighth edition of the TNM classification of the American Joint Committee on Cancer (AJCC) for lung cancer. 13 We collected clinicopathological data from patients' electronic medical record database, including age, gender, smoking history, tumor location, metastasis, pathological TNM stage and the level of serum tumor markers associated with lung cancer (including CEA, CK19, CA125, SCCA, ProGRP and NSE).
Methods of DNA extraction and next‐generation sequencing
All tumor tissue samples were fixed in 10% neutral formalin solution, embedded in paraffin, and 3 μM sections were prepared for hematoxylin and eosin (H&E). There were enough tumor cells for gene detection. We used QIAamp DNA formalin‐fixed paraffin‐embedded (FFPE) tissue kit (Qiagen, Valencia, CA, Germany) to extract genomic DNA from FFPE tissues. The concentration of genomic DNA was detected by the Qubit dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). We used human EGFR/KRAS/BRAF/PIK3CA/ALK/ROS1/HER2/MET/RET/NRAS mutation test kit (Geneis, Beijing, China) to prepare the libraries. PCR products were enriched, purified, concentrated and a gene probe pool was used to capture the libraries. A quantity and quality inspection was performed using the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, USA). Under the condition that the concentration satisfied the requirement, the NGS was conducted on an Illumina MiniSeq system (Illumina, San Diego, USA), utilizing the MiniSeq High Output Reagent Kit (300 cycles) (Illumina) to sequence the genomic DNA with 15–18 pooled libraries, and paired‐end with 2 × 151 bp. We split the original sequencing records to determine whether the sample data was qualified, and removed the inferior quality dates. Alterations were recognized by BWA, Freebayes and Annoval. We used Crest, Factera, ionCOPY, FreeBayes and Annoval to analyze and annotate the mutation information of binary sequence alignment/map files. In the process of verifying the NGS analysis, the background noise cutoff value for the single nucleotide variation was selected as 1%. In this research, the minimum requirement for sequencing depth was 500x. All procedures were conducted following the manufacturers' protocols.
Immunohistochemistry
Immunohistochemical analysis was performed on PPFE sections using the HER2 (Ventana Medical Systems, Tucson, AZ, USA) antibody. HER2 staining was performed on a VENTANA Benchmark XT automated system (Ventana Medical Systems, Inc., Tucson, AZ, USA).
Statistical analysis
The statistical analysis of clinicopathological features was carried out by Chi‐square test or Fisher's exact test to explore the association between 10 targeted gene alterations frequency and clinicopathological profiles. All statistical analyses were evaluated using the SPSS 24.0 software (SPSS, Chicago, IL). A P < 0.05 was considered statistically significant.
Results
Patient characteristics
Detailed clinicopathological information is summarized in Table 1. In this study, 868 patients had not been treated with preoperative chemotherapy or targeted therapy, 10 patients had received targeted treatment, and six patients had received preoperative adjuvant chemo‐radiotherapy. There were 486 (55.0%) females and 398 (45.0%) males, and 266 (30.1%) patients were current or former smokers. The average age was 60.1 years (range, 22–87 years). The pathological analysis identified that 92.1% (814/884) of the samples were from adenocarcinoma, 6.1% (54/884) from squamous cell carcinoma, 1.0% (9/884) from adenosquamous carcinoma and 0.8% (7/884) from large cell carcinoma.
Table 1.
Clinicopathological profiles of 884 patients
| Clinicopathological profiles | No. of patients (%) |
|---|---|
| Sex | |
| Male | 398 (45.0) |
| Female | 486 (55.0) |
| Age | |
| Median | 61.0 |
| Average | 60.1 |
| Range | 22–87 |
| Smoking history | |
| Non‐smoker | 618 (69.9) |
| Former/current smoker | 266 (30.1) |
| Serum tumor markers | |
| Normal | 357 (40.4) |
| Abnormal | 527 (59.6) |
| Tumor location | |
| Left lung | 394 (44.5) |
| Right lung | 488 (55.3) |
| Histologic subtype | |
| AIS | 5 (0.6) |
| MIA | 58 (6.6) |
| Invasive adenocarcinoma | 751 (85.9) |
| Lepidic | 123 (13.9) |
| Acinar | 384 (43.4) |
| Papillary | 89 (12.2) |
| Solid | 56 (10.1) |
| Micropapillary | 7 (0.8) |
| IMA | 20 (2.3) |
| SCC | 54 (6.1) |
| Adenosquamous carcinoma | 9 (1.0) |
| LCC | 7 (0.8) |
| TNM stage | |
| I | 474(53.6) |
| II | 95 (10.7) |
| III | 184 (20.8) |
| IV | 131 (14.8) |
AIS, adenocarcinoma in situ; IMA, invasive mucinous adenocarcinoma; LCC, large cell carcinoma; MIA, minimally invasive adenocarcinoma; SCC, squamous cell carcinoma.
Table 2 shows the comparison of the mutation rate of 10 targeted genes in different genders, age groups, smoking history and histologic types. The mutation rate in females was 86.0% (418/486), which was significantly higher than that in males (66.8%, 266/398) (P < 0.001). There was no statistical difference between the patients over 60 years old (78.5%, 351/447) and those under the age of 60 (76.2%, 333/437) (P = 0.409). Gene mutations were detected in 84.1% (520/618) non‐smokers, which was significantly higher than that in former or current smokers (61.7%, 164/266) (P < 0.001). Overall, 81.7% (672/823) of samples with an adenocarcinoma component, including adenocarcinoma and adenosquamous carcinoma, had gene mutations. In contrast, only 19.7% (12/61) of samples without an adenocarcinoma component, including squamous cell carcinoma and large cell carcinoma, had gene mutations (P < 0.001).
Table 2.
Comparison of the mutation rate of 10 targeted genes in different genders, age groups, smoking history and histologic types
| Group | No (n) | Mutant (%) | Wild‐type (%) | χ2 | P‐value |
|---|---|---|---|---|---|
| Gender (P) | 45.953 | <0.001 | |||
| Male | 398 | 266 (66.8) | 132 (33.2) | ||
| Female | 486 | 418 (86.0) | 68 (14.0) | ||
| Age (P) | 0.681 | 0.409 | |||
| <60 | 437 | 333 (76.2) | 104 (23.8) | ||
| ≥60 | 447 | 351 (78.5) | 96 (21.5) | ||
| Smoking history (P) | 53.721 | <0.001 | |||
| Non‐smokers | 618 | 520 (84.1) | 98 (15.9) | ||
| Former/current smokers | 266 | 164 (61.7) | 102 (38.3) | ||
| Histologic types (P) | 124.625 | <0.001 | |||
| With adenocarcinoma component | 823 | 672 (81.7) | 151 (18.3) | ||
| Without adenocarcinoma component | 61 | 12 (19.7) | 49 (80.3) |
P, P refers to the comparison with different genders, age groups, smoking history and histologic types. With adenocarcinoma component, includes adenocarcinoma and adenosquamous carcinoma; without adenocarcinoma component, includes squamous cell carcinoma and large cell carcinoma.
Frequency of 10 targeted gene alterations
In this study, the mean depth of sequencing was 1000×. Of 884 patients, 684 (77.4%) patients had at least one gene alteration, and 33 (3.7%) samples harbored more than one driver gene alteration. The EGFR, KRAS, HER2, PIK3CA, ALK, BRAF, ROS1, RET, MET, and NRAS gene alteration rates were 57.7% (510/884), 10.3% (91/884), 4.3% (38/884), 3.6% (32/884), 2.4% (21/884), 1.1% (10/884), 0.6% (5/884), 0.6% (5/884), 0.5% (4/884) and 0.1% (1/884), respectively. Moreover, 33 (3.7%) patients had another synchronous gene alteration, EGFR/PIK3CA (19, 2.1%) was the most frequent coalteration, followed by EGFR/HER2 (6, 0.7%), HER2/KRAS (3, 0.3%), EGFR/KRAS (1, 0.1%), EGFR/ROS1 (1, 0.1%), EGFR/NRAS (1, 0.1%), KRAS/PIK3CA (1, 0.1%). Only one case harbored a triple KRAS/PIK3CA/HER2 coalteration (0.1%, 1/884). Detailed information of multiple alteration combinations can be found in Table S1. Multiple alteration EGFR exon 21 L858+PIK3CA exon 9 E545K showed the highest incidence rate (15.2%, 5/33) followed by EGFR exon 19 del +PIK3CA exon 20 H1047R (12.1%, 4/33). Specific mutation frequency of each gene in surgical specimens and biopsy specimens is shown in Table S2. In general, surgical specimens were from patients in early stage disease, and puncture or bronchoscopic biopsy specimens were from patients in advanced stage disease. EGFR mutation occurred mostly in patients with early stage disease (P < 0.001), whereas KRAS and HER2 alterations more frequently occurred in patients in advanced stage disease (P = 0.008, P = 0.005, respectively).
EGFR mutation status and association with clinicopathological features
In this study, 510 (57.7%) patients harbored EGFR mutation, 28 (5.5%) specimens possessed another synchronous gene alteration, EGFR/PIK3CA (19/28) was the most frequent coalteration, followed by EGFR/HER2 (6/28). In addition, double mutations involving the same or different exons in EGFR were observed in 29 patients, accounting for 3.3% (29/884) of all patients. The detailed information of EGFR mutations with clinicopathological characteristics of NSCLC is shown in Table 3. Overall, 31.4% (278/884) of patients had an exon 21 L858R mutation, followed by exon 19 del in 20.8% (184/884) patients. The other four mutation subtypes higher than 1.0% were exon 18 G719X, exon 20 T790M, exon 21 L861Q and exon 20 S768I, and the frequencies were 2.9% (26/884), 1.6% (14/884), 1.5% (13/884), and 1.2% (11/884), respectively. Furthermore, five rare mutations were detected in our study, and the specific mutation phenotype is as follows: exon 18 p.Leu707Trp, exon 18 p.Glu709_Thr710delinsAsp, exon 20 p.Gly796Ser, exon 19 p.Glu749_Ser752delinsAsp, exon 20 p.His773_Val774delinsArgMet. Of the 14 cases with T790M mutation, five cases were accompanied by 19 del mutation, and nine cases were accompanied by 21 L858R mutation. Seven of the patients with 20 T790M mutation had an EGFR‐TKI treatment history.
Table 3.
Associations of EGFR mutation with clinicopathological characteristics of NSCLC
| No. of patients | ||||||||
|---|---|---|---|---|---|---|---|---|
| Clinicopathological characteristics | No | Total EGFR mutation | 18 G719X | 19 del | 20 S768I | 20 T790M | 21 L858R | 21 L861Q |
| No | 510 | 26 | 184 | 11 | 14 | 278 | 13 | |
| Age (P) | 0.575 | 0.846 | 0.995 | 0.120 | 0.116 | 0.352 | 0.812 | |
| <60 | 437 | 248 | 14 | 91 | 8 | 4 | 131 | 6 |
| ≥60 | 447 | 262 | 12 | 93 | 3 | 10 | 147 | 7 |
| Sex(P) | <0.001 | 0.906 | <0.001 | 0.738 | 0.480 | <0.001 | 0.632 | |
| Male | 398 | 157 | 12 | 56 | 6 | 5 | 80 | 5 |
| Female | 486 | 353 | 14 | 128 | 5 | 9 | 198 | 8 |
| Serum tumor markers (P) | <0.001 | 0.543 | 0.032 | 0.033 | 0.045 | 0.004 | 0.669 | |
| Normal | 357 | 240 | 9 | 87 | 1 | 2 | 132 | 6 |
| Abnormal | 527 | 270 | 17 | 97 | 10 | 12 | 146 | 7 |
| Gross type (P) | <0.001 | 0.680 | 0.044 | 0.328 | 0.696 | 0.032 | 0.584 | |
| Central type | 70 | 24 | 1 | 6 | 0 | 2 | 14 | 0 |
| Peripheral type | 814 | 486 | 25 | 176 | 11 | 12 | 264 | 13 |
| Smoking history (P) | <0.001 | 0.939 | <0.001 | 0.900 | 0.111 | 0.020 | 0.802 | |
| Non‐smokers | 618 | 431 | 18 | 156 | 7 | 13 | 238 | 10 |
| Former/current smokers | 266 | 79 | 8 | 28 | 4 | 1 | 40 | 3 |
| Histology (P*) | <0.001 | 0.309 | <0.001 | 0.756 | 0.620 | <0.001 | 0.932 | |
| With adenocarcinoma component | 823 | 509 | 26 | 184 | 11 | 14 | 277 | 13 |
| AIS | 5 | 2 | 0 | 1 | 0 | 0 | 1 | 0 |
| MIA | 58 | 21 | 0 | 6 | 0 | 0 | 15 | 0 |
| Lepidic | 123 | 68 | 1 | 17 | 0 | 0 | 47 | 1 |
| Acinar | 384 | 278 | 15 | 100 | 7 | 6 | 153 | 9 |
| Papillary | 89 | 71 | 6 | 31 | 0 | 1 | 31 | 1 |
| Solid | 56 | 12 | 1 | 3 | 1 | 0 | 7 | 0 |
| Micropapillary | 7 | 4 | 0 | 2 | 0 | 0 | 2 | 0 |
| IMA | 20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Adenosquamous carcinoma | 9 | 6 | 0 | 2 | 0 | 0 | 4 | 0 |
| Without adenocarcinoma component | 61 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| SCC | 54 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| LCC | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TNM stage (P) | <0.001 | 0.599 | 0.336 | 1.000 | 0.158 | 0.001 | 1.000 | |
| I + II | 569 | 358 | 18 | 124 | 7 | 6 | 201 | 8 |
| III + IV | 315 | 152 | 8 | 60 | 4 | 8 | 77 | 5 |
With adenocarcinoma component, includes adenocarcinoma and adenosquamous carcinoma; Without adenocarcinoma component, includes squamous cell carcinoma and large cell carcinoma; P, P refers to the comparison with different age groups, genders, serum tumor markers, gross type, smoking history and TNM stage; P*, P* refers to the comparison between with adenocarcinoma component and without adenocarcinoma component.
AIS, adenocarcinoma in situ; IMA, invasive mucinous adenocarcinoma; LCC, large cell carcinoma; MIA, minimally invasive adenocarcinoma; SCC, squamous cell carcinoma.
As shown in Table 3, the EGFR mutation was significantly associated with women, non‐smoker, and normal serum tumor markers level (both P < 0.001). The patients without lymph node metastasis (P = 0.012) in early stage disease (P < 0.001) exhibited a higher EGFR mutation rate. Patients with adenocarcinoma (P < 0.001) and peripheral tumors (P < 0.001) were found to be more likely to have EGFR mutations.
ALK rearrangement status and association with clinicopathological features
A total of 21 patients (2.4%) with invasive adenocarcinoma had ALK rearrangement, and all demonstrated echinoderm microtubule associated protein‐like 4 (EML4) rearrangement. As shown in Table 4, ALK rearrangement was more frequent in adenocarcinomas with mucus‐producing component (P < 0.001), micropapillary structure (P = 0.004) and invasion of the visceral pleura of the lung (P = 0.048). No difference was detected in age, gender, smoking history, tumor location and TNM stage between patients with and without ALK rearrangement (P > 0.05).
Table 4.
Nine targeted gene alterations status and association with histological characteristics of NSCLC
| No. of patients | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Histology | No | ALK | ROS1 | BRAF | KRAS | HER2 | PIK3CA | RET | MET | NRAS |
| With or without adenocarcinoma component (P*) | 0.207 | 0.699 | 1.000 | 0.062 | 0.684 | 0.001 | 0.542 | 0.715 | 1.000 | |
| With adenocarcinoma component | 823 | 21 | 5 | 9 | 89 | 36 | 25 | 5 | 4 | 1 |
| AIS | 5 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| MIA | 58 | 0 | 0 | 0 | 5 | 10 | 0 | 0 | 1 | 0 |
| Lepidic | 123 | 1 | 0 | 0 | 11 | 11 | 1 | 0 | 2 | 0 |
| Acinar | 384 | 6 | 2 | 1 | 27 | 8 | 14 | 5 | 1 | 1 |
| Papillary | 89 | 3 | 2 | 1 | 6 | 2 | 3 | 0 | 0 | 0 |
| Solid | 56 | 5 | 0 | 0 | 11 | 1 | 1 | 0 | 0 | 0 |
| Micropapillary | 7 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| IMA | 20 | 3 | 0 | 1 | 10 | 1 | 0 | 0 | 0 | 0 |
| Adenosquamous carcinoma | 9 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| Without adenocarcinoma component | 61 | 0 | 0 | 1 | 2 | 2 | 7 | 0 | 0 | 0 |
| SCC | 54 | 0 | 0 | 1 | 2 | 2 | 7 | 0 | 0 | 0 |
| LCC | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mucus‐producing component (P) | <0.001 | 0.033 | 0.440 | <0.001 | 0.643 | 0.848 | 1.000 | 1.000 | 1.000 | |
| Present | 57 | 8 | 2 | 1 | 17 | 3 | 1 | 0 | 0 | 0 |
| Absent | 664 | 11 | 2 | 6 | 49 | 21 | 21 | 4 | 3 | 1 |
| Micropapillary structure (P) | 0.004 | 0.012 | <0.001 | 0.375 | 0.462 | 0.618 | 0.226 | 0.542 | 1.000 | |
| Present | 164 | 10 | 3 | 6 | 12 | 4 | 6 | 2 | 1 | 0 |
| Absent | 553 | 9 | 0 | 0 | 53 | 20 | 16 | 2 | 2 | 1 |
| Invasion of visceral pleura (P) | 0.048 | 0.511 | 0.217 | 0.130 | 0.395 | 0.132 | 1.000 | 1.000 | 1.000 | |
| Present | 152 | 8 | 1 | 3 | 9 | 7 | 8 | 1 | 0 | 0 |
| Absent | 566 | 11 | 2 | 3 | 56 | 18 | 14 | 3 | 3 | 1 |
With adenocarcinoma component, includes adenocarcinoma and adenosquamous carcinoma; without adenocarcinoma component, includes squamous cell carcinoma and large cell carcinoma; P*. P* refers to the comparison between with adenocarcinoma component and without adenocarcinoma component. P, P refers to the comparison of mucus‐producing component, micropapillary structure, and visceral pleura invasion.
AIS, adenocarcinoma in situ; IMA, invasive mucinous adenocarcinoma; LCC, large cell carcinoma; MIA, minimally invasive adenocarcinoma; SCC, squamous cell carcinoma.
ROS1 fusion status and association with clinicopathological features
ROS1 fusion was detected in five patients with adenocarcinoma. The patterns were CD74‐ROS1 (three cases), SDC4‐ROS1 (one case) and GOPC‐ROS1 (one case), respectively. The patient with GOPC‐ROS1 fusion also carried a synchronous EGFR 19 del mutation. As shown in Table 4, ROS1 rearrangement was more frequent in adenocarcinomas with mucus‐producing component (P = 0.033) and micropapillary structure (P = 0.012). No correlation was detected between ROS1 expression and other clinicopathological characteristics (P > 0.05).
BRAF mutation status and association with clinicopathological features
BRAF mutation was detected in 10 (1.1%) cases. Of the 10 cases, there were nine invasive adenocarcinomas and one squamous cell carcinoma. Seven cases demonstrated BRAF V600E mutation in exon 15, and three cases demonstrated BRAF G469A mutation in exon 11. As shown in Tables 4 and 5, nine patients with BRAF mutation had advanced stage III or IV NSCLC . BRAF mutation was significantly associated with advanced disease patients (P < 0.001) and micropapillary structure (P < 0.001).
Table 5.
Nine targeted gene alteration status and association with age, gender, serum tumor markers, tumor location, smoking history, and TNM stage of NSCLC
| No. of patients | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Clinicopathological characteristics | No | ALK | ROS1 | BRAF | KRAS | HER2 | PIK3CA | RET | MET | NRAS |
| Age (P) | 0.247 | 0.356 | 0.723 | 0.509 | 0.162 | 0.014 | 0.383 | 0.632 | 0.506 | |
| <60 | 437 | 13 | 4 | 6 | 42 | 23 | 9 | 1 | 1 | 0 |
| ≥60 | 447 | 8 | 1 | 4 | 49 | 15 | 23 | 4 | 3 | 1 |
| Sex (P) | 0.276 | 1.000 | 0.202 | <0.001 | 0.300 | 0.193 | 1.000 | 0.481 | 1.000 | |
| Male | 398 | 7 | 2 | 7 | 70 | 14 | 18 | 2 | 3 | 0 |
| Female | 486 | 14 | 3 | 3 | 21 | 24 | 14 | 3 | 1 | 1 |
| Serum tumor markers (P) | 0.257 | 0.635 | 1.000 | 0.048 | 0.907 | 0.735 | 0.075 | 0.906 | 0.404 | |
| Normal | 357 | 11 | 1 | 4 | 28 | 15 | 12 | 0 | 1 | 1 |
| Abnormal | 353 | 10 | 4 | 6 | 63 | 23 | 20 | 5 | 3 | 0 |
| Gross type (P) | 0.783 | 0.661 | 0.404 | 0.189 | 0.221 | <0.001 | 0.316 | 0.719 | 1.000 | |
| Central type | 70 | 2 | 0 | 2 | 4 | 5 | 8 | 1 | 0 | 0 |
| Peripheral type | 814 | 19 | 5 | 8 | 87 | 33 | 24 | 4 | 4 | 1 |
| Smoking history (P) | 0.878 | 0.996 | 0.301 | <0.001 | 0.604 | 0.590 | 1.000 | 0.322 | 1.000 | |
| Non‐smokers | 618 | 15 | 4 | 5 | 35 | 28 | 21 | 3 | 4 | 1 |
| Former/current smokers | 266 | 6 | 1 | 5 | 56 | 10 | 11 | 2 | 0 | 0 |
| Site of tumor location (P) | 0.873 | 0.511 | 0.191 | 0.144 | 0.755 | 0.412 | 0.806 | 0.776 | 1.000 | |
| Left lung | 394 | 9 | 1 | 7 | 34 | 16 | 12 | 3 | 1 | 0 |
| Right lung | 488 | 12 | 4 | 3 | 57 | 22 | 20 | 2 | 3 | 1 |
| TNM stage (P) | 0.105 | 0.501 | 0.001 | 0.291 | 0.394 | 0.176 | 1.000 | 1.000 | 1.000 | |
| I + II | 596 | 10 | 2 | 1 | 54 | 22 | 17 | 3 | 3 | 1 |
| III + IV | 315 | 11 | 3 | 9 | 37 | 16 | 15 | 2 | 1 | 0 |
P, P refers to the comparison with different age groups, genders, serum tumor markers, smoking history, site of tumor location, and TNM stage.
KRAS, PIK3CA, HER2, MET, RET and NRAS gene alterations status and association with clinicopathological features
As shown in Tables 4 and 5, KRAS mutations were found more frequently in men (P < 0.001), former or current smokers (P < 0.001), and patients who had a higher level of serum tumor markers (P = 0.048). Moreover, KRAS mutations were correlated with the mucus‐producing component (P < 0.001). PIK3CA mutation was significantly more frequent in elderly patients (P = 0.014) with central invasive adenocarcinoma (P < 0.001). Among the 32 samples with PIK3CA mutation, 19 samples harbored concurrent EGFR mutations, and two samples had KRAS mutations. HER2 gene alterations were detected in 38 cases including gene amplification (19 cases) and mutation located in exon 20 (19 cases). In histological type, 94.7% (36/38) HER2 alterations were adenocarcinoma. We failed to find any significant association between amplification or point mutation with clinicopathological parameters. Synchronous EGFR mutation or KRAS mutation was detected in 10 patients harboring HER2 amplification. RET fusion was found in five patients, three cases had KIF5B‐RET fusion, and two cases CCDC6‐RET fusion. MET gene alteration was observed in four patients, including two cases with gene amplification and two cases with exon 14 skipping mutation. No significant association was observed in any clinicopathological features of HER2 alteration (P > 0.05). The number of samples with rare mutations such as MET, RET and NRAS was too low for statistical analysis.
Discussion
In this study, we investigated the relationship between 10 targeted gene alterations (including EGFR, HER2, BRAF, KRAS, ALK, MET, NRAS, PIK3CA, RET, and ROS1) and the clinicopathological features in 884 patients with NSCLC. Our data showed that approximately 75% of patients had at least one gene alteration. The most popular mutation was EGFR which was detected in more than half of the patients (57.7%), followed by KRAS (10.3%), HER2 (4.3%), PIK3CA (3.6%), ALK (2.4%), BRAF (1.1%). The other four targeted gene alteration rates were less than 1%.
To the best of our knowledge, somatic mutations of EGFR are more common in Asian patients. 14 In our study, the frequency of EGFR mutations was higher than previous reports in Chinese NSCLC cohorts, 15 , 16 but was similar to recent studies. 17 , 18 This may be due to an improvement in medical level and detection methods, whereby more patients are diagnosed in early stage disease which improves the positive detection rate. The higher proportion of adenocarcinoma patients in this study is also a possible cause. EGFR mutation is more likely to be detected in women, non‐smokers, and early stage patients without lymph node and organ metastasis, as previously reported. 19 , 20 In our study, the most common treatment‐sensitive activating mutations were EGFR L858R and 19 del mutations, followed by G719 X and L861Q, which was consistent with previous reports in the literature. 15 , 18 Gefitinib and erlotinib have proved significantly curative effectiveness in the treatment of sensitive EGFR mutation. 21 The 20 T790M mutation is the most common secondary resistance mechanism in EGFR and is associated with poorer survival rates in NSCLC. 22 Therefore, the detection of EGFR gene mutation status is a prerequisite for targeted drugs to be administered to patients. There were seven patients who had a T790M secondary mutation in our study. It is worth noting that we also detected primary T790M mutation in seven patients (0.8%) without any history of TKI therapy. All seven patients had other EGFR activating mutations. Primary T790M mutation has been reported to coexist more commonly with L858R, whereas acquired mutation more likely coexists with 19 del, 23 , 24 indicating different mechanisms are present between the two. Previous studies have shown that whilst patients with primary or acquired T790M mutation show significant differences in certain clinical features, and may benefit from osimertinib therapy, those patients with acquired T790M mutation may have a higher overall survival rate than those with primary T790M mutation. 23 , 24 This may be due to the fact that 19 del could destroy the inactive conformation of the EGFR kinase domain and enhance EGFR‐TKI sensitivity. Searching the databases and reviewing previous studies, the clinical significance of most rare mutations is unclear, and some mutations may possibly be related to TKI resistance. 25
The fusion of ALK, ROS1 and MET are rare genetic alterations in patients with NSCLC who can benefit from crizotinib or vandetanib therapy. 26 , 27 Similarly, these three kinds of fusion occur easily, predominantly in younger non‐smokers, and in lung adenocarcinoma patients. 28 , 29 ALK gene rearrangement has been reported to be present in about 3%–8% of NSCLC patients, 4 , 30 and ROS1 and RET fusion reported to be present in about 1%–2% of patients with NSCLC. 11 In our study, the rate of occurrence of ALK, ROS1 and RET fusion was slightly lower. This may be due to regional or demographic differences. A retrospective analysis by Zhao et al. 31 showed that age, gender, specimen type, histologic type, and smoking history were correlated with ALK status. However, in our study, apart from histologic type, no significant association between ALK status and clinicopathological features was observed. This difference may result from different sample size or geographic differences. ALK rearrangement was more frequent in adenocarcinomas with a mucus‐producing component, micropapillary structure and visceral pleura invasion. This may be the reason for the poor prognosis in patients with ALK rearrangement. Zhao et al. also reported that ALK rearrangement was more common in patients with invasive mucinous adenocarcinoma and solid‐predominant invasive adenocarcinoma. 31 Our data showed that ALK, ROS1, BRAF or KRAS gene alterations were significantly more frequent in adenocarcinomas with a mucus‐producing component or micropapillary component, and these two components could be a nonpredominant component in some cases. This suggests that attention should be paid not only to the predominant component but also to other nonpredominant components in NSCLC. Pathologists should list every histologic component more than 5% in pathological reports as suggested by WHO histologic classification. 3 ALK, ROS1 or RET fusion was not detected in squamous cell carcinomas and large cell carcinomas in our study. The result suggested that these fusions may be very rare in squamous cell carcinomas and large cell carcinomas. 32
The study by Lin et al. revealed that the BRAF mutation rate in Chinese NSCLC patients was 2.8%, and that V600 and G469 were the two most common mutation types. 33 BRAF mutation rate is higher in patients with advanced lung adenocarcinoma. Only V600 and G469 mutations were found in our study, and the mutation rate was only 1.1%. Most of the patients in our study were in early‐stage disease, and therefore the BRAF mutation rate was lower in our study.
HER2 alteration has been reported to be more common in women, non‐smokers, Asians and patients with adenocarcinomas, which is similar to those with EGFR mutation. 9 , 34 , 35 In our patient cohort, we found that HER2 alteration was more frequently observed in adenocarcinoma than other histological types, whereas there was no significant association of HER2 amplification or mutation with gender, age and smoking status in our research, and another study supported our views. 36 Ken et al. 37 reported that HER2 amplification may be a possible mechanism for acquired resistance to EGFR‐TKI without the EGFR T790M mutation. In this study, there was a patient carrying an EGFR 19 del mutation accompanied by primary HER2 amplification, which showed a response to EGFR‐TKI treatment. This case suggested that primary HER2 amplification was not indicative of EGFR‐TKI resistance. Immunohistochemistry for HER2 was performed on the tumor tissue in this case, and immunohistochemical stain showed that the expression of HER2 protein had significant heterogeneity (some of the tumor cells were negative for HER2) (Fig 1). Intratumoral heterogeneity therefore explains the effectiveness of EGFR‐TKI therapy in this patient.
Figure 1.
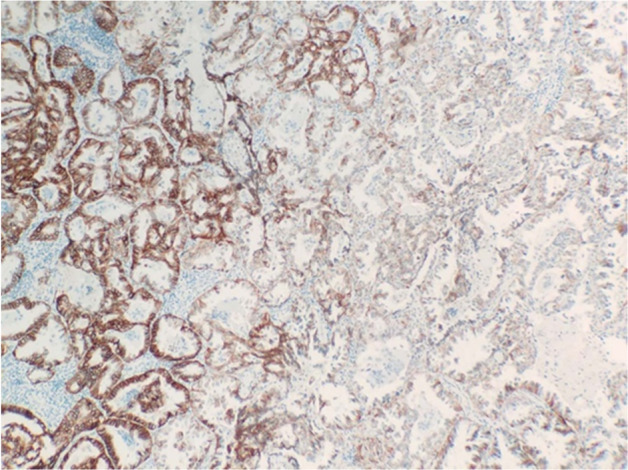
Immunohistochemistry for HER2 protein expression (IHC ×100).
A previous study showed that targetable activating alterations in lung cancer genes, such as EGFR, ALK, RET and ROS1, are mutually exclusive molecular events. 30 Recently, however, some cases of NSCLC with coalteration have been reported in the literature, 38 , 39 , 40 , 41 and these coaltered patients may benefit from treatment with more than one targeted drug. 42 There were 33 cases (3.7%) with coalteration in our study, including EGFR/PIK3CA (2.1%, 19 cases), EGFR/HER2 (0.7%, six cases), HER2/KRAS (0.3%, three cases), EGFR/KRAS (0.1%, one case), EGFR/ROS1 (0.1%, one case), EGFR/NRAS (0.1%, one case), KRAS/PIK3CA (0.1%, one case), and a triple KRAS/PIK3CA/HER2 (0.1%, one case) coalteration. Previous studies revealed that KRAS, BRAF, and PIK3CA mutation have been related to efficacy of EGFR‐TKI, metastasis or overall survival. 43 PIK3CA mutations frequently coexist with EGFR or KRAS mutations. Evidence has suggested that the combination of KRAS and EGFR mutation may have a negative impact on the efficacy of TKI treatment, 41 but EGFR‐TKI may be an effective choice for the treatment of patients with NSCLC with comutation of EGFR/KRAS. 44 Zeng et al. 45 reported a case of adenocarcinoma having acquired GOPC‐ROS1 rearrangement after treatment with osimertinib. These authors suggested that GOPC‐ROS1 rearrangement was a novel acquired resistance mechanism to osimertinib. In our research, a comutation of EGFR 19 del with GOPC‐ROS1 rearrangement was detected in a 60‐year‐old smoking male with an adenocarcinoma. The patient did not undergo anti‐EGFR therapy before a genetic test, indicating it was a coexisting primary EGFR exon 19 del plus GOPC‐ROS1 rearrangement rather than an acquired one. The patient did not undergo targeted therapy after surgery, so we cannot evaluate the efficiency of drug targeted therapy. Moreover, we encountered a unique patient harboring a triple KRAS/PIK3CA/HER2 alteration. The patient was a 60‐year‐old non‐smoking female with an invasive adenocarcinoma (T1bN1M0). With more and more coalterations being found in NSCLC patients, we need to investigate the mechanism of their occurrence, because the efficiency of relevant targeted drugs is unclear in these patients.
In conclusion, our research revealed the status of 10 targeted genes of 884 NSCLCs in eastern China. Most of the patients had at least one gene alteration, and our study confirmed that NGS is a reliable and effective method for gene detection in NSCLCs.
Disclosure
No authors report any conflict of interest.
Supporting information
Table S1 Supporting information.
Acknowledgments
This work was supported by the National Natural Science Foundation of China Grants (No. 81972329, 81672606) and National Major Scientific and Technological Special Project of China for “Significant New Drugs Development” (No. 2020ZX09201‐018).
Contributor Information
Dongliang Lin, Email: lindongliang2008@126.com.
Xiaoming Xing, Email: edithxing@126.com.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68 (6): 394–424. [DOI] [PubMed] [Google Scholar]
- 2. GBD 2017 DALYs and HALE Collaborators . Global, regional, and national disability‐adjusted life‐years (DALYs) for 359 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018; 392 (10159): 1859–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG. WHO Classifcation of Tumours of the Lung, Pleura, Thymus and Heart. IARC Press, Lyon: 2015; 1–412. [DOI] [PubMed] [Google Scholar]
- 4. Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol 2015; 16 (7): e342–51. [DOI] [PubMed] [Google Scholar]
- 5. Dacic S, Nikiforova MN. Present and future molecular testing of lung carcinoma. Adv Anat Pathol 2014; 21 (2): 94–9. [DOI] [PubMed] [Google Scholar]
- 6. Berge EM, Doebele RC. Targeted therapies in non‐small cell lung cancer: Emerging oncogene targets following the success of epidermal growth factor receptor. Semin Oncol 2014; 41 (1): 110–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reckamp KL. Targeted therapy for patients with metastatic non‐small cell lung cancer. J Natl Compr Canc Netw 2018; 16 (5S): 601–4. [DOI] [PubMed] [Google Scholar]
- 8. Bu S, Wang R, Pan Y et al Clinicopathologic characteristics of patients with HER2 insertions in non‐small cell lung cancer. Ann Surg Oncol 2017; 24 (1): 291–7. [DOI] [PubMed] [Google Scholar]
- 9. Shigematsu H, Takahashi T, Nomura M et al Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res 2005; 65 (5): 1642–6. [DOI] [PubMed] [Google Scholar]
- 10. El Osta B, Behera M, Kim S et al Characteristics and outcomes of patients with metastatic KRAS‐mutant lung adenocarcinomas: The lung cancer mutation consortium experience. J Thorac Oncol 2019; 14 (5): 876–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bergethon K, Shaw AT, Ou SH et al ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012; 30 (8): 863–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saito M, Suzuki H, Kono K, Takenoshita S, Kohno T. Treatment of lung adenocarcinoma by molecular‐targeted therapy and immunotherapy. Surg Today 2018; 48 (1): 1–8. [DOI] [PubMed] [Google Scholar]
- 13. Rami‐Porta R, Asamura H, Travis WD, Rusch VW. Lung cancer ‐ major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin 2017; 67 (2): 138–55. [DOI] [PubMed] [Google Scholar]
- 14. Graham RP, Treece AL, Lindeman NI et al Worldwide frequency of commonly detected EGFR mutations. Arch Pathol Lab Med 2018; 142 (2): 163–7. [DOI] [PubMed] [Google Scholar]
- 15. Li S, Li L, Zhu Y et al Coexistence of EGFR with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: A comprehensive mutation profiling from 5125 Chinese cohorts. Br J Cancer 2014; 110 (11): 2812–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu J, He J, Yang H et al Somatic mutation analysis of EGFR, KRAS, BRAF and PIK3CA in 861 patients with non‐small cell lung cancer. Cancer Biomark 2011; 10 (2): 63–9. [DOI] [PubMed] [Google Scholar]
- 17. Jing C, Mao X, Wang Z et al Nextgeneration sequencingbased detection of EGFR, KRAS, BRAF, NRAS, PIK3CA, Her2 and TP53 mutations in patients with nonsmall cell lung cancer. Mol Med Rep 2018; 18 (2): 2191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao M, Zhan C, Li M et al Aberrant status and clinicopathologic characteristic associations of 11 target genes in 1,321 Chinese patients with lung adenocarcinoma. J Thorac Dis 2018; 10 (1): 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li S, Choi YL, Gong Z et al Comprehensive characterization of oncogenic drivers in Asian lung adenocarcinoma. J Thorac Oncol 2016; 11 (12): 2129–40. [DOI] [PubMed] [Google Scholar]
- 20. Jang TW, Oak CH, Chang HK, Suo SJ, Jung MH. EGFR and KRAS mutations in patients with adenocarcinoma of the lung. Korean J Intern Med 2009; 24 (1): 48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martinez‐Marti A, Navarro A, Felip E. Epidermal growth factor receptor first generation tyrosine‐kinase inhibitors. Transl Lung Cancer Res 2019; 8 (Suppl. 3): S235–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gao X, Zhao Y, Bao Y et al Poor prognosis with coexistence of EGFR T790M mutation and common EGFR‐activating mutation in non‐ small cell lung cancer. Cancer Manag Res 2019; 11: 9621–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang S, Yan B, Zhang Y e a. Different characteristics and survival in non‐small cell lung cancer patients with primary and acquired EGFR T790M mutation. Int J Cancer 2019; 144 (11): 2880–6. [DOI] [PubMed] [Google Scholar]
- 24. Li W, Qiu T, Guo L et al Primary and acquired EGFR T790M‐mutant NSCLC patients identified by routine mutation testing show different characteristics but may both respond to osimertinib treatment. Cancer Lett 2018; 423: 9–15. [DOI] [PubMed] [Google Scholar]
- 25. Ou SI, Cui J, Schrock AB et al Emergence of novel and dominant acquired EGFR solvent‐front mutations at Gly796 (G796S/R) together with C797S/R and L792F/H mutations in one EGFR (L858R/T790M) NSCLC patient who progressed on osimertinib. Lung Cancer 2017; 108: 228–31. [DOI] [PubMed] [Google Scholar]
- 26. Falchook GS, Ordonez NG, Bastida CC et al Effect of the RET inhibitor vandetanib in a patient with RET fusion‐positive metastatic non‐small‐cell lung cancer. J Clin Oncol 2016; 34(15): e141–4. [DOI] [PubMed] [Google Scholar]
- 27. Solomon BJ, Mok T, Kim DW et al First‐line crizotinib versus chemotherapy in ALK‐positive lung cancer. N Engl J Med 2014; 371 (23): 2167–77. [DOI] [PubMed] [Google Scholar]
- 28. Lin C, Shi X, Yang S et al Comparison of ALK detection by FISH, IHC and NGS to predict benefit from crizotinib in advanced non‐small‐cell lung cancer. Lung Cancer 2019; 131: 62–8. [DOI] [PubMed] [Google Scholar]
- 29. Lin C, Wang S, Xie W, Chang J, Gan Y. The RET fusion gene and its correlation with demographic and clinicopathological features of non‐small cell lung cancer: A meta‐analysis. Cancer Biol Ther 2015; 16 (7): 1019–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gainor JF, Varghese AM, Ou SH et al ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: An analysis of 1,683 patients with non‐small cell lung cancer. Clin Cancer Res 2013; 19 (15): 4273–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao R, Zhang J, Han Y et al Clinicopathological features of ALK expression in 9889 cases of non‐small‐cell lung cancer and genomic rearrangements identified by capture‐based next‐generation sequencing: A Chinese retrospective analysis. Mol Diagn Ther 2019; 23 (3): 395–405. [DOI] [PubMed] [Google Scholar]
- 32. Zhao W, Choi YL, Song JY et al ALK, ROS1 and RET rearrangements in lung squamous cell carcinoma are very rare. Lung Cancer 2016; 94: 22–7. [DOI] [PubMed] [Google Scholar]
- 33. Lin Q, Zhang H, Ding H et al The association between BRAF mutation class and clinical features in BRAF‐mutant Chinese non‐small cell lung cancer patients. J Transl Med 2019; 17 (1): 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li C, Sun Y, Fang R et al Lung adenocarcinomas with HER2‐activating mutations are associated with distinct clinical features and HER2/EGFR copy number gains. J Thorac Oncol 2012; 7 (1): 85–9. [DOI] [PubMed] [Google Scholar]
- 35. Xu F, Yang G, Xu H, Yang L, Qiu W, Wang Y. Treatment outcome and clinical characteristics of HER2 mutated advanced non‐small cell lung cancer patients in China. Thorac Cancer 2020; 11 (3): 679–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li X, Zhao C, Su C, Ren S, Chen X, Zhou C. Epidemiological study of HER‐2 mutations among EGFR wild‐type lung adenocarcinoma patients in China. BMC Cancer 2016; 16 (1): 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Takezawa K, Pirazzoli V, Arcila ME et al HER2 amplification: A potential mechanism of acquired resistance to EGFR inhibition in EGFR‐mutant lung cancers that lack the second‐site EGFRT790M mutation. Cancer Discov 2012; 2 (10): 922–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tang Z, Zhang J, Lu X et al Coexistent genetic alterations involving ALK, RET, ROS1 or MET in 15 cases of lung adenocarcinoma. Mod Pathol 2018; 31 (2): 307–12. [DOI] [PubMed] [Google Scholar]
- 39. Sweis RF, Thomas S, Bank B, Fishkin P, Mooney C, Salgia R. Concurrent EGFR mutation and ALK translocation in non‐small cell lung cancer. Cureus 2016; 8 (2): e513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qiu T, Zhang F, Li W, Guo L, Ying J. Concurrent presence of ALK rearrangement and MET mutation in lung adenocarcinoma. J Thorac Oncol 2019;14(2): e42–e44.–. [DOI] [PubMed] [Google Scholar]
- 41. Zhuang X, Zhao C, Li J et al Clinical features and therapeutic options in non‐small cell lung cancer patients with concomitant mutations of EGFR, ALK, ROS1, KRAS or BRAF . Cancer Med 2019; 8 (6): 2858–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schmid S, Gautschi O, Rothschild S et al Clinical outcome of ALK‐positive non‐small cell lung cancer (NSCLC) patients with de novo EGFR or KRAS co‐mutations receiving tyrosine kinase inhibitors (TKIs). J Thorac Oncol 2017; 12 (4): 681–8. [DOI] [PubMed] [Google Scholar]
- 43. Hu W, Liu Y, Chen J. Concurrent gene alterations with EGFR mutation and treatment efficacy of EGFR‐TKIs in Chinese patients with non‐small cell lung cancer. Oncotarget 2017; 8 (15): 25046–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tsukumo Y, Naito M, Suzuki T. Influence of EGFR‐activating mutations on sensitivity to tyrosine kinase inhibitors in a KRAS mutant non‐small cell lung cancer cell line. PLOS One 2020; 15 (3): e0229712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zeng L, Yang N, Zhang Y. GOPC‐ROS1 rearrangement as an acquired resistance mechanism to osimertinib and responding to crizotinib combined treatments in lung adenocarcinoma. J Thorac Oncol 2018; 13 (7): e114–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Supporting information.


